Abstract
Transcription from bacteriophage T4 middle promoters uses Escherichia coli RNA polymerase together with the T4 transcriptional activator MotA and the T4 coactivator AsiA. AsiA binds tightly within the C-terminal portion of the σ70 subunit of RNA polymerase, while MotA binds to the 9-bp MotA box motif, which is centered at −30, and also interacts with σ70. We show here that the N-terminal half of MotA (MotANTD), which is thought to include the activation domain, interacts with the C-terminal region of σ70 in an E. coli two-hybrid assay. Replacement of the C-terminal 17 residues of σ70 with comparable σ38 residues abolishes the interaction with MotANTD in this assay, as does the introduction of the amino acid substitution R608C. Furthermore, in vitro transcription experiments indicate that a polymerase reconstituted with a σ70 that lacks C-terminal amino acids 604 to 613 or 608 to 613 is defective for MotA-dependent activation. We also show that a proteolyzed fragment of MotA that contains the C-terminal half (MotACTD) binds DNA with a KD(app) that is similar to that of full-length MotA. Our results support a model for MotA-dependent activation in which protein-protein contact between DNA-bound MotA and the far-C-terminal region of σ70 helps to substitute functionally for an interaction between σ70 and a promoter −35 element.
A programmed cascade of transcriptional events is initiated when bacteriophage T4 infects its host Escherichia coli (reviewed in reference 57). T4 early genes are transcribed immediately after infection by using the existing host RNA polymerase holoenzyme comprising the core (α2ββ′) and the σ70 subunit. Early T4 promoters do not require T4-encoded transcription factors, since they contain excellent matches to the ideal σ70 sequences in their −10 and −35 regions (61; reviewed in reference 60). In contrast, transcription from T4 middle promoters uses two T4 early gene products, the transcriptional activator MotA and the coactivator AsiA (23, 39, 43, 44; reviewed in reference 57). Late promoter utilization requires the replacement of σ70 by the T4 sigma factor, gp55, as well as other phage-encoded activators and coactivators (reviewed in reference 62).
The MotA protein binds as a monomer (6, 33) to a 9-bp element (MotA box) centered at position −30 of middle promoter DNA (3, 19, 23). In addition, MotA forms a complex with σ70 (18). Nuclear magnetic resonance and crystallographic studies indicate that the 211 amino acids of MotA are organized into an N-terminal domain (NTD) and a C-terminal domain (CTD) separated by a small flexible linker (15, 33, 34). Mutations within the NTD of MotA eliminate transcriptional activation and the formation of the MotA-σ70 complex (14, 18), suggesting that the NTD is the activation domain of MotA. Transcriptional activation of middle promoters also requires the T4 coactivator AsiA, which binds very tightly to σ70 (44, 55, 56). Binding sites for AsiA have been mapped within C-terminal amino acids (regions 4.1 and 4.2) of σ70 (8, 49, 50, 53, 59). Residues within region 4.2 normally contact the −35 element of host promoter DNA (5, 9, 17, 29, 54). In the absence of MotA, AsiA binding to σ70 inhibits transcription by polymerase from promoters that require recognition of the −35 canonical sequences (8, 45, 51), suggesting that the presence of AsiA inhibits the σ70 region 4.2-DNA interaction.
In this paper we show that the interaction of a MotA N-terminal peptide (amino acids 1 to 97) with σ70, like the interaction of AsiA with σ70, involves the C-terminal region of σ70. In addition, deletions of the amino acids within the far-C-terminal region of σ70 (amino acids 604 to 613) impair the ability of RNA polymerase to perform MotA-dependent activation in vitro. We also show that a MotA C-terminal peptide, beginning at amino acid 102, binds DNA with an apparent dissociation constant like that of wild-type MotA. Our results support a model for MotA-dependent activation in which the interaction between the DNA-bound MotA and the C-terminal region of σ70 helps to substitute functionally for an interaction between σ70 and a promoter −35 element.
MATERIALS AND METHODS
Strains.
E. coli KS1 (12) contains a chromosomal lacZ reporter gene under the control of a derivative of the lac promoter Plac that carries a lambda operator (OR2) centered at position −62 in place of the binding site for the catabolite receptor protein normally associated with Plac. KS1 also contains an F′ episome bearing lacIq and a gene for kanamycin resistance. E. coli XL1-Blue (Stratagene) was used for transformations during plasmid constructions.
DNA.
Oligonucleotide primers were obtained from Gene Probe Technologies and Cruachem Inc. (Primer sequences are available upon request.) The 5′-32P-labeled 74-bp PuvsX DNA, containing the PuvsX sequences from positions −56 to +18, was obtained as described previously (23). pDKT90, which contains the T4 middle promoter PuvsX, has been described elsewhere (37). Linear templates for transcription, obtained by BsaAI restriction of pDKT90, were purified by phenol extraction followed by ethanol precipitation.
The pBRα-σ70 chimera plasmid (11) contains a ColE1 replication origin, confers carbenicillin resistance, and directs transcription of an α-σ70 chimera gene under the control of the tandem promoters Plpp and PlacUV5. The resulting α-σ70 chimera protein is composed of the NTD of α (amino acids 1 to 248) fused in frame to the C-terminal region of σ70 (amino acids 528 to 613). The pBRα-σ70 derivative encoding α-σ70(R596H) has been described previously (11), and the pBRα-σ70 derivatives encoding α-σ70 chimeras bearing substitutions H600A, H600R, and R608C were constructed similarly. All of these derivatives are identical to pBRα-σ70 except for the indicated changes. The α-σ38 chimera plasmid, pBRα-σ38 (11), is identical to pBRα-σ70 except that all of the σ70 sequences have been replaced with the comparable σ38 sequences (encoding amino acids 243 to 330). The σ70/σ38 hybrid plasmid, pBRα-σ70/σ38, was constructed by replacing the σ70 sequences encoding amino acids 597 to 613 with the comparable σ38 sequences (encoding amino acids 312 to 330) by PCR and standard cloning techniques.
The pACλcI32 plasmid (27), which was used to construct cI-bait fusion proteins, contains a chloramphenicol resistance marker, a p15A replication origin, and a short alanine linker engineered into the λ cI protein gene such that the gene of interest can be fused to the 3′ end of the cI protein by using NotI, AscI, BstYI, or BglII restriction sites. In this construction, the cI fusion is under the control of PlacUV5. Fragments to be cloned into pACλcI32 were obtained as PCR products of pMot58 (to construct pcI-MotAfl, pcI-MotANTD, and pcI-MotA−) (23), pMot21 (to construct pcI-Mot21NTD) (18), or pAsiA (to construct pcI-AsiA) (25) by using PfuTurbo DNA polymerase (Stratagene) and appropriate primers. Primers contained the necessary NotI or BglII sites to allow ligation with pACλcI32 that had been previously cleaved with NotI and BglII. PCR products were purified by using a Wizard PCR purification system (Promega) and cloned into pACλcI32 by using standard techniques. The σ1-570 gene was obtained as a PCR product of pJH62 (gift of V. J. Hernandez, State University of New York at Buffalo), a plasmid that contains the entire rpoD gene (encoding σ70). PCR was performed with Amplitaq polymerase (Perkin-Elmer), a primer that annealed to the start of the σ70 gene and began with an XbaI recognition sequence, and a primer that annealed to sequences surrounding codon no. 571, introduced a stop codon of TAA at that position, and ended with a SacI recognition sequence. After cleavage with XbaI and SacI, the PCR product was ligated into pet21a(+) (Novagen) that had been previously cleaved with XbaI and SacI. The ClaI/SacI fragment, which contained the C-terminal region of the σ70 gene, was then obtained from the resulting plasmid. This fragment was used to replace the corresponding fragment in pLHN12 (22, 42), a plasmid that contains the wild-type σ70 gene downstream of a T7 promoter. The resulting plasmid was designated pσ1-570.
pσfl, which contains an N-terminal His-tagged σ70 gene, has been described previously (63). This plasmid produces full-length σ70 with the amino acid sequence MRGSHHHHHHGSSGLVPRGSGLGTRL at its N terminus (σfl). PCR products encompassing the C-terminal region of σR596H and σR608C were obtained from pBRα-σR596H and pBRα-σR608C, respectively, by using a primer that annealed at the ClaI site within the σ70 sequence and another primer that annealed just downstream of the σ70 gene and introduced a HindIII site. PCR products encompassing the C-terminal region of σ70 with a stop codon at amino acid position 608 or 604 were obtained by using pBRα-σ70, the same upstream primer, and a primer that introduced a TAA at amino acid position 608 or 604, respectively, and introduced a HindIII site. In each case, the products were digested with ClaI and HindIII and then ligated into pσfl that had been previously digested with ClaI and HindIII, resulting in pσR596H, pσR608C, pσΔ608-613, and pσΔ604-613.
DNA sequence analyses (47) of the inserted sequences in each mutant σ70 plasmid confirmed that only the intended changes were present. In some cases, this sequencing was done at the Center for Agricultural Biotechnology, University of Maryland.
Proteins.
AsiA and MotA were purified as described previously (25). Wild-type σ70 and σ1-570 were purified from cultures of pLHN12/pLysS/BL21(DE3) and pσ1-570/pLysS/BL21(DE3), respectively, as described previously (22). σfl, σR596H, σR608C, σΔ608-613, and σΔ604-613 were purified as described previously (63), except that cells were broken by sonication. E. coli core polymerase was purchased from Epicentre Technologies.
The C-terminal MotA fragment (MotAcloned CTD), which contained MotA amino acids 105 to 211, was obtained as follows. BL21(DE3) cells containing a plasmid that expresses the MotAcloned CTD gene under the control of the T7 promoter Φ10 (15) (plasmid was the gift of M. Finnin, S. Porter, and S. White, St. Jude's Children's Hospital, Memphis, Tenn.) were grown in L broth plus 25 μg of kanamycin/ml at 37°C to mid-log phase. The synthesis of MotAcloned CTD was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to 1 mM. Cells were harvested after 2 h and broken by sonication, and highly purified MotAcloned CTD was obtained after phosphocellulose chromatography as described previously (25), except that proteins were eluted from the column by steps of 0.2, 0.5, and 1 M NaCl in sonication buffer. The 0.5 M NaCl eluate, which contained the bulk of the C-terminal MotA protein, was used for subsequent experiments. A control fraction was obtained by the same procedure, except that the cells used were not induced.
Proteolyzed MotA protein (MotAproteolyzed CTD) generated with endogenous proteases as a partially purified fraction of the wild-type protein (0.8 M phosphocellulose fraction) (23) was loaded onto a phosphocellulose column. Fractions containing the 13.5-kDa MotAproteolyzed CTD eluted with a higher salt concentration (peak fractions eluting with 0.47 M NaCl) than that of the wild-type protein (peak fraction eluting with 0.4 M NaCl). To obtain an N-terminal analysis of the proteolyzed fragment, the 13.5-kDa peptide was purified by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (31) and then transferred to a polyvinylidene difluoride membrane (Novex) by using a Novex Western transfer apparatus and a buffer of 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid (pH 11), 1 mM EDTA, and 10% methanol. N-terminal sequence analysis (16 cycles performed at the W. M. Keck Foundation Biotechnology Resource Laboratory, Boyer Center for Molecular Medicine, Yale University) indicated that the 13.5-kDa gel band was composed of approximately equal amounts of two proteins. One protein matched the amino acid sequence of MotA starting at amino acid 102. The other matched the amino acid sequence of E. coli 50S ribosomal protein L24.
The concentrations of MotAproteolyzed CTD and MotAcloned CTD were estimated after SDS-polyacrylamide gel electrophoresis by comparing these proteins with a known amount of wild-type MotA. Levels were corrected for the fact that the MotA fragments were one-half the size of the wild type and that in the case of MotAproteolyzed CTD, only one-half of the band seen on the gel corresponded to the MotA peptide.
β-Galactosidase assays.
β-Galactosidase assays were performed by a modification of the procedure of Jain (28). Because the synthesis of either AsiA or MotA is toxic for E. coli (25), overnight cultures were always started from single colonies obtained by streaking the −80°C culture stock onto Luria-Bertani plates containing 50 μg of kanamycin/ml, 50 μg of carbenicillin/ml, and 25 μg of chloramphenicol/ml. The plates were then incubated at 37°C, and single colonies were used to inoculate liquid cultures of M9 plus Casamino Acids (Quality Biological) supplemented with the same antibiotics. Cultures were grown overnight at 37°C with aeration, diluted 1:100 in fresh media plus antibiotics, and then grown to mid-log phase. IPTG either was present throughout growth (for the assays in Fig. 3) or was added at the indicated concentration once the cells reached mid-log phase (for the assays in Fig. 4 and 5), and then the cells were grown for another 60 min. A final optical density at 600 nm (OD600-final) of the cells was determined, and the cell pellets from 5 ml of culture were harvested by centrifugation at 1,880 × g at 4°C and then resuspended on ice in 0.2 ml of lacZ buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM 2-mercaptoethanol). Cell extracts, obtained after the addition of 20 μl each of 0.1% (wt/vol) SDS and CHCl3 and vigorous vortexing of the cell suspension for 10 s, were kept on ice until needed. A 20-μl aliquot of the extract was then added to 0.98 ml of the lacZ buffer, and the mixture was incubated for 5 min at 28°C. The reaction was started by the addition of 0.2 ml of a solution containing 4 mg of o-nitrophenyl-β-d-galactopyranoside (ONPG) per ml of lacZ buffer. β-Galactosidase activity was measured by the hydrolysis of ONPG. After the solution turned yellow, the reaction was stopped by the addition of 0.5 ml of 1 M Na2CO3, and the time needed for the reaction (ΔT) was noted. Reaction mixtures were briefly vortexed and then centrifuged at 830 × g. The OD420 of the clear supernatant was then measured. The β-galactosidase activity in Miller units (40) was determined as (2,000 × OD420)/(ΔT × OD600-final).
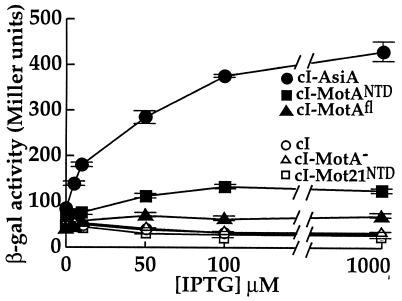
FIG. 3.
AsiA, MotA, and MotANTD each interact with the C-terminal region of σ70 in the E. coli two-hybrid assay. β-Galactosidase (β-gal) activity is plotted versus IPTG concentration for KS1 cells containing pBRα-σ70 and pcI-AsiA (•), pcI-MotANTD (▪), pcI-MotAfl (▴), pcI-Mot21NTD (□), pcI-MotA− (▵), or pACλcI32 (○). Cultures were grown continuously in the presence of the indicated concentrations of IPTG. Points and standard deviations (indicated by error bars) represent the averages of the results of three assays. In this assay, in which cultures were grown continuously in the presence of IPTG, MotANTD gave higher levels of β-galactosidase activity than did MotAfl. However, in assays in which cultures were grown for only 1 h with IPTG, the activities seen with MotANTD and MotAfl were similar (data not shown).
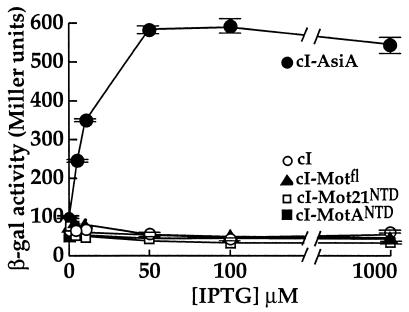
FIG. 4.
MotANTD does not interact with the C-terminal region of σ38. β-Galactosidase (β-gal) activity is plotted versus IPTG concentration for KS1 cells containing pBRα-σ38 and pcI-AsiA (•), pcI-MotANTD (▪), pcI-MotAfl (▴), pcI-Mot21NTD (□), or pACλcI32 (○). Cultures were grown to mid-log phase in the absence of IPTG and then grown in the presence of the indicated IPTG concentrations for 1 h. Points and standard deviations (indicated by error bars) represent the averages of the results of two assays.
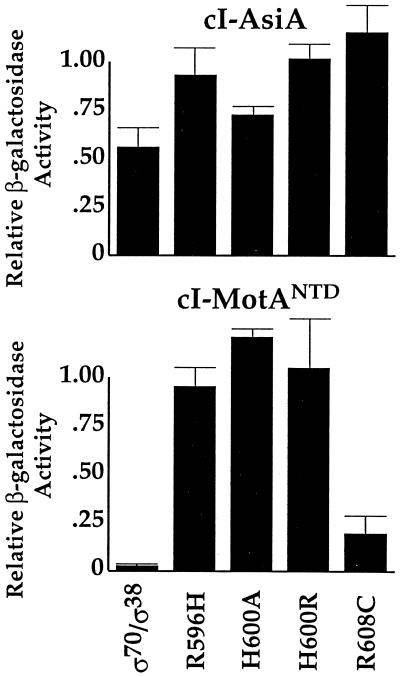
FIG. 5.
The MotA-σ70 interaction in the E. coli two-hybrid assay requires the last 17 amino acids of σ70. Relative β-galactosidase activity is shown for assays with cI-AsiA (top panel) or cI-MotANTD (bottom panel) with α-σ70/σ38, α-σ70(R596H), α-σ70(H600A), α-σ70(H600R), or α-σ70(R608C). (See Materials and Methods for the determination of relative β-galactosidase activity.) Points and standard deviations (indicated by error bars) represent the averages of the results of two to eight assays.
Relative β-galactosidase activities at a 100 μM IPTG concentration (Fig. 5) were calculated as follows:
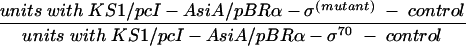 |
where the control was the units obtained with KS1/pACλcI32/pBRα-σ70, and
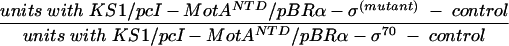 |
where the control was the units obtained with KS1/pcI-Mot21NTD/pBRα-σ70.
Native protein gels.
Protein complexes were assayed by electrophoresis on native polyacrylamide gels as described previously (18).
In vitro transcriptions.
Incubation buffer I contained 44 mM Tris-Cl (pH 8), 50 mM NaCl, 42% glycerol, 1 mM EDTA, 0.18 mM dithiothreitol, 0.008% Triton X-100, and 0.2 mM 2-mercaptoethanol. Incubation buffer II contained 5 mM Tris-Cl (pH 7.5), 69 mM Tris-acetate (pH 7.9), 25 mM NaCl, 5% glycerol, 0.18 mM EDTA, 0.27 mM dithiothreitol, 260 mM potassium glutamate, 6.9 mM magnesium acetate, and 173 μg of bovine serum albumin/ml. DNA buffer contained 7.4 mM Tris-Cl (pH 7.9), 51 mM Tris-acetate (pH 7.9), 57 mM NaCl, 2.1% glycerol, 0.66 mM EDTA, 0.13 mM dithiothreitol, 0.21 mM 2-mercaptoethanol, 190 mM potassium glutamate, 5.1 mM magnesium acetate, 130 μg of bovine serum albumin/ml, 220 μM ATP, 220 μM GTP, 220 μM CTP, and 11 μM [α-32P]UTP (6.5 × 105 dpm/pmol). Finally, 1× Tris-borate-EDTA (TBE) contained 2.5 mM EDTA and 89 mM Tris-borate (pH 8.3).
Proteins were preincubated and mixed with the DNA template and other transcription components as indicated in the figure legend. Reaction mixtures were then placed at 37°C for 20 s before the addition of 0.5 μl of rifampin at 300 μg/ml. After an additional 7.5 min at 37°C, reaction mixtures were collected on dry ice. Twenty-five microliters of gel loading solution (1× TBE containing 7 M urea, 0.1% bromophenol blue, and 0.1% xylene cyanol FF) was added, and the samples were heated at 95°C for 2 min. An 8-μl sample was then subjected to electrophoresis in a 4% polyacrylamide, 7 M urea denaturing gel run in 0.5× TBE.
Protein-DNA gel retardation assays.
Gel retardation assays were performed by using the procedure of Hinton (23). KD(app) was determined as the protein concentration needed to retard 50% of a 5′-32P-labeled 74-bp fragment containing PuvsX in a 10-μl reaction mixture that contained 0.05 pmol of PuvsX DNA and 1.25 pmol of Ptac competitor DNA.
Quantitation of autoradiograms.
After autoradiography, films were scanned with a Scanmaster III Plus from Howtek, Inc. Quantification was performed with Diversity One software from Protein-Databases, Inc.
RESULTS
A σ70 lacking the last 43 amino acids (σ1-570) fails to make a discrete complex with MotA.
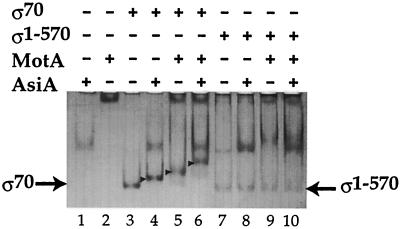
The T4 MotA and AsiA proteins each form a complex with σ70 that can be distinguished from the free proteins on native protein gels (18) (Fig. 1, lanes 4 and 5). σ70 contains 613 amino acids, which are divided into regions 1 through 4 based on sequence similarity among the various members of the sigma protein family (35). Binding sites for AsiA have been found within the C-terminal region of σ70 in both region 4.1 and region 4.2 (amino acids 567 to 600) (8, 49, 50, 53, 59). To test whether the last 43 residues of σ70, which include region 4.2, were also important for MotA-σ70 complex formation, we assayed the formation of complexes by using σ1-570, a truncated σ70 that contains amino acids 1 through 570. As expected, AsiA did not form a complex with σ1-570 (Fig. 1, lane 8). MotA also did not form the discrete complex with σ1-570 that was seen with σ70 (Fig. 1, compare lanes 9 and 5). Instead, a very diffuse band migrating behind the position of free AsiA was observed. These results indicate that a σ70 missing region 4.2 is not capable of forming a discrete complex with MotA that is stable under the conditions of electrophoresis and thus implicate region 4.2 in the MotA-σ70 interaction.
FIG. 1.
Formation of discrete complexes of AsiA-σ70 and MotA-σ70 requires the last 43 amino acids of σ70. A 7.25-μl mixture containing 17 mM Tris-Cl (pH 7.9), 280 mM NaCl, 22% glycerol, 0.7 mM EDTA, 0.7 mM 2-mercaptoethanol, 0.04 mM dithiothreitol, and (as indicated by + or −) 80 pmol of AsiA, 50 pmol of MotA, 14 pmol of σ70, and 14 pmol of σ1-570 was incubated at 37°C for 5 min. Samples were then subjected to electrophoresis in a 6% polyacrylamide native protein gel, and proteins were detected after staining with colloidal Coomassie blue (Invitrogen). The positions of free σ70 and free σ1-570 are indicated. (Both the σ70 and the σ1-570 preparations contain a slow-moving band that is consistent with the presence of σ dimer.) The locations of the AsiA-σ70 complex, the MotA-σ70 complex, and the AsiA-σ70-MotA complex are indicated by arrowheads.
The C-terminal region of σ70 interacts with the N-terminal domain of MotA in an E. coli two-hybrid assay.
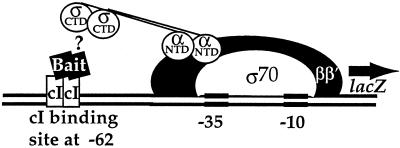
To investigate further the possibility of an interaction between MotA and the C-terminal region of σ70, we used an E. coli-based two-hybrid system (11, 27). In this system (Fig. 2), a chromosomal reporter lacZ gene lies downstream of a promoter that has a binding site for λ cI protein at position −62. The bait is created by the fusion of a protein or domain of interest to the 3′ end of cI. The prey consists of σ70 amino acids 528 to 613, which start within region 3.2 and then include all of region 4 plus the far-C-terminal region (35). This prey is fused in frame to the NTD of the α subunit of RNA polymerase. The addition of IPTG induces the synthesis of both the α-σ70 chimera and the cI-bait protein. An interaction between the bait protein positioned at −62 and the C-terminal region of σ70, which is available in the pool of RNA polymerase that contains the α-σ70 chimera, then increases lacZ transcription.
FIG. 2.
E. coli two-hybrid system for detecting interactions between the C-terminal region of σ70 and other proteins. The cartoon depicts the positions of RNA polymerase subunits β, β′, and σ70 and the α-σ70 chimera, which consists of the N-terminal domain of α fused to the C terminus of σ70, at a promoter upstream of the reporter lacZ gene. The cI-bait fusion protein is located at position −62. (See text for details.)
As has been previously reported (10), there was a large increase in β-galactosidase activity upon the addition of IPTG to cells containing pcI-AsiA and pBRα-σ70 (Fig. 3). To assay an interaction between MotA and the σ70 prey, we tested a bait consisting of cI fused to the entire MotA gene (cI-MotAfl) and a bait in which the motA gene was positioned out of frame with cI (cI-MotA−). The addition of IPTG resulted in an increase in the level of β-galactosidase activity in cells containing the cI-MotAfl fusion compared to the activity seen in the presence of cI-MotA− or cI alone (Fig. 3). Although this was only a twofold increase over background, it was highly reproducible. In addition, in control assays, cells containing pcI-MotA but lacking pBRα-σ70 expressed background levels of lacZ (data not shown), indicating that by itself pcI-MotA was not responsible for the increase in β-galactosidase activity.
Residues within the NTD of MotA are required for MotA activation (14, 18). Induced synthesis of a cI-MotANTD fusion, which contained MotA amino acids 1 to 97, in cells containing the α-σ70 chimera resulted in a significant increase in β-galactosidase activity (Fig. 3). As a control, we constructed a plasmid containing a fusion of cI with the comparable NTD of Mot21, a mutant of MotA in which the first 8 amino acids of MotA have been replaced with 11 different amino acids. Mot21 is a positive control mutant of MotA (18). Full-length Mot21 binds DNA like wild-type MotA but fails to activate transcription or form a complex with σ70 in a native protein gel (18). Production of cI-Mot21NTD in cells containing the α-σ70 chimera resulted in a β-galactosidase activity curve that was coincident with that observed with cI alone (Fig. 3). In addition, the presence of pcI-MotANTD alone (in the absence of pBRα-σ70) resulted only in background levels of β-galactosidase activity (data not shown). Taken together, these results suggest that MotA interacts with the C-terminal region of σ70 and that the NTD of MotA is sufficient for this interaction.
The MotANTD-σ70 interaction in the two-hybrid assay involves the far-C-terminal region of σ70.
σ38, an alternative sigma factor for E. coli RNA polymerase that is used during stationary phase and under certain conditions of stress (58; reviewed in references 20 and 21), has a region 4 which is similar in amino acid sequence to that of σ70 (35). Replacement of the σ70 residues in the α-σ70 chimera with the comparable region of σ38 (amino acids 243 to 330) resulted in background levels of β-galactosidase activity when tested with either cI-MotANTD or cI-MotAfl but resulted in nearly 600 Miller units of β-galactosidase activity when tested with cI-AsiA (Fig. 4). Furthermore, an α-σ70/σ38 hybrid chimera, in which only the last 17 amino acids of σ70 (597 to 613) were replaced with comparable σ38 residues (amino acids 312 to 330), also gave background levels of β-galactosidase activity with cI-MotANTD (Fig. 5). In contrast, this chimera gave only twofold less activity with the cI-AsiA fusion than did the α-σ70 chimera (Fig. 5). These results suggest that the far-C-terminal region of σ70 is involved in the σ70-MotA interaction.
To investigate the need for specific σ70 C-terminal residues, we tested α-σ70 chimeras containing the single amino acid substitutions R596H, H600A, H600R, or R608C in the σ70 moiety (Fig. 5). The R596H, H600A, and H600R substitutions had little effect on the level of β-galactosidase activity observed with either cI-MotANTD or cI-AsiA. In addition, the R608C substitution had no significant effect on the level of β-galactosidase activity observed with cI-AsiA. However, this substitution caused a significant defect when the mutant chimera was tested with cI-MotANTD. Varying the IPTG concentration from 5 μM to 1 mM gave results similar to those obtained with 100 mM IPTG (data not shown), indicating that the low level of β-galactosidase activity observed with cI-MotANTD and α-σR608C could not be improved by increasing the levels of these proteins. In summary, these two-hybrid assays suggested that a residue(s) within the far-C-terminal region of σ70 is important for an interaction with MotA but is relatively unimportant for the σ70-AsiA interaction.
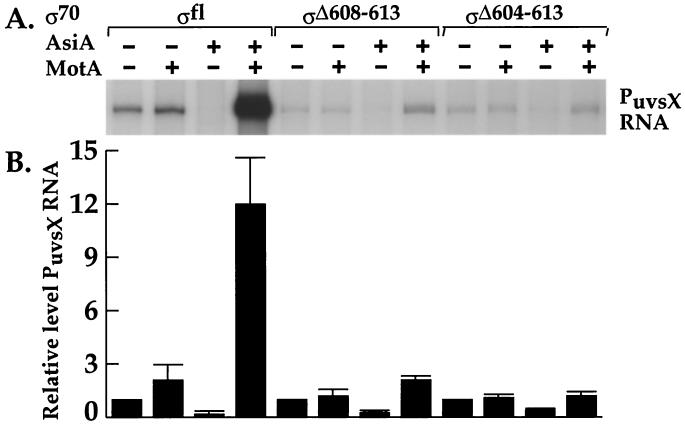
Polymerase reconstituted with a σ70 lacking either amino acids 608 to 613 or amino acids 604 to 613 is defective for MotA-dependent transcription in vitro.
To investigate the involvement of the far-C-terminal amino acids of σ70 in MotA-dependent transcription, we tested σ70 mutant proteins in single-round in vitro transcription reactions by using a DNA template containing the T4 middle promoter PuvsX. Control reactions contained either wild-type σ70 (σfl) or σR596H, a σ70 whose mutation had no effect in the two-hybrid assays. With both of these σ70s, transcription from PuvsX was observed in the presence of polymerase alone, was inhibited by the addition of AsiA, and was greatly activated by MotA/AsiA (Fig. 6 and data not shown). MotA alone resulted in a small (approximately twofold) increase (Fig. 6 and data not shown). Previous work has indicated that MotA alone has no effect on PuvsX transcription in multiple-round transcription reactions (24) but gives this small effect in single-round reactions (D. M. Hinton, unpublished data).
FIG. 6.
Polymerase reconstituted with σΔ608-613 or σΔ604-613 is defective for MotA-dependent transcription at PuvsX. AsiA (23 pmol) and the indicated σ70 (1.3 pmol) were incubated for 10 min at 37°C in 2.5 μl of incubation buffer I. The mixture was placed on ice, and then 0.5 pmol of core polymerase in 2.89 μl of incubation buffer II was added. Transcription was initiated by adding an aliquot (2.16 μl) of the resulting solution to 2.35 μl of DNA buffer containing 0.02 pmol of linearized pDKT90 DNA with or without 1.9 pmol of MotA. (A) The 32P-labeled PuvsX RNA obtained after a set of single-round transcription reactions; (B) quantitation of the results from three independent reactions. For each σ70, the values shown for +AsiA, +MotA, and +AsiA/MotA were normalized relative to a value of 1 for that polymerase alone.
Despite the fact that in the two-hybrid assays the σ70 R608C substitution behaved as if it impaired the σ70-MotA interaction, polymerase containing this mutation was fully active in the in vitro transcription reactions (data not shown). This result suggested either that the MotA-σ70 interaction inferred from the two-hybrid assays was not relevant or that in the context of the transcription complex, this single mutation was not sufficient to impair MotA activity. Thus, we investigated whether σ70 C-terminal deletions of 6 (σΔ608-613) or 10 (σΔ604-613) amino acids affected transcription from PuvsX. Although the level of transcription was lower than that of the wild type, polymerase containing either deletion was able to transcribe from PuvsX in the absence of MotA/AsiA. Thus, the deletions did not destroy polymerase activity (Fig. 6A). When polymerase with σΔ608-613 or σΔ604-613 was used, AsiA alone inhibited transcription significantly or partially, respectively (Fig. 6). Thus, polymerases with these deletions were still susceptible to AsiA inhibition, although the larger deletion was more resistant to AsiA action. In contrast, polymerase with either σ70 deletion was significantly impaired in MotA/AsiA activation at PuvsX (Fig. 6). The transcription results support the idea that the far-C-terminal region of σ70 is important for MotA to function effectively as an activator. We speculate that the single R608C substitution was not sufficient to interfere with MotA activation in vitro because other stabilizing contacts within the transcription complex compensated for this mutation.
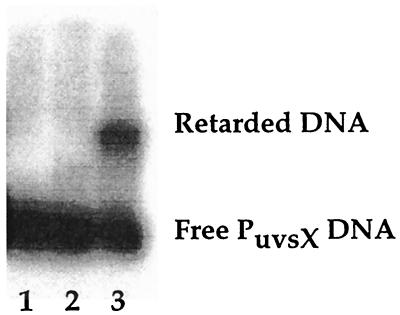
The CTD of MotA binds DNA.
We tested MotAcloned CTD, a protein containing amino acids 105 to 211, and MotAproteolyzed CTD, a proteolyzed fraction of MotA starting at amino acid 102, for their abilities to bind a DNA fragment containing the T4 middle promoter PuvsX in a gel retardation assay. Both peptides bound PuvsX DNA (Fig. 7 and data not shown). The KD(app) determined for MotAproteolyzed CTD was 400 nM, a value that is similar to the previously reported values of 220 nM (6) and 130 nM (52) for wild-type MotA. For MotAcloned CTD, the determined KD(app) was fourfold higher (2,000 nM). These results suggest that the CTD of MotA is sufficient to bind DNA. As expected, MotAproteolyzed CTD, which lacks the MotANTD that is required for activation, did not support activated transcription from PuvsX in vitro (data not shown).
FIG. 7.
A C-terminal peptide of MotA binds DNA. Gel retardation assays, which contained 0.5 pmol of the 32P-labeled 74-bp PuvsX DNA, 26 ng of poly(dI-dC) competitor DNA, and buffer (lane 1), protein fraction from uninduced BL21(DE3) cells containing the plasmid with MotAcloned CTD (lane 2), or 16 pmol of MotAcloned CTD protein in a purified fraction from induced BL21(DE3) cells containing the plasmid with MotAcloned CTD (lane 3), are shown. The fraction used in lane 2 was purified in a manner similar to that of the fraction used in lane 3.
DISCUSSION
Bacteriophage T4 middle promoters represent a hybrid of host and phage promoter sequences, having an excellent match to the σ70 −10 recognition sequence but lacking a good match to σ70 recognition sequences at −35 (3, 19, 38). It is common for a promoter that lacks canonical −35 sequences to require an activator(s) for transcription initiation by RNA polymerase. Such activators fall into two general categories (reviewed in references 4, 26, and 46). Class I activators bind to sites located upstream of the promoter sequences (at −61.5 or farther), while class II activators bind to sites centered near position −41.5, immediately adjacent to core promoter sequences. In both cases, however, these proteins appear to work by contacting polymerase directly and stabilizing the interaction of σ70 region 4.2 with noncanonical sequences within the −35 region of the promoter (2, 11, 30; reviewed in references 4, 26, and 46).
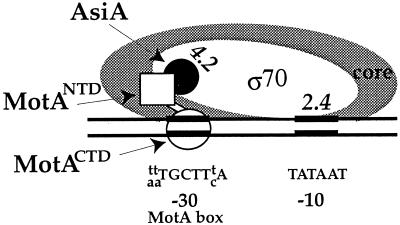
Evidence indicates that MotA/AsiA-dependent activation does not fit either class I or class II. The MotA binding site lies within the core promoter sequence rather than adjacent to it or farther upstream (3, 19, 38). In addition, within the MotA-AsiA-RNA polymerase-middle promoter complex, σ70 retains its contacts with the −10 region of the DNA, but the upstream protein-promoter contacts are significantly rearranged (1, 24). We have previously proposed a model to explain this middle promoter architecture (52). In this model (Fig. 8), AsiA interacts with residues within region 4 of σ70 (8, 49, 50, 53, 59), MotA binds to the MotA box (23, 48) and interacts with σ70 (18), and σ70 region 2.4 retains its contacts with the −10 element of the promoter DNA (24). We speculated that positioning an interaction between MotA and the C-terminal region of σ70 would be reasonable, given that residues within region 4.2 of σ70 normally interact with the −35 sequences of the DNA and that the MotA binding site is centered at −30 (52). The work here demonstrates that the NTD of MotA, which contains residues needed for activation (14, 18), can interact with the C-terminal region of σ70. In addition, we have shown that a C-terminal peptide of MotA starting at amino acid 102 can bind DNA with a binding constant similar to that of the full-length protein. These results are consistent with the idea that the two physical domains of MotA (15), an NTD that is formed by five α-helices and a short β-ribbon (34) and a CTD that is composed of three α-helices interspersed with six β-strands (33), represent two functionally distinct domains. We suggest that MotA belongs to a class of activators that is distinct from both class I and class II. Instead of stabilizing the typical contacts between region 4.2 of σ70 and the DNA, an activator in this third class functionally replaces such contacts by serving as a molecular bridge between σ70 and the DNA.
FIG. 8.
Model of MotA/AsiA activation at a T4 middle promoter. The cartoon depicts the positions of σ70, AsiA, and MotA at a T4 middle promoter. MotACTD interacts with the MotA box motif (5′ [T/A][T/A]TGCTT[T/C]A 3′) centered at −30. Both AsiA and MotANTD interact with the C-terminal region of σ70. The positions of σ70 regions 2.4 and 4.2 are shown. (See text for details.)
The amino acid sequence of region 4 of σ38 is similar to that of region 4 of σ70 (35) and recognizes the same −35 canonical promoter element (13, 16). We found that AsiA interacted with the C-terminal region of σ38 in the two-hybrid assay. AsiA binds to a broad surface of σ70 region 4.2 (8), and AsiA can tolerate amino acid changes throughout this binding surface (41). In addition, in the two-hybrid assay, the AsiA-σ70 interaction was not significantly affected by mutations at residues R596, H600, or R608 (reference 10 and this paper). Thus, the simple explanation for an AsiA-σ38 interaction is that this interaction occurs because region 4.2 of σ38 is very similar to that of σ70. However, this result is surprising, since AsiA neither inhibits transcription by polymerase containing σ38 nor forms a complex with full-length σ38 in a native protein gel (8). Our results suggest that AsiA is indeed able to interact with the C-terminal region of σ38 but that some feature of the full-length protein prevents this interaction.
In contrast to the results seen with AsiA, replacement of even the last 17 amino acids of σ70 with comparable σ38 residues eliminated the stimulation of β-galactosidase activity observed with cI-MotANTD. This effect is specific for MotA, since the σ70/σ38 chimera worked both with cI-AsiA (this paper) and with a cI that had been fused to the E. coli anti-sigma protein Rsd (S. L. Dove and A. Hochschild, unpublished data). In addition, a substitution of R608C in α-σ70 also reduced the interaction with cI-MotANTD but had no effect on the interaction with cI-AsiA. However, not all substitutions within the C-terminal region of σ70 weakened the interaction of σ70 with cI-MotANTD. In particular, a substitution at R596 or H600 had no significant effects. In contrast, the substitution R596H significantly reduced the interaction between the fused σ70 moiety and either Rsd from E. coli or the related regulator, AlgQ, from Pseudomonas aeruginosa (10). The specific effects of these various substitutions argue that the defects seen with cI-MotANTD and the mutant α-σ70 chimeras arise from a loss of the MotA-σ70 interaction rather than a misfolding of the mutant chimeras. Furthermore, our in vitro transcription experiments indicated that a σ70 lacking 6 or 10 C-terminal amino acids is defective for MotA activation of transcription. Taken together, our results are consistent with the idea that the far-C-terminal region of σ70 contacts MotA and that this contact is necessary for MotA to work as an activator. Previous work has indicated that an amino acid substitution at position 604 can partially suppress the growth defect of a T4 motA-positive control mutant in vivo (7), a result that is compatible with this conclusion.
The far-C-terminal region of σ70 lies within an alpha helix (amino acids 603 to 613) (5, 36) at the very end of the protein. This region is just C-terminal of residues that interact with the −35 region of DNA (residues 584, 585, and 588) (5, 9, 17, 29, 54) and of residues that have been implicated in the interactions of σ70 with E. coli class II activators (residues 590 to 603) (32, 36). Thus, the C terminus of σ70 appears to be involved both in class II activation and in the architecturally different activation achieved by MotA/AsiA. Further studies will be needed to determine exactly how the region is configured in class II- versus MotA/AsiA-dependent activation.
Acknowledgments
We thank Jeffrey Gerber for construction of pσ1-570 and Neelowfar Wais, Xanthia Johnson, and Madhavi Vuthoori for help with β-galactosidase assays. We also thank Mike Finnin, Stephanie Porter, and Steve White for the plasmid that expresses the MotAcloned CTD gene.
This work was supported in part by National Institutes of Health grant GM44025, an established investigatorship from the American Heart Association (to A.H.), and a Charles A. King Trust postdoctoral fellowship (to S.L.D.).
REFERENCES
- 1.Adelman, K., E. Brody, and M. Buckle. 1998. Stimulation of bacteriophage T4 middle transcription by the T4 proteins MotA and AsiA occurs at two distinct steps in the transcription cycle. Proc. Natl. Acad. Sci. USA 95:15247-15252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bown, J., A. Kolb, C. Meares, A. Ishihama, S. Minchin, and S. Busby. 2000. Positioning of region 4 of the Escherichia coli RNA polymerase σ70 subunit by a transcription activator. J. Bacteriol. 182:2982-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brody, E., D. Rabussay, and D. Hall. 1983. Regulation of transcription of prereplicative genes, p. 174-183. In C. K. Mathews, E. M. Kutter, G. Mosig, and P. B. Berget (ed.), Bacteriophage T4. American Society for Microbiology, Washington, D.C.
- 4.Busby, S., and R. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, E. A., O. Muzzin, M. Chlenov, J. L. Sun, C. A. Olson, O. Weinman, M. L. Trester-Zedlitz, and S. A. Darst. 2002. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell 9:527-539. [DOI] [PubMed] [Google Scholar]
- 6.Cicero, M., K. Alexander, and K. Kreuzer. 1998. The MotA transcriptional activator of bacteriophage T4 binds to its specific DNA site as a monomer. Biochemistry 37:4977-4984. [DOI] [PubMed] [Google Scholar]
- 7.Cicero, M., M. Sharp, C. Gross, and K. Kreuzer. 2001. Substitutions in bacteriophage T4 AsiA and Escherichia coli σ70 that suppress T4 motA activation mutations. J. Bacteriol. 183:2289-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colland, F., G. Orsini, E. Brody, H. Buc, and A. Kolb. 1998. The bacteriophage T4 AsiA protein: a molecular switch for sigma 70-dependent promoters. Mol. Microbiol. 27:819-829. [DOI] [PubMed] [Google Scholar]
- 9.Dombroski, A., W. Walter, M. Record, Jr., D. Siegele, and C. Gross. 1992. Polypeptides containing highly conserved regions of transcription initiation factor σ70 exhibit specificity of binding to promoter DNA. Cell 70:501-512. [DOI] [PubMed] [Google Scholar]
- 10.Dove, S., and A. Hochschild. 2001. Bacterial two-hybrid analysis of interactions between region 4 of the σ70 subunit of RNA polymerase and the transcriptional regulators Rsd from Escherichia coli and AlgQ from Pseudomonas aeruginosa. J. Bacteriol. 183:6413-6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dove, S., F. Huang, and A. Hochschild. 2000. Mechanism for a transcriptional activator that works at the isomerization step. Proc. Natl. Acad. Sci. USA 97:13215-13220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dove, S., J. Joung, and A. Hochschild. 1997. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature 386:627-630. [DOI] [PubMed] [Google Scholar]
- 13.Espinosa-Urgel, M., C. Chamizo, and A. Tormo. 1996. A consensus structure for σS-dependent promoters. Mol. Microbiol. 21:657-659. [DOI] [PubMed] [Google Scholar]
- 14.Finnin, M., M. Cicero, C. Davies, S. Porter, S. White, and K. Kreuzer. 1997. The activation domain of the MotA transcription factor from bacteriophage T4. EMBO J. 16:1992-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finnin, M., D. Hoffman, K. Kreuzer, S. Porter, R. Schmidt, and S. White. 1993. The MotA protein from bacteriophage T4 contains two domains. Preliminary structural analysis by X-ray diffraction and nuclear magnetic resonance. J. Mol. Biol. 232:301-304. [DOI] [PubMed] [Google Scholar]
- 16.Gaal, T., W. Ross, S. T. Estrem, L. H. Nguyen, R. R. Burgess, and R. L. Gourse. 2001. Promoter recognition and discrimination by EσS RNA polymerase. Mol. Microbiol. 42:939-954. [DOI] [PubMed] [Google Scholar]
- 17.Gardella, T., H. Moyle, and M. Susskind. 1989. A mutant Escherichia coli σ70 subunit of RNA polymerase with altered promoter specificity. J. Mol. Biol. 206:579-590. [DOI] [PubMed] [Google Scholar]
- 18.Gerber, J., and D. Hinton. 1996. An N-terminal mutation in the bacteriophage T4 motA gene yields a protein that binds DNA but is defective for activation of transcription. J. Bacteriol. 178:6133-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guild, N., M. Gayle, T. Sweeney, T. Hollingsworth, T. Modeer, and L. Gold. 1988. Transcriptional activation of bacteriophage T4 middle promoters by the motA protein. J. Mol. Biol. 199:241-258. [DOI] [PubMed] [Google Scholar]
- 20.Hengge-Aronis, R. 1993. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell 72:165-168. [DOI] [PubMed] [Google Scholar]
- 21.Hengge-Aronis, R. 1996. Back to log phase: sigma S as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol. Microbiol. 21:887-893. [DOI] [PubMed] [Google Scholar]
- 22.Hernandez, V., L. Hsu, and M. Cashel. 1996. Conserved region 3 of Escherichia coli σ70 is implicated in the process of abortive transcription. J. Biol. Chem. 271:18775-18777. [DOI] [PubMed] [Google Scholar]
- 23.Hinton, D. 1991. Transcription from a bacteriophage T4 middle promoter using T4 motA protein and phage-modified RNA polymerase. J. Biol. Chem. 266:18034-18044. [PubMed] [Google Scholar]
- 24.Hinton, D., R. March-Amegadzie, J. Gerber, and M. Sharma. 1996. Characterization of pre-transcription complexes made at a bacteriophage T4 middle promoter: involvement of the T4 MotA activator and the T4 AsiA protein, a σ70 binding protein, in the formation of the open complex. J. Mol. Biol. 256:235-248. [DOI] [PubMed] [Google Scholar]
- 25.Hinton, D., R. March-Amegadzie, J. Gerber, and M. Sharma. 1996. The bacteriophage T4 middle transcription system: T4-modified RNA polymerase, AsiA (sigma-70 binding protein), and the transcriptional activator MotA. Methods Enzymol. 274:43-57. [DOI] [PubMed] [Google Scholar]
- 26.Hochschild, A., and S. Dove. 1998. Protein-protein contacts that activate and repress prokaryotic transcription. Cell 92:597-600. [DOI] [PubMed] [Google Scholar]
- 27.Hu, J., M. Kornacker, and A. Hochschild. 2000. Escherichia coli one- and two-hybrid systems for the analysis and identification of protein-protein interactions. Methods 20:80-94. [DOI] [PubMed] [Google Scholar]
- 28.Jain, C. 1999. An Escherichia coli-based genetic strategy for characterizing RNA binding proteins, p. 161-175. In S. Haynes (ed.), RNA-protein interaction protocols. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 29.Keener, J., and M. Nomura. 1993. Dominant lethal phenotype of a mutation in the −35 recognition region of Escherichia coli σ70. Proc. Natl. Acad. Sci. USA 90:1751-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuldell, N., and A. Hochschild. 1994. Amino acid substitutions in the −35 recognition motif of σ70 that result in defects in phage λ repressor-stimulated transcription. J. Bacteriol. 176:2991-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laemmli, U. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 32.Li, M., H. Moyle, and M. Susskind. 1994. Target of the transcriptional activation function of phage λ cI protein. Science 263:75-77. [DOI] [PubMed] [Google Scholar]
- 33.Li, N., E. A. Sickmier, R. Zhang, A. Joachimiak, and S. W. White. 2002. The MotA transcription factor from bacteriophage T4 contains a novel DNA-binding domain: the ‘double wing’ motif. Mol. Microbiol. 43:1079-1088. [DOI] [PubMed] [Google Scholar]
- 34.Li, N., W. Zhang, S. White, and R. Kriwacki. 2001. Solution structure of the transcriptional activation domain of the bacteriophage T4 protein, MotA. Biochemistry 40:4293-4302. [DOI] [PubMed] [Google Scholar]
- 35.Lonetto, M., M. Gribskov, and C. Gross. 1992. The σ70 family: sequence conservation and evolutionary relationships. J. Bacteriol. 174:3843-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lonetto, M., V. Rhodius, K. Lamberg, P. Kiley, S. Busby, and C. Gross. 1998. Identification of a contact site in region 4 of the Escherichia coli RNA polymerase σ70 subunit. J. Mol. Biol. 284:1353-1365. [DOI] [PubMed] [Google Scholar]
- 37.March-Amegadzie, R., and D. M. Hinton. 1995. The bacteriophage T4 middle promoter PuvsX: analysis of regions important for binding of the T4 transcriptional activator MotA and for activation of transcription. Mol. Microbiol. 15:649-660. [DOI] [PubMed] [Google Scholar]
- 38.Marshall, P., M. Sharma, and D. Hinton. 1999. The bacteriophage T4 transcriptional activator MotA accepts various base pair changes within its binding sequence. J. Mol. Biol. 285:931-944. [DOI] [PubMed] [Google Scholar]
- 39.Mattson, T., J. Richardson, and D. Goodin. 1974. Mutant of bacteriophage T4D affecting expression of many early genes. Nature 250:48-50. [DOI] [PubMed] [Google Scholar]
- 40.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Minakin, L., J. Camarero, M. Holford, C. Parker, T. Muir, and K. Severinov. 2001. Mapping the molecular interface between the σ70 subunit of E. coli RNA polymerase and T4 AsiA. J. Mol. Biol. 306:631-642. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen, L. 1996. Ph.D. thesis. University of Wisconsin, Madison.
- 43.Ouhammouch, M., K. Adelman, S. Harvey, G. Orsini, and E. Brody. 1995. Bacteriophage T4 MotA and AsiA proteins suffice to direct Escherichia coli RNA polymerase to initiate transcription at T4 middle promoters. Proc. Natl. Acad. Sci. USA 92:1415-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ouhammouch, M., G. Orsini, and E. Brody. 1994. The asiA gene product of bacteriophage T4 is required for middle mode RNA synthesis. J. Bacteriol. 176:3956-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pahari, S., and D. Chatterji. 1997. Interaction of bacteriophage T4 AsiA protein with Escherichia coli σ70 and its variant. FEBS Lett. 411:60-62. [DOI] [PubMed] [Google Scholar]
- 46.Rhodius, V., and S. Busby. 1998. Positive activation of gene expression. Curr. Opin. Microbiol. 1:152-159. [DOI] [PubMed] [Google Scholar]
- 47.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt, R. P., and K. N. Kreuzer. 1992. Purified MotA protein binds the −30 region of a bacteriophage T4 middle-mode promoter and activates transcription in vitro. J. Biol. Chem. 267:11399-11407. [PubMed] [Google Scholar]
- 49.Severinov, K., and T. Muir. 1998. Expressed protein ligation, a novel method for studying protein-protein interactions in transcription. J. Biol. Chem. 273:16205-16209. [DOI] [PubMed] [Google Scholar]
- 50.Severinova, E., K. Severinov, D. Fenyo, M. Marr, E. N. Brody, J. W. Roberts, B. T. Chait, and S. A. Darst. 1996. Domain organization of the Escherichia coli RNA polymerase sigma 70 subunit. J. Mol. Biol. 263:637-647. [DOI] [PubMed] [Google Scholar]
- 51.Severinova, E., K. Severinov, and S. Darst. 1998. Inhibition of Escherichia coli RNA polymerase by bacteriophage T4 AsiA. J. Mol. Biol. 279:9-18. [DOI] [PubMed] [Google Scholar]
- 52.Sharma, M., P. Marshall, and D. Hinton. 1999. Binding of the bacteriophage T4 transcriptional activator, MotA, to T4 middle promoter DNA: evidence for both major and minor groove contacts. J. Mol. Biol. 290:905-915. [DOI] [PubMed] [Google Scholar]
- 53.Sharma, U., S. Ravishankar, R. Shandil, P. Praveen, and T. Balganesh. 1999. Study of the interaction between bacteriophage T4 asiA and Escherichia coli σ70, using the yeast two-hybrid system: neutralization of asiA toxicity to E. coli cells by coexpression of a truncated σ70 fragment. J. Bacteriol. 181:5855-5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siegele, D., J. Hu, W. Walter, and C. Gross. 1989. Altered promoter recognition by mutant forms of the σ70 subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 206:591-603. [DOI] [PubMed] [Google Scholar]
- 55.Stevens, A. 1976. A salt-promoted inhibitor of RNA polymerase isolated from T4 phage-infected E. coli, p. 617-627. In R. Losick and M. Chamberlin (ed.), RNA polymerase. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 56.Stevens, A., and J. Rhoton. 1975. Characterization of an inhibitor causing potassium chloride sensitivity of an RNA polymerase from T4 phage-infected Escherichia coli. Biochemistry 14:5074-5079. [DOI] [PubMed] [Google Scholar]
- 57.Stitt, B., and D. Hinton. 1994. Regulation of middle-mode transcription, p. 142-160. In J. Karam, J. Drake, K. Kreuzer, G. Mosig, D. Hall, F. Eiserling, L. Black, E. Spicer, E. Kutter, K. Carlson, and E. Miller (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, D.C.
- 58.Tanaka, K., Y. Takayanagi, N. Fujita, A. Ishihama, and H. Takahashi. 1993. Heterogeneity of the principal σ factor in Escherichia coli: the rpoS gene product, σ38, is a second principal σ factor of RNA polymerase in stationary-phase Escherichia coli. Proc. Natl. Acad. Sci. USA 90:3511-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Urbauer, J. L., M. F. Simeonov, R. J. Urbauer, K. Adelman, J. M. Gilmore, and E. N. Brody. 2002. Solution structure and stability of the anti-sigma factor AsiA: implications for novel functions. Proc. Natl. Acad. Sci. USA 99:1831-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilkens, K., and W. Rüger. 1994. Transcription from early promoters, p. 132-141. In J. Karam, J. Drake, K. Kreuzer, G. Mosig, D. Hall, F. Eiserling, L. Black, E. Spicer, E. Kutter, K. Carlson, and E. Miller (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, D.C.
- 61.Wilkens, K., B. Tiemann, F. Bazan, and W. Rüger. 1997. ADP-ribosylation and early transcription regulation by bacteriophage T4, p. 71-82. In F. Haag and F. Koch-Nolte (ed.), ADP-ribosylation in animal tissue. Plenum Press, New York, N.Y. [DOI] [PubMed]
- 62.Williams, K., G. Kassavetis, D. Herendeen, and E. Geiduschek. 1994. Regulation of late-gene expression, p. 161-175. In J. Karam, J. Drake, K. Kreuzer, G. Mosig, D. Hall, F. Eiserling, L. Black, E. Spicer, E. Kutter, K. Carlson, and E. Miller (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, D.C.
- 63.Wilson, C., and A. J. Dombroski. 1997. Region 1 of σ70 is required for efficient isomerization of transcription by Escherichia coli RNA polymerase. J. Mol. Biol. 267:60-74. [DOI] [PubMed] [Google Scholar]