Abstract
Rhizobium bacteria synthesize signal molecules called Nod factors that elicit responses in the legume root during nodulation. Nod factors, modified N-acylated β-(1,4)-N-acetylglucosamine, are synthesized by the nodulation (nod) gene products. We tested the ability of three Sinorhizobium meliloti nod gene products to modify Nod factor analogs with thio linkages instead of O-glycosidic bonds in the oligosaccharide backbone.
Nod factors are lipochitooligosaccharides synthesized by Rhizobium bacteria that signal the legume root during nodulation. The biosynthesis of Nod factors is dependent upon the nodulation (nod) genes, many of which have been identified and characterized previously (6). nodABC are common to all Rhizobium spp. and encode proteins responsible for synthesizing the core Nod factor (Fig. 1A). NodC is proposed to synthesize the β-(1,4)-N-acetylglucosamine oligomers (N-acetyl chitooligosaccharide). NodA and NodB are required for the N acylation of the oligosaccharide backbone. NodB deacetylates the nonreducing-end glucosamine to allow for the attachment of the fatty acyl group by the acyltransferase NodA. Some of the nod genes confer host specificity by modifying the core structure. For example, the Sinorhizobium meliloti host-specific NodH is a sulfotransferase which catalyzes the sulfation of the reducing-end C-6. The presence of the sulfate on the Nod factor produced by S. meliloti is necessary for its activity on Medicago host plants.
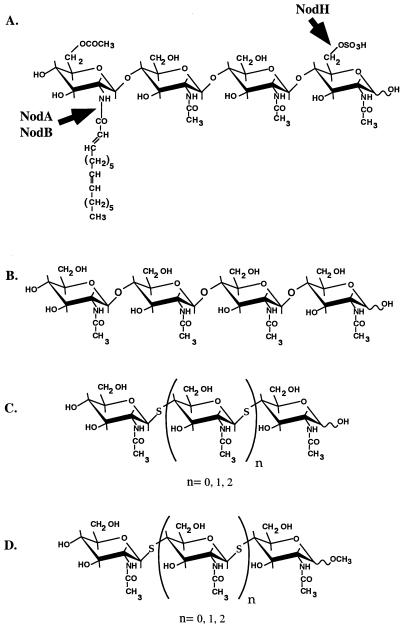
FIG. 1.
(A) Structure of the major S. meliloti Nod factor and the biosynthetic role of specific nod genes. The Nod factor β-(1,4)-N-acetylglucosamine oligosaccharide backbone is synthesized by NodC. NodB deacetylates the nonreducing-end glucosamine to accept the fatty acyl group from the acyltransferase NodA. NodH is a sulfotransferase which catalyzes the sulfation of the reducing-end C-6 position. (B to D) Structures of N-acetyl chitotetraose (B); N-acetyl thiochitobiose (n = 0), thiochitotriose (n = 1), and thiochitotetraose (n = 2) (C); and the α- and β-methyl glycosides of N-acetyl thiochitooligosaccharides (D).
The action of nod gene products has been defined by several approaches. In some cases, direct protein purification has established the activity of enzymes encoded by nod genes, such as the sulfotransferase NodH (8, 27), the O-acetyltransferase NodL (3), the deacetylase NodB (14, 20), and the fucosyltransferase NodZ (22). An alternative approach is to use permeabilized cells or cell extracts, in which varied genotypes allow the inference of gene action, to test specific biochemical steps in Nod factor biosynthesis. This has been used to study NodA and NodC, for example (2, 18, 30).
The use of permeabilized cells for in vitro Nod factor synthesis also allows for the introduction of novel exogenous substrates. We were interested in testing the activity of the Nod proteins for the purpose of creating novel Nod factor analogs built on thio-linked N-acetylglucosamine backbones (thiochitooligosaccharides) rather than chitooligosaccharide backbones (Fig. 1B and C). Authentic Nod factors are susceptible to degradation in vitro by chitinases (24) and lysozyme (A. Southwick, unpublished results). Chitinases in the roots of host plants have been shown to hydrolyze Nod factors, and this is proposed to have a role in Nod factor specificity and/or signaling (31, 32). The thio linkage is resistant to hydrolysis by chitinases; thus, the successful synthesis of thiooligosaccharide Nod factor analogs could test the suggestion that the host plant uses chitinase degradation in the process of specific Nod factor recognition and response.
It is increasingly apparent that oligosaccharides are factors in cell recognition in various animal development systems as well as in plant systems. There are several examples where specific modifications such as sulfation are important for the processing of oligosaccharides or for oligosaccharide or glycoprotein activity. One example is the 2-O or 6-O sulfation of sialyl-Lewis x ligand required for high-affinity selectin binding by neutrophils to the vascular endothelium in the inflammation response (26). Another is the multiple sulfations associated with heparan modification in carbohydrate side chains in developing Drosophila melanogaster embryos; pipe, a gene which encodes a protein homologous to a heparan sulfate-2-O-sulfotransferase, is required for the correct establishment of dorsoventral polarity in Drosophila (28). The synthesis of analogs and the development of inhibitors for basic carbohydrate modification reactions such as sulfation may provide useful probes for development in several organisms.
We have established that NodA, NodB, and NodH are active in modifying thiochitooligosaccharide backbones, thus allowing the synthesis of chitooligosaccharide Nod factor analogs. Tests of substrate requirements for these enzymes provide a guide for their use in modifying other compounds.
Bacterial cultures and strains.
S. meliloti strains were grown in tryptone-yeast extract medium at 30°C under antibiotic selection to an optical density at 600 nm of 1.0 to 1.2. Transposon Tn5 insertion strains were grown in medium with neomycin (50 mg/ml). We used 3 μM luteolin and strains containing the plasmid pRmE65 for the overexpression of NodD3 (10) to maximize nod gene expression. The Rhizobium strains used in this study include wild-type S. meliloti (1021/pE65), a nodC::Tn5 mutant (TJ170/pE65) (13), and the nod gene deletion strain SL44, which lacks nodDABC (10).
Escherichia coli cells were grown in ACH medium (7) with ampicillin (50 mg/ml) at 30°C to an optical density at 600 nm of 1.0 to 1.2. E. coli strain HB101 was used as the host strain for plasmids expressing nodA and nodB (pE40), nodA (pE45), or nodB (pE41) or for the expression vector alone (pAD10) (7).
Oligosaccharide substrates.
Chitooligosaccharide (β-[1,4]-[GlcNAc]n) substrates include N-acetyl-chitotriose and N-acetyl-chitotetraose (Seikagaku Chemicals). Thiooligosaccharides were synthesized as previously described (33). Thiooligosaccharide substrates include N-acetyl-thiochitobiose, N-acetyl-thiochitotriose, N-acetyl-thiochitotetraose, the α-methyl glycosides of the same oligosaccharide series, and the β-methyl glycoside of N-acetyl-thiochitotriose. (Fig. 1C and D).
NodH transfers sulfate to reducing thiooligosaccharide acceptors.
Both chitooligomers and thiochitooligomers were sulfated with NodH and [35S]PAPS (3′-phosphoadenosine-5′-phosphosulfate). The sulfation reactions were performed using the S. meliloti sulfotransferase NodH as previously described (8) but with the following modifications. The sulfate donor, [35S]PAPS, was generated with carrier-free [35S]Na2SO4 (∼43 Ci/mg of S; ICN Pharmaceuticals) by use of the S. meliloti adenosine-5′-phosphosulfate kinase NodQ purified from E. coli (M. Willits, unpublished data). The sulfation reaction products were analyzed by thin-layer chromatography (TLC) on polyethyleneimine (PEI)-cellulose (J. T. Baker) developed with 0.9 M LiCl (16). NodH is active on chitotetraose and on reducing thiooligosaccharides (Fig. 2). The sulfated oligosaccharides migrate at or near the solvent front.
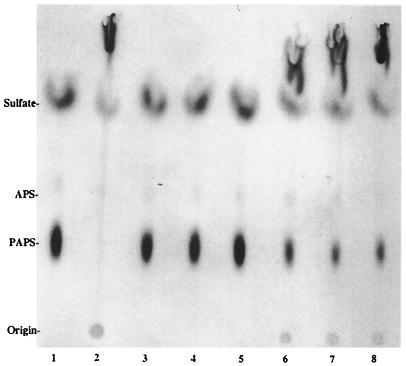
FIG. 2.
NodH sulfation activity on reducing thiooligosaccharides. Chitooligosaccharides and thiochitooligosaccharides were incubated with NodH and [35S]PAPS. The reaction products were analyzed by TLC on PEI-cellulose and by autoradiography and were identified as previously described (8). Lanes: 1, no substrate; 2, chitotetraose; 3 to 5, α-methyl glycosides (3, thiochitobiose; 4, thiochitotriose; 5, thiochitotetraose); 6 to 8, reducing thio-oligosaccharides (6, thiochitobiose; 7, thiochitotriose; 8, thiochitotetraose).
However, no product was detected with the α-methyl glycosides of the thiochitooligosaccharides. The importance of the methyl group configuration for sulfation was tested with the thiochitotriose β-methyl glycoside substrate (Fig. 3). In this case as well, NodH catalyzed the sulfation of chitotetraose and thiochitotetraose but not of the α-methyl or β-methyl glycoside of thiochitotriose. There was no appreciable difference in the sulfations of chitotriose and chitotetraose (data not shown).
FIG. 3.
The α- and β-methyl glycosides of thiochitotriose are not substrates for NodH. The oligosaccharides were incubated with NodH and [35S]PAPS. The reaction products were analyzed by TLC on PEI-cellulose and by autoradiography. Lanes: 1, no substrate; 2, thiochitotriose α-methyl glycoside; 3, thiochitotriose β-methyl glycoside; 4, thiochitotetraose; 5, chitotetraose.
NodH sulfation activity on chitotriose is not inhibited by thiochitotriose methyl glycosides.
To determine if the methyl glycosides were competitive inhibitors, chitotriose and thiochitotriose methyl glycosides were incubated with NodH and [35S]PAPS. For the competition-inhibition sulfation reactions, half the amount of NodH was used. The reaction mixtures were incubated for 15 min, and the reactions were terminated by boiling. The reaction conditions with limited NodH resulted in incomplete sulfation of the chitotetraose control reaction mixture and excess [35S]PAPS (data not shown). The reaction products were analyzed by TLC on PEI-cellulose (Fig. 4). Reactions were performed with substrates separately and as mixtures in 1:1 and 1:10 ratios of chitotriose to α- or β-thiochitotriose methyl glycoside. The data showed that the methyl glycosides did not act as competitive inhibitors of NodH activity on reducing chitooligosaccharides (Fig. 4).
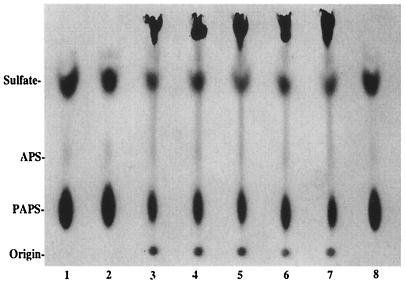
FIG. 4.
NodH sulfation activity on chitotriose is not inhibited by thiochitotriose methyl glycosides. Chitotriose and thiochitotriose methyl glycosides were incubated with NodH and [35S]PAPS. The reaction products were analyzed by TLC on PEI-cellulose. Lanes: 1, thiochitotriose β-methyl glycoside (β-MTG3); 2, thiochitotriose α-methyl glycoside (α-MTG3); 3, chitotriose (C3); 4, β-MTG3:C3 (1:1); 5, α-MTG3:C3 (1:1); 6, β-MTG3:C3 (10:1); 7, α-MTG3:C3 (10:1); 8, no substrate.
The activity of Nod enzymes on the thiooligosaccharides and related substrates provides clues about their substrate specificity and tolerance. NodH, as a sulfotransferase active on a saccharide acceptor, is a representative of a diverse family of enzymes. Sulfated molecules have important biological functions and are found in a variety of organisms (9, 12). In general, the sulfotransferase reaction mechanism is not well understood (15). Two interpretations emerged about NodH sulfotransferase action based on its substrate preference. First, while NodH modifies only the C-6 of the reducing-end N-acetylglucosamine (8, 27), it apparently requires some recognition of the overall substrate backbone: an unmodified dimer is sulfated, but monomers or acylated dimers are not (8, 27). We found that NodH is active on substrates containing thioglycosidic linkages, although this is predicted to affect the conformation of the oligosaccharide backbone due to differences in bond lengths (carbon—sulfur at ∼1.8 Å compared to carbon—oxygen at ∼1.4 Å) and bond angles (the C—S—C bond angle is more acute at ∼105° than that of C—O—C at ∼110°) (17). Thus, while NodH may require recognition of sugar residues and modifying side groups, as well as recognition of the target C—OH group, there is apparently no requirement for recognition of the glycosidic C—O—C linkages.
Second, we found that NodH was inactive on C-1 methyl acceptor derivatives of the chitooligosaccharide backbone, suggesting that the reducing-end residue must be able to attain an open-ring form in order for NodH-mediated sulfate transfer to occur. However, we also observed that the C-1 methyl derivatives did not act as competitive inhibitors of successful NodH sulfotransfer to reducing chitooligosaccharide acceptors. This presents the alternative possibility that the C-1 methyl derivatives do not bind to enzymes due to steric hindrance. Mutagenic studies, as well as tests of additional alternative substrates, may further reveal the basis for the lack of action of the NodH enzyme on the nonreducing substrate. The accessibility of NodH to genetic and biochemical manipulation makes it an attractive candidate for tests of the sulfotransferase mechanism.
NodA and NodB carry out N acylation of thiochitotetraose backbones.
Acylation of 35S-labeled chitotetraose or thiochitotetraose was assayed using semipermeabilized S. meliloti cells (2). Briefly, cells were washed twice with ice-cold 70 mM Tris, pH 8.2, and resuspended in 0.01 volume of 70 mM Tris (pH 8.2)-2.5 mM EDTA. S. meliloti cells were frozen at −80°C. MgCl2 was added to a concentration of 5 mM, followed by addition of oligosaccharide (5 μCi/100 μl of cells). The cells were subjected to three cycles of freeze-thaw in liquid nitrogen and then were incubated at 15°C for 2 to 16 h. Cells were pelleted and extracted with chloroform-methanol-water (10:20:3).
The introduction of a radiolabeled substrate for Nod factor biosynthetic enzymes expressed in the cells allowed the products to be detected. For this study, the products of interest were the lipid-linked products generated by NodA and NodB. The S. meliloti cells used were a nodC mutant strain (TJ170/pE65) (13) and the common nod gene deletion strain SL44. The nodC mutant does not produce the chitin backbone needed for endogenous Nod factor. The lack of activity in the deletion strain confirmed that the lipid modification of sulfated chitotetraose depends on the presence of nodA and nodB (2). The reaction products were analyzed on 10-cm-square Silica-Gel 60 high-performance TLC (HPTLC) plates (Merck) with a chloroform-methanol-water-acetic acid (25:15:4:2) solvent system (2) (Fig. 5). Radiolabeled compounds were detected on dried plates by autoradiography after exposure to X-ray film.
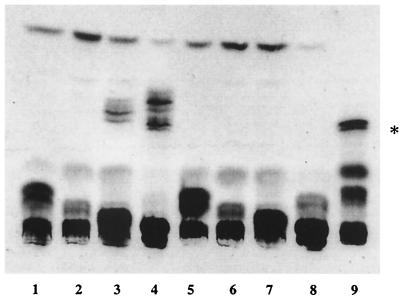
FIG. 5.
Acylation of thiochitotetraose. Acylation of 35S-labeled chitotetraose or thiochitooligosaccharides was assayed using permeabilized S. meliloti cells. The S. meliloti cells used were from a nodC::Tn5 mutant (TJ170/pE65) and the common nod gene deletion strain SL44. The reaction products were extracted and analyzed on Silica-Gel 60 HPTLC plates. Multiple bands near the origin may represent variable anomeric and/or acetylated substrate forms (see the text). The TJ170 extracts assayed included thiochitobiose (lane 1), thiochitotriose (lane 2), thiochitotetraose (lane 3), and chitotetraose (lane 4). The SL44 extracts assayed included thiochitobiose (lane 5), thiochitotriose (lane 6), thiochitotetraose (lane 7), and chitotetraose (lane 8). The 35S-labeled Nod factor (indicated by asterisk) is also shown (lane 9).
The sulfated chitotetraose and thiochitotetraose substrates were converted into hydrophobic products showing mobilities similar to that of the Nod factor standard (Fig. 5). The multiple hydrophobic products generated with sulfated chitotetraose may represent acyl substituents that are different from the specific acyl group present on the authentic Nod factor standard (2). NodA and NodB carry out the acylation but do not determine the structure of the acyl substituent to be attached to the oligosaccharide (2). Variable acetylation at the nonreducing end of the oligosaccharide by an O-acetyltransferase could also contribute to the heterogeneity of the products. The unacylated oligosaccharide substrates, which remain near the origin, have multiple bands, indicating possible resolution of anomeric forms in addition to variable acetylation.
We observed that the shorter thiochitobiose and thiochitotriose oligosaccharides were not substrates for the acylation reaction. This preference for a tetrasaccharide substrate was previously established for chitooligosaccharide acceptors (2). To confirm the identities of the products, the assay products with the tetrasaccharide substrates were also analyzed by high-performance liquid chromatography and eluted similarly to Nod factor standards (data not shown) (29).
Thiochitotetraose methyl glycoside is a poor substrate for NodAB.
To test for the dependence of the N acylation reaction on the configuration at the reducing-end C-1 position of thiochitotetraose, a 14C-radiolabeled form of the thiooligosaccharide substrate was generated by N acetylation with [14C]acetic anhydride by use of a protocol modified from that of Röhrig et al. (data not shown) (25, 29).
Permeabilized-cell assays were performed, and extracts were analyzed on Silica-Gel 60 HPTLC plates. The extracts containing the [14C]methyl glycoside of thiochitotetraose generated complex chromatograms with either solvent system (data not shown). To simplify the product mixture, the assay was reduced to its component reactions by use of permeabilized E. coli cells expressing nodA and nodB separately and together (2, 23). Extracts were analyzed on Silica-Gel 60 HPTLC plates with an n-butanol-ethanol-water (5:3:2) solvent system used for NodB analysis (14). Additionally, to simplify the chromatogram, the assay products from all of the extracts were first purified on BioBeads SM16 absorbent matrix (1). Extracts from Rhizobium cells were used, such that any products would have the Rhizobium acyl group for comparison to Nod factor standards. Product analysis indicated that the thiochitotetraose methyl glycoside can be acylated but is a poor substrate for the acylation reaction (data not shown) (29).
The activity of NodA and NodB on the reducing thiochitooligosaccharides showed a preference for tetrameric substrates rather than the shorter oligomers observed with natural substrates (2, 14, 25). The poor acylation of the methyl glycosides appears primarily to be the result of decreased activity by NodA, since NodB activity was detected independently in the cell extracts. The relative inactivity of NodAB acylation on the C-1 methyl derivatives implies that the reducing end of the oligosaccharide may be an important determinant for the substrate specificity of modifications at the nonreducing end. Alternatively, it is possible that the C-1 methyl glycosides resemble Nod factor biosynthetic intermediates. It is not known whether Nod factors are synthesized from lipid-linked intermediates (5), but if there were such an intermediate, as occurs in the synthesis of LPS, then the C-1 position is the logical position where such a linkage would occur (4).
Conclusions.
NodA, NodB, and NodH modify the chitooligosaccharide backbone during Nod factor biosynthesis in S. meliloti. These enzymes were also able to modify the alternative thiochitooligosaccharide substrates used in this study. The end product of reactions with the reducing thiochitotetramer is a novel Nod factor analog with an oligosaccharide backbone predicted to be resistant to chitinase enzymatic hydrolysis.
The success in synthesizing a Nod factor analog from the thiochitotetramer demonstrates the potential for the enzymatic synthesis of derivatives. Preliminary bioassays of the thiooligomeric products generated by semipermeabilized cells suggest that they were active on plants, but the assays were complicated by the background activities of other materials produced by permeabilized cells and even of the nod gene deletion strains of Rhizobium. Based on these considerations, we suggest that approaches to Nod factor derivatization that are completely in vitro enzymatic or chemienzymatic should be used to provide material for further activity studies. This has been accomplished for NodH (27), NodB (14), NodL (3), NodZ (22), NoeE (21), and NodS (11). The particular challenge for Nod factor derivatization will probably be the acylation reaction. Either direct in vitro activity of both NodA and NodB or a chemoenzymatic synthetic scheme (2, 19, 25) should be the future method of choice.
Acknowledgments
We thank Morrey Atkinson, David Ehrhardt, and other members of our laboratory for helpful discussions and advice on enzymes and chromatography.
A.M.S. was supported by an NIH training grant to Stanford University. Support for this project was provided by the Howard Hughes Medical Institute, by the Department of Energy Division of Energy Biosciences grant DE-FG03-90ER20010 to S.R.L., and by NIH grant DK09970 to Y.C.L.
REFERENCES
- 1.Atkinson, E. M. 1994. Structure and biosynthesis of Rhizobium meliloti lipo-oligosaccharide nodulation signals. Ph.D. thesis. Stanford University, Stanford, Calif.
- 2.Atkinson, E. M., M. M. Palcic, O. Hindsgaul, and S. R. Long. 1994. Biosynthesis of Rhizobium meliloti lipooligosaccharide Nod factors: NodA is required for an N-acyltransferase activity. Proc. Natl. Acad. Sci. USA 91:8418-8422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloemberg, G. V., J. E. Thomas-Oates, B. J. J. Lugtenberg, and H. P. Spaink. 1994. Nodulation protein NodL of Rhizobium leguminosarum O-acetylated lipo-oligosaccharides, chitin fragments and N-acetylglucosamine in vitro. Mol. Microbiol. 11:793-804. [DOI] [PubMed] [Google Scholar]
- 4.Carlson, R. W., N. P. J. Price, and G. Stacey. 1994. The biosynthesis of rhizobial lipo-oligosaccharide nodulation signal molecules. Mol. Plant-Microbe Interact. 7:684-695. [DOI] [PubMed] [Google Scholar]
- 5.Dénarié, J., F. Debellé, and J.-C. Promé. 1996. Rhizobium lipo-chitooligosaccharide nodulation factors. Annu. Rev. Biochem. 65:503-535. [DOI] [PubMed] [Google Scholar]
- 6.Downie, A. J. 1998. Functions of rhizobial nodulation genes, p. 387-402. In H. P. Spaink, A. Kondorosi, and P. J. J. Hooykaas (ed.), The Rhizobiaceae: molecular biology of model plant-associated bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 7.Egelhoff, T. T., and S. R. Long. 1985. Rhizobium meliloti nodulation genes: identification of nodDABC gene products, purification of nodA protein, and expression of nodA in Rhizobium meliloti. J. Bacteriol. 164:591-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrhardt, D. W., E. M. Atkinson, K. F. Faull, D. I. Freedberg, D. P. Sutherlin, R. Armstrong, and S. R. Long. 1995. In vitro sulfotransferase activity of NodH, a nodulation protein of Rhizobium meliloti required for host-specific nodulation. J. Bacteriol. 177:6237-6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falany, C. N. 1997. Sulfation and sulfotransferases. Introduction: changing view of sulfation and the cytosolic sulfotransferases. FASEB J. 11:1-2. [DOI] [PubMed] [Google Scholar]
- 10.Fisher, R. F., T. T. Egelhoff, J. T. Mulligan, and S. R. Long. 1988. Specific binding of proteins from Rhizobium meliloti cell-free extracts containing NodD to DNA sequences upstream of inducible nodulation genes. Genes Dev. 2:282-293. [DOI] [PubMed] [Google Scholar]
- 11.Geelen, D., B. Leyman, P. Mergaert, K. Klarskov, M. Van Montagu, R. Geremia, and M. Holsters. 1995. NodS is an S-adenosyl-l-methionine-dependent methyltransferase that methylates chitooligosaccharides deacetylated at the non-reducing end. Mol. Microbiol. 17:387-397. [DOI] [PubMed] [Google Scholar]
- 12.Hooper, L. V., S. M. Manzella, and J. U. Baenziger. 1996. From legumes to leukocytes: biological roles for sulfated carbohydrates. FASEB J. 10:1137-1147. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs, T. W., T. T. Egelhoff, and S. R. Long. 1985. Physical and genetic map of a Rhizobium meliloti nodulation gene region and nucleotide sequence of nodC. J. Bacteriol. 162:469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.John, M., H. Röhrig, J. Schmidt, U. Wieneke, and J. Schell. 1993. Rhizobium NodB protein involved in nodulation signal synthesis is a chitooligosaccharide deacetylase. Proc. Natl. Acad. Sci. USA 90:625-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kakuta, Y., L. G. Pedersen, C. W. Carter, M. Negishi, and L. C. Pederson. 1997. Crystal structure of estrogen sulfotransferase. Nat. Struct. Biol. 4:904-908. [DOI] [PubMed] [Google Scholar]
- 16.Leyh, T. S., J. C. Taylor, and G. D. Markham. 1988. The sulfate activation locus of Escherichia coli K12: cloning, genetic, and enzymatic characterization. J. Biol. Chem. 263:2409-2416. [PubMed] [Google Scholar]
- 17.Lide, D. R. 1990. Handbook of chemistry and physics, 71st ed. CRC Press, Boca Raton, Fla.
- 18.Mergaert, P., M. Van Montagu, and M. Holsters. 1997. Molecular mechanisms of Nod factor diversity. Mol. Microbiol. 25:811-817. [DOI] [PubMed] [Google Scholar]
- 19.Nicolaou, K. C., N. J. Bockovich, D. R. Caranague, C. W. Hummel, and L. F. Even. 1992. Total synthesis of the NodRM-IV factors, the Rhizobium nodulation signals. J. Am. Chem. Soc. 114:8701-8702. [Google Scholar]
- 20.Quesada-Vincens, D., R. Felley, T. Nasim, V. Viprey, U. Burger, J.-C. Prome, W. J. Broughton, and S. Jabbouri. 1997. Rhizobium sp. strain NGR234 NodZ protein is a fucosyltransferase. J. Bacteriol. 179:5087-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quesada-Vincens, D., M. Hanin, W. J. Broughton, and S. Jabbouri. 1998. In vitro sulfotransferase activity of NoeE, a nodulation protein of Rhizobium sp. NGR234. Mol. Plant-Microbe Interact. 11:592-600. [DOI] [PubMed] [Google Scholar]
- 22.Quinto, C., A. H. M. Wijfjes, G. V. Bloemberg, L. Blok-Tip, I. M. Lopez-Lara, B. J. Lugtenberg, J. E. Thomas-Oates, and H. P. Spaink. 1997. Bacterial nodulation protein NodZ is a chitin oligosaccharide fucosyltransferase which can also recognize related substrates of animal origin. Proc. Natl. Acad. Sci. USA 94:4336-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reuber, T. L., and G. Walker. 1993. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell 74:269-280. [DOI] [PubMed] [Google Scholar]
- 24.Roche, P., P. Lerouge, C. Ponthus, and J.-C. Promé. 1991. Structural determination of bacteria nodulation factors involved in the Rhizobium meliloti-alfalfa symbiosis. J. Biol. Chem. 266:10933-10940. [PubMed] [Google Scholar]
- 25.Röhrig, H., J. Schmidt, U. Wieneke, E. Kondorosi, I. Barlier, J. Schell, and M. John. 1994. Biosynthesis of lipooligosaccharide nodulation factors: Rhizobium NodA protein is involved in N-acylation of the chitooligosaccharide backbone. Proc. Natl. Acad. Sci. USA 91:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosen, S. D., and C. R. Bertozzi. 1996. Leukocyte adhesion: two selectins converge on sulphate. Curr. Biol. 6:261-264. [DOI] [PubMed] [Google Scholar]
- 27.Schultze, M., C. Staehelin, H. Röhrig, M. John, J. Schmidt, E. Kondorosi, J. Schell, and A. Kondorosi. 1995. In vitro sulfotransferase activity of Rhizobium meliloti NodH protein: lipochitooligosaccharide nodulation signals are sulfated after synthesis of the core structure. Proc. Natl. Acad. Sci. USA 92:2706-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sen, J., J. S. Goltz, L. Stevens, and D. Stein. 1998. Spatially restricted expression of pipe in the Drosophila egg chamber defines embryonic dorsal-ventral polarity. Cell 95:471-481. [DOI] [PubMed] [Google Scholar]
- 29.Southwick, A. M. 1998. Investigations into the biosynthesis of Rhizobium meliloti Nod factors and the molecular basis of their perception in Medicago. Ph.D. thesis. Stanford University, Stanford, Calif.
- 30.Spaink, H. P., A. H. Wijfjes, K. M. van der Drift, J. Haverkamp, J. E. Thomas-Oates, and B. J. Lugtenberg. 1994. Structural identification of metabolites produced by the NodB and NodC proteins of Rhizobium leguminosarum. Mol. Microbiol. 13:821-831. [DOI] [PubMed] [Google Scholar]
- 31.Staehelin, C., M. Schultze, E. Kondorosi, and A. Kondorosi. 1995. Lipo-chitooligosaccharide nodulation signals from Rhizobium meliloti induce their rapid degradation by the host plant alfalfa. Plant Physiol. 108:1607-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staehelin, C., M. Schultze, E. Kondorosi, R. B. Mellor, T. Boller, and A. Kondorosi. 1994. Structural modifications in Rhizobium meliloti Nod factors influence their stability against hydrolysis by root chitinases. Plant J. 5:319-330. [Google Scholar]
- 33.Wang, L.-X., and Y. C. Lee. 1996. Stereoselective synthesis of N-acetyl thiochitooligosaccharides. Different behaviours of methyl N-acetyl-α- and -β-thiochitobiosides during acetolysis. J. Chem. Soc. Perkin Trans. I 1:581-591. [Google Scholar]