Abstract
In previous studies we identified an 18-kb region of the Bacteroides conjugative transposon CTnDOT that was sufficient for mobilization of coresident plasmids and unlinked integrated elements, as well as self-transfer from Bacteroides to Escherichia coli. When this 18-kb region was cloned on a plasmid (pLYL72), the plasmid transferred itself constitutively in the absence of a coresident conjugative transposon. However, when this plasmid was present in a Bacteroides strain containing a coresident conjugative transposon, conjugal transfer was repressed in the absence of tetracycline and enhanced in the presence of tetracycline. These results suggested that a negative and a positive regulator of conjugal transfer were encoded outside the transfer region of the CTnDOT element. In this work, a minimal and inducible transfer system was constructed and used in transfer and Western blot analyses to identify the differentially regulated genes from CTnDOT responsible for the enhancement and repression of pLYL72 conjugal transfer. Both of these regulatory functions have been localized to a region of the CTnDOT element that is essential for CTn excision. In the presence of tetracycline, the regulatory protein RteC activates the expression of a putative topoisomerase gene, exc, which in turn results in an increase in transfer protein expression and a concomitant 100- to 1,000-fold increase in the frequency of pLYL72 transfer. Our results also suggest that since exc alone cannot result in enhancement of transfer, other factors encoded upstream of exc are also required. Conversely, in the absence of tetracycline, a gene located near the 3′ end of exc is responsible for the repression of transfer protein expression and also results in a 100- to 1,000-fold decrease in the frequency of pLYL72 transfer.
Bacteroides spp. are obligate anaerobic bacteria and account for about 25 to 30% of the bacteria in the human intestinal tract (15). Although Bacteroides spp. play a number of roles as part of the normal intestinal microflora, some strains are also opportunistic pathogens. Bacteroides spp. are the anaerobes most frequently isolated from clinical specimens (9).
Bacteroides spp. are hosts to a variety of transmissible elements, including plasmids, transposons, mobilizable transposons, and conjugative transposons (CTns) (18, 20, 21). The results of a recent survey suggest that it is this last group of elements, the CTns, that is responsible for the significant increase in tetracycline and macrolides-lincosamides-streptogramin-B-type resistance in the Bacteroides group (25).
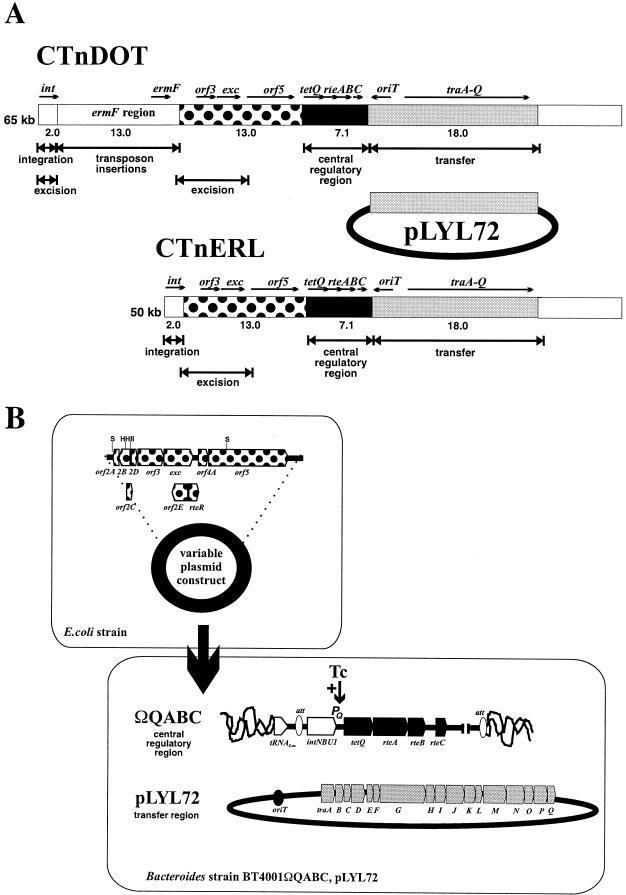
Bacteroides CTns range in size from 50 to 150 kb (1). The two best-characterized Bacteroides CTns are CTnERL and CTnDOT, which are almost identical except that CTnDOT contains a 13-kb insertion which contains an ermF resistance determinant (Fig. 1A) (34). A novel feature of the Bacteroides CTnDOT/CTnERL family of CTns is that self-transfer and the mobilization of coresident plasmids and mobilizable transposons are all stimulated 100- to 1,000-fold by pregrowth in a medium containing tetracycline. A regulatory region that includes four genes designated tetQ, rteA, rteB, and rteC mediates this tetracycline enhancement effect. This region is located near the middle of CTnDOT (31). In the presence of tetracycline, tetQ, rteA, and rteB are transcribed, and RteB directly or indirectly activates transcription of rteC (31), which subsequently controls the excision and transfer of the CTn.
FIG. 1.
Comparison of the Bacteroides CTns, CTnERL and CTnDOT, and the construction of the CTnDOT minimal transfer system. (A) CTnDOT is distinguished from CTnERL by a 13-kb insertion designated the ermF region, which is present in CTnDOT but absent from CTnERL (34). Modules of these Bacteroides CTns that are known to be necessary for integration, excision, transfer, and regulation are indicated. The 18-kb region contained in pLYL72, which contains sequences sufficient for constitutive self-transfer from Bacteroides to E. coli, is also shown. Various sequences from the region between the ermF region and the central regulatory region of CTnDOT (dotted box) were cloned into a Bacteroides shuttle vector, creating a variable plasmid. (B) These variable plasmid constructs were subsequently transferred from E. coli strain S17-1 into B. thetaiotaomicron strain 4001ΩQABC(pLYL72). This strain of Bacteroides contains a site-specifically integrated copy of the central regulatory region of CTnDOT (tetQ, rteA, rteB, and rteC) that mediates the tetracycline (Tc+) induced expression of the tetQ promoter (PQ) that results in the tetracycline induced transfer characteristic of the CTnDOT/CTnERL family of CTns and a plasmid sufficient for self-transfer (pLYL72) from Bacteroides to E. coli (13).
Previously, an 18-kb region of CTnDOT that is sufficient for self-transfer from Bacteroides to Escherichia coli (10−5 to 10−6 transconjugants per recipient) was identified (13). The plasmid containing this 18-kb region (pLYL72) transferred constitutively in the absence of a coresident CTn. However, in the presence of a coresident CTn, transfer of pLYL72 was not constitutive. Instead, transfer of pLYL72 was repressed 100- to 1,000-fold in the absence of tetracycline. In the presence of tetracycline, the transfer frequency of pLYL72 was enhanced 100- to 1,000-fold above the constitutive level of pLYL72 transfer. These results suggested that a negative and a positive regulator of conjugal transfer were encoded outside the transfer region of the CTnDOT element (13).
RteC is known to play an important role in the excision of the CTnDOT/CTnERL family of CTns. When a plasmid containing constitutively expressed rteC is provided in trans with a coresident CTnERL element, the CTn excises and transfers constitutively, even in a strain in which rteB has been disrupted (13). Although RteC is necessary for excision and was known to have some undetermined role in transfer of the CTn, RteC alone is not sufficient for either process (6, 13). This suggested that sequences encoded outside the transfer region and the central regulatory region of CTnDOT are required for repression and enhancement of conjugal transfer. The link between RteC and CTn excision has recently been determined (6), but the link between RteC and the modulation of transfer was, until now, unknown.
In this work, a minimal transfer system for CTnDOT was constructed in an effort to localize the regions of CTnDOT responsible for the repression of pLYL72 transfer in the absence of tetracycline and RteC-mediated enhancement of pLYL72 transfer in the presence of tetracycline.
MATERIALS AND METHODS
Strains, plasmids, and DNA manipulation.
The strains and plasmids used in this study are listed in Table 1. Two Bacteroides CTns, CTnERL and CTnDOT, were used in this study. CTnERL and CTnDOT sequences cross hybridize on Southern blots performed under high-stringency conditions, though it is known that there are some sequence differences between them in some regions of the CTns (2). In experiments to assay for tetracycline-induced enhancement of various functions, Bacteroides strains were cultured in TYG (Trypticase-yeast extract-glucose [24]) medium (uninduced) or in TYG medium containing tetracycline (1 μg/ml; induced). E. coli DH5αMCR (Bethesda Research Laboratories Inc., Gaithersburg, Md.) or HB101 (4) was grown in Luria broth or Luria broth agar. Antibiotic concentrations (in micrograms per milliliter) were as follows: ampicillin, 100; cefoxitin, 20; chloramphenicol, 10; erythromycin, 10; gentamicin, 200; kanamycin, 50; tetracycline, 1; thymidine, 100; and trimethoprim, 100. The methods utilized for DNA extraction, cloning, and Southern analysis have been described previously (17, 22).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant phenotypea | Source or reference and/or description |
|---|---|---|
| E. coli | ||
| DH5αMCR | RecA | Gibco BRL |
| HB101 | RecA Strr | 4 |
| BW19851 | RecA Tra+ Tpr Strr Pir+ | E. coli S-17 (28) with RK6 pir in uidA (14) |
| B. thetaiotaomicron 5482A | ||
| BT4100 | (Thy− Tpr) | Spontaneous thymidine-requiring mutant of BT5482A (27) |
| BT4104 | (Thy− Tpr Tcr) | BT4100 with CTnERL inserted in the chromosome (27) |
| BT4100N1S1 | (Thy− Tpr) | BT4100 containing an integrated copy of NBU1 (27) |
| BT4001ΩQABC | (Rifr Tcr) | BT4001 with site-specific insertion of tetQ, rteA, rteB, rteC in the chromosome (this study) |
| BT4100N1S1ΩQAB | (Thy− Tpr Tcr) | BT4100N1S1 with site-specific insertion of tetQ, rteA, and rteB in the chromosome (G.-R. Wang, unpublished). |
| Plasmids | ||
| pAMS7 | Apr (Tcr) | pNFD13-2 (32) with a deletion of a 2.1-kb EcoRI fragment and an insertion of a 7.4-kb fragment containing tetQ, rteA, rteB, and rteC from CTnDOT (30) |
| pLYL72 | Knr Cmr (Cmr Tra+) | Subclone containing an 18-kb tra region fragment from CTnDOT that is sufficient for self-transfer from Bacteroides to E. coli (13) |
| pLYL01 | Apr (Tcr) | 2.6-kb SstI fragment (containing the tetQ gene) from pNFD13-2 (32) cloned into the aatII site of pFD160R (13, 29) |
| pLYL05 | Apr (Cefr) | E. coli-Bacteroides shuttle vector containing cfxA cloned into the aatII site of pFD160 (16; L.-Y. Li, unpublished data) |
| pKSO1 | Apr (Cefr) | 7.6-kb SspI fragment from CTnDOT cloned into the SmaI site of pLYL05 containing sequences necessary for CTn excision (6); this fragment contains nine putative open reading frames, orf2A, -2B, -2C, and -2D; orf3; exc; 2E; rteR; and orf4A |
| pKSO4 | Apr (Cefr) | 4.9-kb Erase-a-Base clone of pKSO1 containing orf2A, -2B, -2C, and -2D; orf3; and a truncated copy of exc in which the last 840 bp of exc and all downstream regions have been deleted (6) |
| 2.5::pGEMT | Apr | 2.5-kb PCR fragment containing exc and putative orf2E and rteR cloned into pGEMT (6) |
| 2.1::pGEMT | Apr | 2.1-kb PCR fragment containing orf2A, -2B, -2C, and -2D and 227 bp from the amino-terminal region of orf3 cloned into pGEMT (6) |
| pKSO5 | Apr (Cefr) | 2.5-kb PstI/SstI fragment from 2.5::pGEMT containing exc and putative orf2E and rteR cloned into pLYL05 (6) |
| pKSO6 | Apr (Cefr) | 0.7-kb PCR fragment containing orf2d and 227 bp from the amino-terminal region of orf3 ligated to a 2.5-kb fragment from 2.5::pGEMT containing exc only cloned into pLYL05 (6) |
| pKSO7 | Apr (Cefr) | 2.1-kb PCR fragment from 2.1::pGEMT containing orf2A, -2B, -2C, and -2D and 227 bp from the amino-terminal region of orf3 ligated to a 2.5-kb fragment from 2.5::pGEMT containing exc, orf2E, and rteR; this clone contains an in-frame deletion of orf3 (6) |
| pGW41 | Apr (Cefr) | 2.1-kb fragment from 2.1::pGEMT containing orf2A, -2B, -2C, and -2D and 227 bp from the amino-terminal region of orf3 cloned into pLYL05 (this study) |
| pGW45 | Apr (Cefr) | 1.6-kb PCR fragment amplified using primers GW17F and JFLR/U10592F, containing orf2B, -2C, and -2D and 227 bp from the amino-terminal region of orf3 directionally ligated to a 2.5-kb fragment from 2.5::pGEMT containing exc, orf2E, and rteR cloned into pLYL05 (this study) |
| pGW46 | Apr (Cefr) | 1.2-kb PCR fragment amplified using primers GW16F and JFLR/U10592F, containing orf2C and -2D and 227 bp from the amino-terminal region of orf3 directionally ligated to a 2.5-kb fragment from 2.5::pGEMT containing exc, orf2E, and rteR cloned into pLYL05 (this study) |
| pYS41 | Apr (Cefr) | 1.5-kb PCR fragment containing orf2A and 764 bp of orf2B ligated to a 2.5-kb fragment from 2.5::pGEMT containing exc, orf2E, and rteR cloned into pLYL05 (Y. Sutanto, unpublished data) |
| pGW59 | Apr (Cefr) | 1.4-kb PCR fragment amplified with primers GW19F and GW20R and containing orf2E and 192 bp from the amino-terminal end of rteR cloned into pLYL05 (this study) |
| pGW60 | Apr (Cefr) | 1.7-kb PCR fragment amplified with primers GW19F and GW18R containing orf2E, rteR, and 296 bp from the amino-terminal region of orf4A cloned into pLYL05 (this study) |
| pGW88 | Apr (Cefr) | 1.2-kb SstI/DdeI blunted fragment from pGW60 rteR and 296 bp from the amino-terminal region of orf4A cloned into the SmaI site of pLYL05 (this study) |
| pGW87 | Apr (Cefr) | 0.78-kb SstI/FspI fragment from pGW60 containing rteR cloned into SmaI/SstI sites of pLYL05 (this study) |
Bacteroides phenotypes are shown in parentheses, and E. coli phenotypes are shown without parentheses. Abbreviations: Tra+, self-transmissible or able to mobilize plasmids in trans. Abbreviations of antibiotic resistances are as follows: Ap, ampicillin; Cef, cefoxitin; Cm, chloramphenicol; Em, erythromycin; Kn, kanamycin; Nal, Naladixic acid; Rif, rifampin; Str, streptomycin; Tc, tetracycline; Tp, trimethoprim; Thy−, thymidine auxotroph.
Construction of a Bacteroides strain carrying the regulatory region of CTnDOT integrated in the chromosome.
A 7.8-kb fragment from pAMS7 containing the central regulatory region of CTnDOT (tetQ, rteA, rteB, and rteC) was cloned into the EcoRV site of the pir-based suicide vector pNPR-IA46 containing the attachment site and integrase gene from NBU1 (23), generating pGWA35(1.3). pGWA35(1.3) was transformed into the pir+ E. coli strain BW19851 (14) and mobilized from E. coli BW19851 into Bacteroides thetaiotaomicron strain 4001. In Bacteroides spp., pGWA35(1.3) is unable to replicate, but because pGWA35(1.3) encodes NBU1 sequences sufficient for site-specific integration into the Bacteroides chromosome, transconjugants could result from site-specific recombination of pGWA35(1.3) into the Bacteroides chromosome. Since this construct lacks the genes necessary for NBU1 excision, integration is irreversible. Southern blot analysis was performed to confirm that pGWA35(1.3), and therefore the central regulatory region of CTnDOT, had been integrated site specifically into the 3′ end of a tRNALeu gene and was intact, generating the B. thetaiotaomicron strain BT4001ΩQABC. A similar method was employed to introduce single copies of tetQ, rteA, and rteB (ΩQAB) into B. thetaiotaomicron strain BT4100N1S1 (G.-R. Wang, unpublished data).
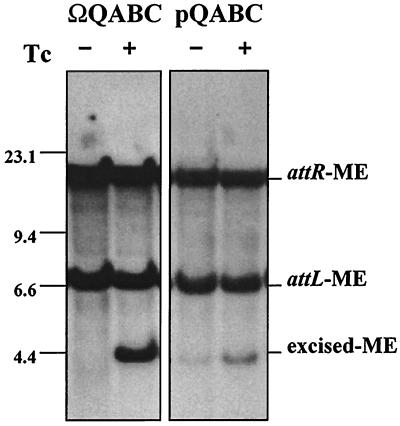
Recently, it was shown that a strain of B. thetaiotaomicron 4001 containing an integrated minielement (ME) (BT4001Ω2.6pGERM) that carried only the CTnDOT int and a small open reading frame, orf2, was able to excise from the Bacteroides chromosome if two other plasmids were provided in trans (6). One of these plasmids, pAMS9, contained the central regulatory region of the CTn (tetQ, rteA, rteB, and rteC), while the other plasmid (pKSO1) contained a gene, exc, shown to be essential for CTn excision (6). Consequently, to check that the regulatory region cloned into pGWA35(1.3) was functional, the construct was put into B. thetaiotaomicron strain BT4001Ω2.6pGERM(pKSO1), which contains a site-specifically integrated copy of the CTnDOT integrase and exc. If the integrated copy of QABC were functional, then the ME would be able to excise from the chromosome. Excision of the ME in BT4001Ω2.6pGERMΩQABC(pKSO1) was observed by Southern blot analysis (Fig. 2).
FIG. 2.
Comparison of the excision levels of the CTnDOT ME in strains containing the integrated versus the plasmid form of the central regulatory region. Southern blot analysis of Bacteroides strains containing the ME, a plasmid containing excision functions (pKSO1), and a single integrated copy of the central regulatory region (ΩQABC) are shown in the left panel. Strains containing the ME, a plasmid containing excision functions (pKSO1), and a multiple copy of the central regulatory region (pQABC) are shown in the right panel. The strains were grown in the absence (−) and presence (+) of tetracycline (Tc). Genomic DNA was prepared and digested with HindIII, and the Southern blot was then probed with a 1.1-kb probe that contains the left and right joined ends of the circular transfer intermediate of CTnDOT. The positions of the right (attR) and left (attL) attachment sites of the integrated ME and the joined junctions of the excised ME are indicated. The positions of lambda HindIII standard markers are shown on the left.
In the strain containing a single integrated copy of the central regulatory region, there is no detectable excision of the ME without tetracycline induction (Fig. 2, left panel), unlike the strain containing the plasmid form of the central regulatory region (Fig. 2, right panel). Also, under induced conditions, the level of excision is greater for the integrated central regulatory region (Fig. 2, left panel) than for the plasmid form of the central regulatory region (Fig. 2, right panel). These results clearly show that the level of excision of the ME was higher and more tightly regulated in this system than in the one previously reported (6), in which the regulatory system was encoded on a plasmid (Fig. 2). After confirmation that the central regulatory region was functional in strain BT4001ΩQABC, other constructs were introduced into BT4001ΩQABC or BT4100N1S1ΩQAB for transfer assays.
Mobilization and conjugal-transfer experiments.
E. coli HB101 containing IncPα plasmid RP1 was utilized to mobilize plasmids transformed into E. coli DH5αMCR into Bacteroides strains using a triparental mating procedure described elsewhere (24). Transfers of pLYL72 from B. thetaiotaomicron to E. coli were performed as described previously (13). The transfer frequency is expressed as the number of transconjugants per recipient.
Western blot analyses.
To detect the expression of transfer proteins, TraG, TraN, and TraP, the membrane fraction (11, 12) was isolated from the appropriate B. thetaiotaomicron strains (Table 1) and the protein concentration was determined using the DC protein assay (Bio-Rad). Between 100 and 150 μg of each membrane sample was loaded into each well of a 10% sodium dodecyl sulfate-polyacrylamide gel for separation and subsequently electrotransferred to Trans-blot nitrocellulose membranes (Bio-Rad). Western blots were incubated with polyclonal antibodies to TraG, TraN, or TraP (3) diluted 500- to 1,000-fold in TTBS (20 mM Tris [pH 7.5], 0.2% Tween 20, 0.5 M NaCl) with 1% bovine serum albumin at room temperature for 1 to 16 h, depending upon the titer of the antibody. The TraG, TraN, and TraP proteins were detected using the colorimetric Opti-4CN substrate and detection kit (Bio-Rad) and subsequently scanned using a Bio-Rad gel documentation system and Quantity One software.
PCR cloning.
In some experiments, derivatives of pKSO1 were generated by using PCR to construct deletions of selected open reading frames (Table 1 and Fig. 3). These constructs were assembled by amplification of fragments of various sizes from a region upstream of orf3 or including the 5′ end of orf3 and amplification of a second fragment from inside the 5′ end of orf3 to a region 200 bp downstream of exc. These amplicons were subsequently ligated together using restriction sites engineered into the oligonucleotide sequences. The primers utilized for these experiments are summarized in Table 2.
FIG. 3.
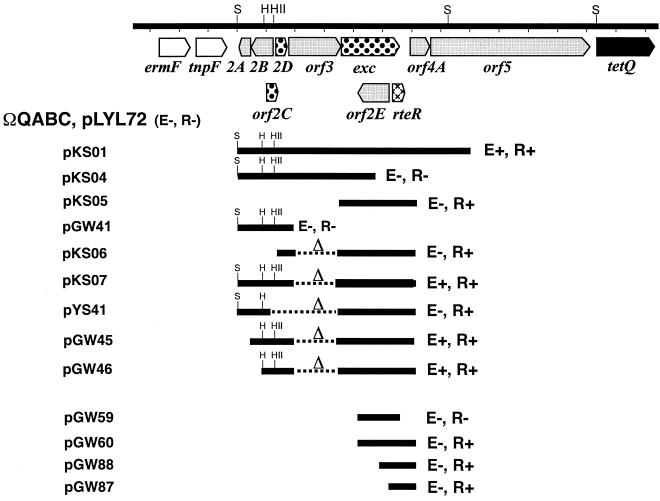
Localization of positive and negative regulators of conjugal transfer. A restriction map of sequences between the ermF region (open arrows) and tetQ (solid arrow) is shown. The horizontal lines below the restriction map show the DNA fragments from this region that were cloned into pLYL05 and transferred into B. thetaiotaomicron strain BT4001ΩQABC(pLYL72), as indicated above the names of the plasmid subclones. The name of the plasmid carrying the cloned DNA fragment is indicated to the left of each line. Deleted regions are shown as dashed lines. At the right of each line, the presence (+) or absence (−) of the positive regulator (E; enhancer) or the negative regulator (R; repressor) is indicated, based on the results of conjugal-transfer experiments and Western blot analyses. The restriction sites shown include HindIII (H), HincII (HII), and SspI (S). The smallest clone sufficient for enhancement of transfer is pGW46. The smallest clone sufficient for repression of conjugal transfer is pGW87. Open reading frames that potentially have a role in the positive regulation of transfer (dotted arrows) or in the negative regulation of conjugal transfer (cross-hatched arrow) or which are not involved in either positive or negative regulation (shaded arrows) are distinguished.
TABLE 2.
Oligonucleotides used for construction of subclones
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| GW16F | TTTCTCGATAGATACTATTTCCAT |
| GW17F | ATTTCGAGCCTTTGCGCATATTCTGCA |
| JFLR/U10592F | GTAGAAAAGCTTGCGACTGGGTTC |
| GW18R | AAGCCGAGCTCTCTCATACG |
| GW19F | TGCGACGGCGAAGTCGTGAAATTCTCATCC |
| GW20R | GAACAAGTGGCAAAGTTAATACTATTT |
RESULTS
Construction of a Bacteroides strain to identify modulators of conjugal transfer.
In an effort to identify the regions outside of the transfer and central regulatory regions of the CTn responsible for the modulation of pLYL72 transfer, it was necessary to construct a new strain of B. thetaiotaomicron 4001 [BT4001ΩQABC(pLYL72)]. This Bacteroides strain contains the transfer region (pLYL72) of CTnDOT and a single copy of the tetracycline-induced central regulatory region (tetQ, rteA, rteB, rteC, or QABC) that was introduced site specifically into the chromosome (BT4001ΩQABC) using the NBU1 integrase and attP (Fig. 1B) (26). Other regions of CTnDOT that were potentially involved in the modulation of pLYL72 transfer could then be introduced into strain BT4001ΩQABC(pLYL72) and tested for the ability to stimulate or repress transfer (Fig. 1B).
Localization of sequences responsible for repression and enhancement of the conjugal transfer of pLYL72.
Plasmid constructs containing DNA fragments from a 13.0-kb region located between the ermF region and tetQ of the central regulatory region of CTnDOT were tested for the ability to repress or enhance conjugal transfer (Fig. 1A). This region was chosen because it is outside the transfer and central regulatory regions and is present in both CTnDOT and CTnERL. Fragments from this 13.0-kb region (Fig. 3) were transferred to Bacteroides host strain BT4001ΩQABC(pLYL72), and the subsequent frequency of pLYL72 transfer was determined in the presence and absence of tetracycline induction. The results of these transfer experiments are summarized in Table 3.
TABLE 3.
Effect of CTnDOT sequences between the ermF region and tetQ on the frequency of pLYL72 self-transfer from Bacteroides to E. coli
| B. thetaiotaomicron 4001 strain | Frequency of pLYL72 transfera
|
Presence of enhancer and repressorb | |
|---|---|---|---|
| −Tc | +Tc | ||
| (pLYL72, pLYL01) | 10−5-10−6 | 10−5-10−6 | E−, R− |
| ΩCTnERL(pLYL72) | 10−7-10−8 | 10−2-10−3 | E+, R+ |
| ΩQABC(pLYL72) | 10−5-10−6 | 10−5-10−6 | E−, R− |
| ΩQABC(pLYL72, pKSO1)c | 10−8-<10−9 | 10−2-10−3 | E+, R+ |
| ΩQABC(pLYL72, pKSO4) | 10−5-10−6 | 10−5-10−6 | E−, R− |
| ΩQABC(pLYL72, pKSO5) | 10−8-<10−9 | 10−8-<10−9 | E−, R+ |
| ΩQABC(pLYL72, pKSO6) | 10−8-<10−9 | 10−8-<10−9 | E−, R− |
| ΩQABC(pLYL72, pKSO7) | 10−8-<10−9 | 10−2-10−3 | E+, R+ |
| ΩQABC(pLYL72, pGW41) | 10−5-10−6 | 10−5-10−6 | E−, R− |
| ΩQABC(pLYL72, pGW45) | 10−8-<10−9 | 10−2-10−3 | E+, R+ |
| ΩQABC(pLYL72, pGW46) | 10−8-<10−9 | 10−2-10−3 | E+, R+ |
| ΩQABC(pLYL72, pGW59) | 10−5-10−6 | 10−5-10−6 | E−, R− |
| ΩQABC(pLYL72, pGW87) | 10−8-<10−9 | 10−8-<10−9 | E−, R+ |
| ΩQAB(pLYL72) | 10−5-10−6 | 10−5-10−6 | E−, R− |
| ΩQAB(pLYL72, pKSO1) | 10−8-<10−9 | 10−8-<10−9 | E−, R+ |
Frequencies of conjugal transfer are expressed as transconjugants per recipient cell observed when donors were induced (+Tc) or not induced (−Tc) by tetracycline. The range of values reflects the experiment-to-experiment variation and represents at least three separate mating experiments. Transfer experiments were from a Bacteroides donor to an E. coli HB101 recipient.
E, enhancer; R, repressor; +, present; −, absent.
The results showed that a plasmid conferring tetracycline resistance (pLYL01) or the central regulatory region (ΩQABC) alone were not sufficient for either repression or enhancement of pLYL72 transfer, since transfer of pLYL72 was constitutive in these backgrounds (Table 3). However, a plasmid containing a 7.6-kb region (pKSO1) caused a 100- to 1,000-fold decrease in the frequency of pLYL72 transfer in the absence of tetracycline and a 100- to 1,000-fold increase in pLYL72 transfer in the presence of tetracycline (Table 3 and Fig. 3). Hence, sequences cloned on pKSO1 were sufficient for both repression and enhancement of pLYL72 transfer.
pKSO1 contains nine possible open reading frames (Fig. 3) (GenBank accession number AJ431573). It was previously reported that the putative protein encoded by exc had significant amino acid similarity to proteins encoded by some conjugal plasmids and a CTn, Tn1549 (10), from gram-positive organisms and to chromosomally encoded topoisomerases (6). None of the other putative open reading frames contained by pKSO1 had significant similarity to sequences available in the GenBank database, nor did they contain any motifs that could be indicative of function.
In an attempt to further localize the determinants responsible for the repression and enhancement of conjugal transfer, pKSO1 was subcloned further. In one of these subclones, pKSO4, the 3′ end of the exc gene had been truncated, and this neither positively nor negatively affected pLYL72 transfer. This indicated that both repressor and enhancer functions were located in the region of pKSO1 that has been truncated in pKSO4 (Table 3 and Fig. 3). The results showed that although the central regulatory region of CTnDOT was not sufficient for the enhancement of pLYL72 transfer, the central regulatory region is important for the activation of the positive activator(s) of conjugal transfer and that exc is required for enhancement of conjugal transfer. Interestingly, this region of CTnDOT, and exc in particular, has recently been shown to be required for the tetracycline-dependent excision of the CTnDOT ME (6).
Localization of sequences responsible for the repression of pLYL72 conjugal transfer.
A 2.5-kb region containing exc, orf2E, and rteR (pKSO5) was sufficient for repression of conjugal transfer. In an effort to further localize the repressor of pLYL72 transfer, subclones of the 2.5-kb fragment containing repressor activity were constructed (Fig. 3). In pGW59, the 7 bp from the C-terminal region of rteR has been deleted, and no repression of pLYL72 transfer was detected. Instead, transfer of pLYL72 appeared to be constitutive. In contrast, the repressor appeared to be functional in subclones pGW60 and pGW88 (data not shown) and in the smallest subclone, pGW87, which contained a fragment of only 778 bp (Table 3 and Fig. 3). This fragment contains a putative open reading frame, designated rteR, which may encode the repressor of conjugal transfer.
Localization of sequences responsible for the enhancement of pLYL72 conjugal transfer.
Although pKSO5 contained sequences sufficient for repression of pLYL72 transfer, sequences present in pKSO5 were not sufficient for the enhancement of conjugal transfer. This suggested that sequences upstream of exc were required in conjunction with exc for enhancement of pLYL72 transfer. This was not surprising, since exc was previously shown to be in an operon with orf3 (6). A subclone containing orf2ABCD, an in-frame deletion of orf3, and an intact copy of the exc gene (pKSO7) resulted in enhancement of transfer, indicating that orf3 and orf4A are not required for enhancement of pLYL72 transfer (Fig. 3). Further truncation of the upstream region showed that orf2A (pGW45) and orf2B (pGW46) were also not required for enhancement of pLYL72 transfer.
Deletion of sequences containing putative genes orf2C and orf2D (pYS41 and pKSO6) from the upstream region abolished enhancement of transfer (Table 3). However, since pYS41 was able to complement an exc disruption in CTnERL (CTnERLΩexc), exc was still produced from this construct, and hence the promoter was still intact (Y. S. Sutanto, N. B. Shoemaker, J. F. Gardner, and A. A. Salyers, submitted for publication). These results suggest that other trans factors in this region are required to work in conjunction with exc to result in enhancement of conjugal transfer.
The expression of the pLYL72 transfer proteins TraG, TraN, and TraP is modulated in the presence and absence of tetracycline.
Transfer assays led to the identification of sequences responsible for the 100- to 1,000-fold enhancement of pLYL72 transfer in the presence of tetracycline and the 100- to 1,000-fold repression of pLYL72 transfer in the absence of tetracycline. However, it was unknown whether the modulation of pLYL72 transfer was associated with a change in transfer protein expression or otherwise. Western blot analyses of TraG, TraN, and TraP were undertaken to determine whether the expression of transfer proteins differed when the conjugal transfer of pLYL72 was repressed, constitutive, or enhanced. Antibodies against these three transfer proteins were used because traG, traN, and traP represent three of four operons thought to make up the transfer region of CTnDOT (2), and we wanted to know whether all three operons were regulated similarly.
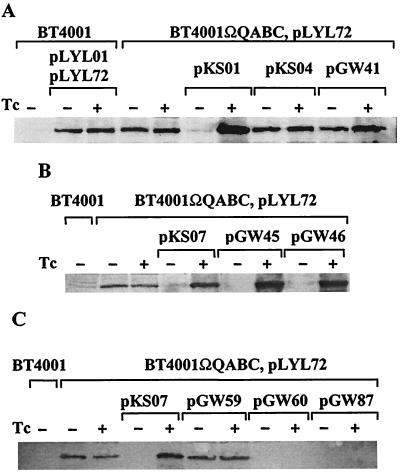
Results of Western blots with antibodies that detect TraG, TraN, or TraP yielded identical profiles. Thus, the three tra operons are regulated similarly. The results for TraG production show that in the absence of a coresident CTn, the expression of TraG from pLYL72 is constitutive (Fig. 4A). Similarly, when pLYL72 was present in a strain containing the regulatory region from CTnDOT (ΩQABC), the expression of transfer proteins was also constitutive, indicating that the central regulatory region alone does not encode the functions necessary for repression or enhancement of transfer (Fig. 4A). In contrast, when pKSO1, which contains both the repressor and enhancer of transfer, was present in host strain BT4001ΩQABC(pLYL72), TraG expression was repressed in the absence of tetracycline and enhanced above the constitutive level in the presence of tetracycline (Fig. 4A). When exc was truncated, as in pKSO4, or when the sequences upstream of exc alone (pGW41) were present in B. thetaiotaomicron strain BT4001ΩQABC(pLYL72), expression of TraG was no longer repressed or enhanced (Fig. 4A). These results are consistent with transfer experiments that suggest that exc, and sequences downstream of exc, are important for both positive and negative regulatory activities.
FIG. 4.
TraG expression levels in the presence and absence of modulators of conjugal transfer. For Western blot analysis of TraG (96.6 kDa), total membrane fractions were isolated from the indicated Bacteroides strains. Each lane contains 100 μg of protein prepared from cells induced (+) or not induced (−) with tetracycline (Tc). (A) Sequences responsible for the repression and enhancement of conjugal transfer repress and enhance the expression of TraG in the absence and presence of tetracycline, respectively. (B) Further localization of sequences sufficient for enhancement of TraG expression. (C) Further localization of sequences sufficient for repression of TraG expression.
Further Western blot analysis showed that the deletion of orf4A and orf3 (pKSO7), or deletion of orf2A (pGW45) or orf2B (pGW46), does not have a deleterious effect on the level of negative or positive regulation of TraG (Fig. 4B). These results are consistent with transfer experiments that indicated that exc and the orf2C-orf2D region present in pGW46 are sufficient for enhanced transfer of pLYL72, since they are also sufficient for an enhanced level of TraG expression.
In the absence of sequences involved in the repression of conjugal transfer, TraG is expressed constitutively from pLYL72, while in the presence of the enhancer and repressor (pKSO7), TraG expression is repressed in the absence of tetracycline and enhanced in the presence of tetracycline (Fig. 4C). Western blot analysis of a strain containing pKSO4 (Fig. 4A) had already suggested that the repressor of transfer was located near the 3′ end of exc, and so Western analysis was used to further localize the repressor of transfer. When rteR is truncated (pGW59), TraG production is no longer negatively regulated in the absence of tetracycline, and instead, TraG is produced constitutively. In contrast, when sequences upstream of rteR are deleted and the 250 bp downstream of rteR is still present, the repressor is active (pGW60) and the expression of the pLYL72 transfer protein, TraG, is repressed in both the presence and absence of tetracycline (Fig. 4C).
Since the repression of transfer protein gene expression occurs in the absence of tetracycline, it was possible that the repressor of pLYL72 transfer is expressed in the absence of tetracycline. Similarly, the higher level of transfer proteins only occurs in the presence of tetracycline, and so the positive effector of transfer is only produced under induced conditions. These results are consistent with two differentially regulated factors being involved in the modulation of conjugal transfer. Our results also showed that an increase in conjugal transfer is associated with an increase in the level of transfer proteins detectable by Western blotting. Whether the increase in transfer protein levels detected by Western blotting is attributable to an increase in the stability of the transfer proteins, to more efficient localization and therefore protection of the membrane-associated transfer proteins, or to an increase in expression at the transcriptional or translational level remains to be determined.
RteC is required for production of the enhancer of pLYL72 transfer.
Previous results suggested that RteC plays an important role in conjugal transfer (13). To determine whether RteC is also required for enhancement of conjugal transfer, pKSO1 was introduced into a strain of Bacteroides that contains pLYL72 and tetQ, rteA, and rteB from the central regulatory region but no rteC [BT4100N1S1ΩQAB(pLYL72)] (G.-R. Wang, unpublished). In the absence of RteC, no enhancement of pLYL72 transfer or increase in transfer protein production was detected. Repression of transfer and transfer protein expression, however, was constitutive. In the absence of pKSO1, there is no repression of constitutive pLYL72 transfer (Table 3). These results show that RteC is required for the positive regulation of conjugal transfer.
In the presence of sequences required for enhancement, transfer of pLYL72 between Bacteroides strains is detectable.
Prior to this work, pLYL72 had been shown to contain sequences sufficient for self-transfer from Bacteroides to E. coli (13). However, pLYL72 appeared not to transfer at a detectable level from Bacteroides to Bacteroides (<10−9). The reason for this was not known. In light of results presented in this work, in which a region of CTnDOT shown to enhance pLYL72 transfer has been identified, the ability of pLYL72 to transfer from a Bacteroides donor to a Bacteroides recipient (BT4100) was reassessed. In the presence of the central regulatory region alone [BT4001ΩQABC(pLYL72)], a 10- to 100-fold increase in the transfer frequency of pLYL72 was observed (Table 4). This suggests that for conjugal transfer between Bacteroides strains, the central regulatory region alone is able to elicit a positive effect that we were not able to detect in Bacteroides-to-E. coli matings. In the presence of sequences known to enhance pLYL72 transfer from Bacteroides to E. coli [BT4001ΩQABC(pLYL72, pKSO7)], transfer of pLYL72 was increased at least 1,000- to 10,000-fold (Table 4). These results suggest that the enhancement of transfer is important for transfer between Bacteroides strains.
TABLE 4.
Effect of CTnDOT sequences between the ermF region and tetQ on the frequency of pLYL72 self-transfer from a Bacteroides donor to a Bacteroides recipient
| B. thetaiotaomicron 4001 strain | Frequency of pLYL72 transfera
|
|
|---|---|---|
| −Tc | +Tc | |
| (pLYL72) | <10−9 | <10−9 |
| ΩQABC(pLYL72) | <10−9 | 10−7-10−8 |
| ΩQABC(pLYL72, pKSO7) | <10−9 | 10−5-10−6 |
Frequencies of conjugal transfer are expressed as transconjugants per recipient cell observed when donors were induced (+Tc) or not induced (−Tc) by tetracycline. The range of values reflects the experiment-to-experiment variation and represents at least three separate mating experiments. Transfer experiments were from a Bacteroides donor to a Bacteroides recipient.
DISCUSSION
Previous studies had suggested that the central regulatory region of CTnDOT, and RteC in particular, played a role in the initiation of excision and some undetermined role in transfer of the integrated CTn. In this work, the link between RteC and the enhanced levels of pLYL72 transfer in the presence of a coresident CTn has been identified and localized to a region of CTnDOT that is known to be required for the excision of the CTnDOT element (6). In addition, the factor responsible for the repression of pLYL72 transfer has been localized to the same region of the CTnDOT element as the positive regulator of transfer.
Although these two factors have been localized to the same region of the CTn, they are clearly separate from each other. The enhancer is induced in the presence of tetracycline, is transcribed from a promoter region located upstream of orf2C, and includes Exc plus another trans factor or factors encoded within a 1.0-kb region upstream of orf3. In contrast, the repressor of transfer is expressed in the absence of tetracycline and encoded by a gene that is not dependent on the Exc promoter region for expression.
It is perhaps not surprising that the regulations of excision and transfer functions, both positive and negative, are linked, because these processes need to be tightly coordinated to ensure that the excision of the integrated CTn takes place before transfer. If transfer was able to occur prior to excision, this might, as in the case of E. coli strains containing an integrated F plasmid, result in the formation of Hfr-like strains of Bacteroides. This is because after the internal oriT of CTnDOT is nicked, the entire 6-Mb Bacteroides chromosome would have to be transferred before the part of the element attached to the 3′ end of the oriT. Consequently, premature nicking of oriT could result in deletions of part of the CTn.
The gram-positive CTn Tn916 solves this coordination problem structurally, since excision and circularization of the transfer intermediate are required for the expression of transposon-encoded transfer functions (5). In Tn916, the expression of tra functions in the circular intermediate is due to transcription from a promoter that runs through the attachment site. For the larger Bacteroides CTns of the CTnDOT/CTnERL family, such a coordination strategy is not feasible. The transfer genes of CTnDOT are not located near an end of the transposon; in fact, the first transfer gene in the tra operon, traA, is 40 kb from the attachment site in the circular intermediate from which such a promoter would be derived (Fig. 1A). These results suggest that for the CTnDOT/CTnERL family of transposons, multiple regulatory circuits have been recruited to ensure that excision and transfer are coordinated appropriately.
One explanation for the apparent increase in the amount of transfer protein detected by Western blot analyses after induction with tetracycline might be that the accumulation is due to an increase in the copy number of circular forms of the CTn. Such a time-dependent accumulation of the circular form of the mobilizable transposon NBU1 was observed recently (33). However, in the transfer system [BT4001ΩQABC(pLYL72)] used in these studies, the CTn is essentially already in a circular form, since the region being transferred is located on a plasmid. This plasmid contains a pB8-51 replicon that is maintained at a copy number of approximately 8 to 10 per copy of the Bacteroides chromosome (19).
Results also show that the enhancement of transfer is important for detection of pLYL72 transfer between Bacteroides strains. The frequency of transfer for a single copy of a CTnDOT/CTnERL element between Bacteroides strains is 10−5 to 10−6 transconjugants per recipient, the same frequency at which pLYL72 transferrs from Bacteroides to Bacteroides in the presence of the enhancer sequences. It should be noted, however, that an integrated CTn differs from pLYL72 in that transfer involves three steps. The element must excise, transfer, and integrate into the recipient chromosome. In contrast, transfer of pLYL72 involves only two steps, transfer and the establishment of a plasmid. With the omission of the excision and integration steps, one might expect that the multiple-copy plasmid pLYL72 would transfer at a higher frequency than the CTnDOT element. Our results show that this is not the case.
Also, the transfer frequency of pLYL72 from Bacteroides to E. coli is about 1,000- to 10,000-fold higher than the transfer frequency of pLYL72 between Bacteroides spp. There are many possible explanations for this phenomenon, one of which could be that the oriV (RSF1010-base) utilized in E. coli is not the same as the one utilized in Bacteroides (pB8-51). Consequently, the efficiency with which the plasmid can be established and the stability of the plasmid in the recipient strain may be influencing the observed transfer frequency.
The previous report that the protein encoded by exc had significant homology to proteins encoded by some conjugal plasmids and a CTn, Tn1549 (10), and to chromosomally encoded topoisomerases (6) is of significant interest. This is because there are several examples of topoisomerase alleles being implicated in the global modulation of gene expression by altering DNA topology (7, 8). Consequently, it is possible that exc, like other topoisomerase alleles, is able to enhance conjugal transfer and transfer protein production by altering local supercoiling. Hence, investigations are in progress to characterize the Exc protein to determine whether it is a topoisomerase and whether this topoisomerase activity is required for the enhancement of conjugal transfer and of transfer protein expression. The nature of the interaction between Exc and the other trans factors encoded near the exc promoter region is an area that we also intend to investigate in the future. Similarly, the mechanism by which the repressor factor modulates conjugal transfer will also be investigated.
Acknowledgments
We thank Jorge Frias and Song Ok Kang for assistance in the preliminary cloning of this region of CTnDOT.
This work was supported by grant AI 22383 from the National Institutes of Health.
REFERENCES
- 1.Bedzyk, L. A., N. B. Shoemaker, K. E. Young, and A. A. Salyers. 1992. Insertion and excision of Bacteroides conjugative chromosomal elements. J. Bacteriol. 174:166-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonheyo, G., D. Graham, N. B. Shoemaker, and A. A. Salyers. 2001. Transfer region of a Bacteroides conjugative transposon, CTnDOT. Plasmid 45:41-51. [DOI] [PubMed] [Google Scholar]
- 3.Bonheyo, G. T., B. B. Hund, N. B. Shoemaker, and A. A. Salyers. 2001. Transfer region of a Bacteroides conjugative transposon contains regulatory as well as structural genes. Plasmid 46:202-209. [DOI] [PubMed] [Google Scholar]
- 4.Boyer, H. B., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification system of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 5.Celli, J., and P. Trieu-Cuot. 1998. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol. Microbiol. 28:103-117. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, Q., Y. Sutanto, N. B. Shoemaker, J. F. Gardner, and A. A. Salyers. 2001. Identification of genes required for excision of CTnDOT, a Bacteroides conjugative transposon. Mol. Microbiol. 41:625-632. [DOI] [PubMed] [Google Scholar]
- 7.Dorman, C. J. 1991. DNA supercoiling and environmental regulation of gene expression in pathogenic bacteria. Infect. Immun. 59:745-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorman, C. J. 1996. Flexible response: DNA supercoiling, transcription and bacterial adaptation to environmental stress. Trends Microbiol. 4:214-216. [DOI] [PubMed] [Google Scholar]
- 9.Finegold, S. M., and W. L. George. 1989. Anaerobic infections in humans. Academic Press, San Diego, Calif.
- 10.Garnier, F., S. Taourit, P. Glaser, P. Courvalin, and M. Galimand. 2000. Characterization of transposon Tn1549, conferring VanB-type resistance in Enterococcus spp. Microbiology 146:1481-1489. [DOI] [PubMed] [Google Scholar]
- 11.Kotarski, S. F., J. Linz, D. M. Braun, and A. A. Salyers. 1985. Analysis of outer membrane proteins which are associated with growth of Bacteroides thetaiotaomicron on chondroitin sulfate. J. Bacteriol. 163:1080-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotarski, S. F., and A. A. Salyers. 1984. Isolation and characterization of outer membranes of Bacteroides thetaiotaomicron grown on different carbohydrates. J. Bacteriol. 158:102-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, L. Y., N. B. Shoemaker, and A. A. Salyers. 1995. Location and characteristics of the transfer region of a Bacteroides conjugative transposon and regulation of transfer genes. J. Bacteriol. 177:4992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metcalf, W. W., W. Jiang, and B. L. Wanner. 1994. Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6K gamma origin plasmids at different copy numbers. Gene 138:1-7. [DOI] [PubMed] [Google Scholar]
- 15.Moore, W. E., E. P. Cato, and L. V. Holdeman. 1978. Some current concepts in intestinal bacteriology. Am. J. Clin. Nutr. 31:S33-S42. [DOI] [PubMed]
- 16.Parker, A. C., and C. J. Smith. 1993. Genetic and biochemical analysis of a novel Ambler class A beta-lactamase responsible for cefoxitin resistance in Bacteroides species. Antimicrob. Agents Chemother. 37:1028-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saito, H., and K. I. Miura. 1963. Preparation of transforming deoxy-ribonucleic acid by phenol treatment. Biochim. Biophys. Acta 72:619-629. [PubMed] [Google Scholar]
- 18.Salyers, A. A., N. B. Shoemaker, G. Bonheyo, and J. Frias. 1999. Conjugative transposons: transmissible resistance islands, p. 331-346. In J. B. Kaper and J. Hacker (ed.), Pathogenicity islands and other mobile virulence elements. ASM Press, Washington, D.C.
- 19.Salyers, A. A., N. B. Shoemaker, A. Cooper, J. D'Elia, and J. A. Shipman. 1999. Genetic methods for Bacteroides species. Methods Microbiol. 29:229-249. [Google Scholar]
- 20.Salyers, A. A., N. B. Shoemaker, and L. Y. Li. 1995. In the driver's seat: the Bacteroides conjugative transposons and the elements they mobilize. J. Bacteriol. 177:5727-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salyers, A. A., N. B. Shoemaker, A. M. Stevens, and L. Y. Li. 1995. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59:579-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 23.Shoemaker, N., G. Wang, and A. Salyers. 1996. NBU1, a mobilizable site-specific integrated element from Bacteroides spp., can integrate nonspecifically in Escherichia coli. J. Bacteriol. 178:3601-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shoemaker, N. B., C. Getty, E. P. Guthrie, and A. A. Salyers. 1986. Regions in Bacteroides plasmids pBFTM10 and pB8-51 that allow Escherichia coli-Bacteroides shuttle vectors to be mobilized by IncP plasmids and by a conjugative Bacteroides tetracycline resistance element. J. Bacteriol. 166:959-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shoemaker, N. B., H. Vlamakis, K. Hayes, and A. A. Salyers. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shoemaker, N. B., G. R. Wang, and A. A. Salyers. 1996. The Bacteroides mobilizable insertion element, NBU1, integrates into the 3′ end of a Leu-tRNA gene and has an integrase that is a member of the lambda integrase family. J. Bacteriol. 178:3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shoemaker, N. B., G. R. Wang, A. M. Stevens, and A. A. Salyers. 1993. Excision, transfer, and integration of NBU1, a mobilizable site-selective insertion element. J. Bacteriol. 175:6578-6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon, R., U. Prkseifer, and A. Puhler. 1983. A broad-host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 29.Smith, C. J. 1985. Development and use of cloning systems for Bacteroides fragilis: cloning of a plasmid-encoded clindamycin resistance determinant. J. Bacteriol. 164:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevens, A. M., J. M. Sanders, N. B. Shoemaker, and A. A. Salyers. 1992. Genes involved in production of plasmidlike forms by a Bacteroides conjugal chromosomal element share amino acid homology with two-component regulatory systems. J. Bacteriol. 174:2935-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stevens, A. M., N. B. Shoemaker, L. Y. Li, and A. A. Salyers. 1993. Tetracycline regulation of genes on Bacteroides conjugative transposons. J. Bacteriol. 175:6134-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens, A. M., N. B. Shoemaker, and A. A. Salyers. 1990. The region of a Bacteroides conjugal chromosomal tetracycline resistance element which is responsible for production of plasmidlike forms from unlinked chromosomal DNA might also be involved in transfer of the element. J. Bacteriol. 172:4271-4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, J., G. R. Wang, N. B. Shoemaker, and A. A. Salyers. 2001. Production of two proteins encoded by the Bacteroides mobilizable transposon NBU1 correlates with time-dependent accumulation of the excised NBU1 circular form. J. Bacteriol. 183:6335-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whittle, G., B. D. Hund, N. B. Shoemaker, and A. A. Salyers. 2001. Characterization of the 13 kb ermF region of Bacteroides conjugative transposon, CTnDOT. Appl. Environ. Microbiol. 67:3488-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]