Abstract
Photorhabdus is an entomopathogenic bacterium symbiotically associated with nematodes of the family Heterorhabditidae. Bacterial hemolysins found in numerous pathogenic bacteria are often virulence factors. We describe here the nucleotide sequence and the molecular characterization of the Photorhabdus luminescens phlBA operon, a locus encoding a hemolysin which shows similarities to the Serratia type of hemolysins. It belongs to the two-partner secretion (TPS) family of proteins. In low-iron conditions, a transcriptional induction of the phlBA operon was observed by using the chloramphenicol acetyltransferase reporter gene, causing an increase in PhlA hemolytic activity compared to iron-rich media. A spontaneous phase variant of P. luminescens was deregulated in phlBA transcription. The phlA mutant constructed by allelic exchange remained highly pathogenic after injection in the lepidopteran Spodoptera littoralis, indicating that PhlA hemolysin is not a major virulence determinant. Using the gene encoding green fluorescent protein as a reporter, phlBA transcription was observed in hemolymph before insect death. We therefore discuss the possible role of PhlA hemolytic activity in the bacterium-nematode-insect interactions.
Members of the family Enterobacteriaceae and the genus Photorhabdus (6) are associated with the entomopathogenic nematodes of the family Heterorhabditidae. These bacteria, which are highly pathogenic to insects, are transported by their nematode hosts into the hemocoel of the insect prey, which is killed probably via a combination of toxin action and septicemia. The bacterial symbionts contribute to the symbiotic relationship by establishing and maintaining suitable conditions for nematode reproduction. During the final stages of development, the bacteria and the nematode reassociate and subsequently leave the insect carcass in search of a new insect host (26). Recently, isolations of some nonsymbiotic Photorhabdus strains from infected humans in Australia and the United States were reported (21, 40).
The wild-type bacterium that is normally isolated from symbiotic infective-stage nematodes is referred to as phase I. Like many pathogenic bacteria, Photorhabdus strains spontaneously produce colonial variants in vitro which have been called phase II variants (5, 26). Both variants of the bacteria have generally been shown to be equally pathogenic for the larvae of the greater wax moth, Galleria mellonella (26).
The bacterial factors involved in killing the insect or in overcoming the insect's immune reactions are still under investigation. After invasion of the insect host by the nematodes, bacteria produce potential virulence factors, including lipase, protease, and lipopolysaccharides in the hemocoel (for a review, see reference 26). It was shown that purified lipopolysaccharide or Photorhabdus protease fractions had no toxic effect after injection into insect hemocoel (7, 13). Recently, a novel toxin complex (Tc) with oral toxicity to a wide range of insects was identified in a supernatant of Photorhabdus luminescens W14 (8). Purified toxin complex a (Tca) has specific effects on the midgut epithelium of the insect (4). It has recently been shown that Tca, Tcd, and an RTX-like metalloprotease are expressed in vivo during insect infection (17).
Hemolysins are extracellular toxic proteins produced by many gram-negative (e.g., Escherichia coli, Vibrio spp.) and gram-positive (e.g., Streptococcus spp., Listeria spp.) bacteria and are known to function as virulence factors. Most of them are active against a wide range of nucleated cell types and thus are also called cytolysins. Bacterial hemolysins are usually recognized on blood agar plates where a transparent zone appears around colonies. Hemolysis production in some Photorhabdus strains was first described by Farmer et al. (21). These bacteria have been shown to display an unusual reaction on sheep blood agar plate, designated annular hemolysis (1); this reaction was considered to be a marker in the identification of Photorhabdus asymbiotica isolated from clinical specimens (21, 25). Recently, we described the production of zones of hemolysis on sheep blood agar around several strains of Photorhabdus and Xenorhabdus, another symbiotic bacterium of nematodes (11). We also identified the presence of a cytolysin active on insect hemocytes and on sheep erythrocytes in Xenorhabdus culture supernatants, whereas such activity could not be detected in Photorhabdus supernatants. In Photorhabdus spp., other factors might be responsible for the hemolysis observed on blood agar medium. Therefore, we investigated hemolysin production by Photorhabdus.
The recently published partial genome sequence of P. luminescens W14 revealed a diverse array of genes that putatively encodes potential virulence factors (23). These factors include RTX-like toxins, hemolysin and cytotoxin homologs (23). Lately, the complete genome of P. luminescens subsp. laumondii TT01 was sequenced (E. Duchaud, unpublished data), confirming the presence of genes with similarities to various hemolysins.
In order to evaluate hemolysin contribution in P. luminescens TT01 to insect virulence, we characterized a hemolysin locus homologous to the loci of the pore-forming, calcium-independent hemolysins from Serratia marcescens, Proteus mirabilis, Edwardsiella tarda, and Haemophilus ducreyi.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
All bacterial strains and plasmids used in this study and their sources are listed in Table 1. P. luminescens was grown for 48 h at 28°C and strains of Escherichia coli for 24 h at 37°C, on Luria-Bertani (LB) broth or LB supplemented with 1.5% agar. When required, ampicillin, colistin, kanamycin, chloramphenicol (CHL), or tetracycline were added to the media at final concentrations of 100, 50, 20, 15, and 10 μg/ml, respectively. When necessary, growth media were supplemented with FeSO4 to a final concentration of 100 μM or were modified to be iron deficient by addition of 0.3 mM 2,2′-dipyridyl. Colistin-resistant mutants arose at a frequency of ca. 10−9 (A. Givaudan, unpublished data). These strains behaved similarly to the parent strain in terms of growth rates, insect pathology, hemolysin production, and all other phenotypes that we tested. Phase variation was spontaneously obtained after long-term growth in LB broth as previously described (5). Wild type is referred to as phase I. Phase I colonies are blue when grown on NBTA (i.e., nutrient agar supplemented with 25 mg of bromothymol blue per liter and 40 mg of triphenyltetrazolium chloride per liter), are bioluminescent, and produce antimicrobial activity against Micrococcus luteus (from the culture collection of the Institut Pasteur, Paris, France), while phase II colonies are red, are not bioluminescent, and produce reduced or no antibacterial activity. Phase II of TT01 is indicated by addition of the suffix “/2” to the strain designation.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotypea | Source or reference |
|---|---|---|
| Strains | ||
| P. luminescens TT01 | Wild type, phase I variant | CIP 105565 |
| P. luminescens TT01/2 | Phase II variant | Laboratory collection |
| P. luminescens TT01c | Colr natural mutant | Laboratory collection |
| P. luminescens TT01c-phlA::Ω | TT01c phlA::ΩCm | This work |
| E. coli XL1-Blue | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 | Stratagene |
| MRF′ | thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn5 (Kmr)] | |
| E. coli S17-1 | pro r− n+ Tpr Smr RP4-2-Tc::Mu::Tn7 recA thi | 49 |
| E. coli HB101 | F−mcrB mrr hsdS20(rB− mB−) recA13 leuB6 ara-14 proA2 lacY1 galK2 xyl-5 mtl-1 rpsL20 (strR) supE44λ− | 9 |
| Plasmids | ||
| pUC19 | Apr cloning vehicle | Biolabs |
| pJT02 | 1.2-kb HindIII internal fragment of phlA in the HincII site of pUC19 | This work |
| pHP45-ΩCm | Apr Cmr ΩCm | 22 |
| pJT02Ω | 3.8-kb BamHI fragment from pHP45-ΩCm in the NsiI site of pJT02 | This work |
| pJQ200KS | GmrsacRB mob oriV (p15A replicon) | S. Forst |
| pJT421Ω | 5-kb BamHI-PstI fragment of pJT02Ω in the BamHI-PstI sites of pJQ200KS | This work |
| pPLG-11B9 | Bacterial artificial chromosome of P. luminescens genome (contains a 40-kb fragment overlapping phlBA) | This work |
| pRK404 | Tcr low-copy mobilizable plasmid | 19 |
| pJT95 | 9.7-kb BamHI fragment of pPLG-8C6 in the BamHI site of pRK404 (contains phlBA) | This work |
| pRK2013 | Kmr (Tn903), ColE1 replicon, tra+ (RK2); helper plasmid | 24 |
| pBBR1MAC2 | Medium-copy mobilizable vector; Apr; cat reporter gene | 38 |
| pJT-PB-MAC | 500-bp region overlapping phlB start codon cloned in BamHI-KpnI sites of pBBR1MAC2 | This work |
| pBBR1KGFP | Medium-copy mobilizable vector; Kmr; gfpmut3 reporter gene | 33 |
| pJT-PB-KGFP | 500-bp region overlapping phlB start codon cloned in BamHI-KpnI sites of pBBR1KGFP | This work |
Colr, colicin resistant; Kmr, kanamycin resistant; Apr, ampicillin resistant; Tpr, trimethoprim resistant; Cmr, chloramphenicol resistant; Gmr, gentamicin resistant; Tcr, tetracycline resistant.
Sequence analysis.
The genome of the strain TT01 of P. luminescens has been sequenced by using the whole-genome shotgun strategy (Duchaud, unpublished). Cloning, sequencing, and assembly were done as described by Frangeul et al. (27). Annotation was performed during the finishing phase by using GMP-Tool-Box (L. Frangeul and E. Duchaud, unpublished data). Coding sequences (CDS) were defined by combining Genemark predictions (30) with visual inspection of each open reading frame for the presence of a start codon preceded by a ribosome-binding site and BLASTP similarity searches on the Nrprot database (2).
Molecular genetic techniques.
Genomic DNA from P. luminescens TT01 was prepared according to a method described previously (44). Endonucleases (Roche Molecular Biochemicals), DNA polymerase, calf intestine phosphatase, and T4 DNA ligase (Q-Biogene) were used according to the recommendations of the manufacturers. In order to control the phlA mutant, Southern blotting experiments were performed as previously described (44).
Construction of a phlA-null mutant.
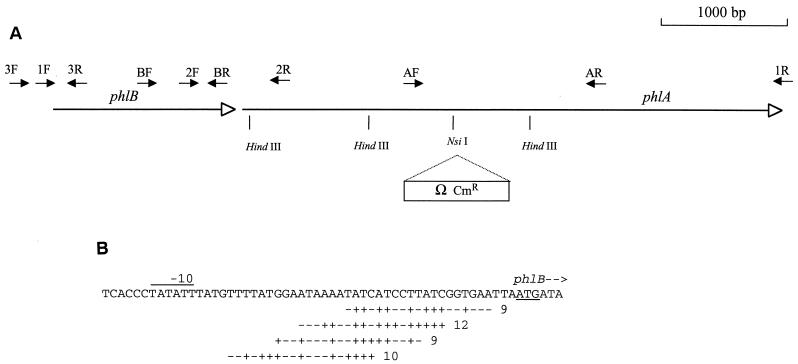
PCR was performed with the primers 1F (5′-CCACTCGTTAATCAGAGCA-3′) and 1R (5′-CCCTTTATGCTAACCTGTGA-3′) (Fig. 1A) to amplify the phlBA locus by using the Tfu polymerase, a high-fidelity polymerase. The 7-kb amplified fragment was subsequently digested with HindIII, and the fragments were separated on a 0.8% agarose gel. Fragments of 1.2 kb corresponding to the internal phlA region were collected by using Nucleotrap (Macherey-Nagel), blunt ended by using Klenow polymerase, ligated to HincII-digested and dephosphorylated pUC19 to yield to pJT02 (Table 1), and cloned in E. coli XL1MRF′. The CHL-resistant Ω interposon, which has been described previously (22), was used for phlA disruption. Construction of pJT02Ω was performed by insertion of the BamHI-digested and blunt-ended CHL-resistant Ω cassette ligated to blunt-ended, dephosphorylated NsiI-digested pJT02. The NsiI site is located approximately in the middle of the 1.2-kb phlA fragment of pJT02. Thereafter, the Ω cassette-interrupted phlA fragment was isolated by BamHI-PstI digestion and ligated to corresponding sites of the mobilizable plasmid pJQ200KS to create pJT421Ω (Table 1), which was transformed in E. coli S17-1. Conjugations between S17-1(pJT421Ω) and TT01c, followed by allelic exchange, were performed as described previously (28). Briefly, mating between bacterial cells was performed overnight on a nitrocellulose filter. After we made sure that transconjugants harbored pJT421Ω, we selected clones with allelic exchange on medium containing saccharose (4% [wt/vol]) and CHL. Insertion of the Ω cassette in the phlA gene of the resulting mutant (Fig. 1A) was confirmed by PCR amplification and Southern blot analysis. This mutant was termed TT01c phlA::Ω.
FIG. 1.
Map of the P. luminescens phlBA hemolysin locus. (A) Transcription orientation of phlB and phlA (open arrows) are represented with the positions of the restriction endonucleases recognition sites used. The mutation obtained by Ω interposon insertion is indicated. Primers used and their orientation are represented by solid arrows. (B) Possible Fur-binding sites in the promoter region of the phlBA operon. The identities with the 19-nucleotide sequence of the Fur-binding consensus sequence reported previously (18) are indicated below the sequence, with matches represented by “+” and mismatches represented by “−”; the total number of identities is indicated at the end. The putative −10 box is overlined, and start codon is underlined.
Complementation of phlA mutant.
In the framework of the genome project, we have end sequenced 1,200 BAC clones, constructed in pBeloBAC11 (32). One BAC spanning the phlBA locus has been selected. This BAC (59 kb), called pPLG-11B9, was digested by BamHI in order to isolate a 9.7-kb fragment containing phlBA and 1,000 bp upstream of phlB and then cloned in the BamHI dephosphorylated site of the mobilizable plasmid pRK404, yielding pJT95 (Table 1). Because pJT95 lacks the tra genes, mating with TT01c phlA::Ω was performed with E. coli S17-1 harboring pJT95 and E. coli HB101 harboring pRK2013, a helper plasmid.
Hemolytic assays.
Based on the description of an hemolytic assay to reveal the presence of ShlA activity in Serratia marcescens (10), we developed a liquid hemolytic assay with horse erythrocytes. It was performed with horse defibrinated blood (BioMérieux, Marcy l'Etoile, France), and hemolysis was expressed as the percentage of the total erythrocytes lysed. Phosphate-buffered saline (PBS)-washed erythrocytes were diluted to a final concentration of 25% (vol/vol). Aliquots of 1 ml of exponentially growing Photorhabdus cells were mixed with 50 μl of an erythrocyte suspension and then incubated for 3 h at 28°C. After centrifugation (3,000 × g for 15 min), the A540 of the resulting supernatant was measured to evaluate the hemoglobin release. Hemolytic activity on blood agar plates was determined as previously described (11).
Detection of hemolysin mRNA by reverse transcription-PCR (RT-PCR).
Total RNA from exponential-phase bacteria growing in low-iron conditions was extracted by using the High Pure RNA Isolation Kit (Roche). cDNA synthesis from 2 μg of total RNA was performed by using AMV-RT polymerase according to the instructions given by the Access RT-PCR System kit (Promega). Specific amplifications were performed with the primers BF (5′-GCCATGAACTGACTCATCTT-3′) and BR (5′-ACTACCAAACCCACTGAATG-3′) for the phlB region, AF (5′-GGAAATGCGAGCTTAAATGC-3′) and AR (5′-CCGATATTGAGTGCCCTGAT-3′) for the phlA region, and 2F (5′-CTCGCCAACAGCACATTATC-3′) and 2R (5′-GTTATGTGACAGCCCGGAAG-3′) for a region overlapping phlB and phlA. This step was coupled with 30 cycles of PCR amplification with Tfl polymerase (Promega) as follows: 30 s of denaturation at 95°C, 30 s of annealing at 55°C, and 1 min of extension at 72°C.
Construction of cat transcriptional fusion.
Transcriptional fusion between phlB and cat genes were constructed by using plasmid pBBR1MAC2 provided by S. Köhler (38). This plasmid was derived from pBBR1-MCS (34): T3 and T7 promoters were deleted, and the antibiotic resistance gene was replaced by a β-lactamase gene bla encoding ampicillin resistance and a promotorless reporter gene encoding chloramphenicol acetyltransferase (CAT). PCR amplification of a 500-bp fragment corresponding to the 350 bp upstream of phlB ATG and the 150 bp of the 5′ end of phlB was performed with primers 3F (5′-GTCTTGGTACCTCTTCCTAGTACAACCCAT-3′) and 3R (5′-GATGTGGATCCTTAGCCTGCTGCTTTAGTT-3′) containing BamHI and KpnI sites, respectively. BamHI-KpnI digestion of the PCR product was performed for adequate orientation when introduced into the corresponding sites of pBBR1MAC2 to yield pJT-PB-MAC. Because cat expression confers CHL resistance to the bacteria, the absence of such expression in TT01c harboring pBBR1MAC2 was controlled by determination of the CHL MIC compared to TT01c without plasmid. Values were comparable in both strains (<7.5 and <2 μg of CHL/ml, respectively); thus, pBBR1MAC2 without an insert was used as a negative control for transcriptional analysis of phlB.
Determination of CAT expression in P. luminescens.
Intracellular fractions of exponentially growing P. luminescens harboring pBBR1MAC2 or pJT-PB-MAC were collected in order to determine their CAT concentrations. When a bacterial culture reached an optical density of ca. 1.0, a growth phase for which the presence of hemolytic activity could be observed, the bacterial pellet of 500 μl of culture was resuspended in PBS and immediately frozen at −80°C. Lysis of bacterial cells was performed by repeated brief freezing-thawing cycles (at −80°C and 37°C), and the lysate was centrifuged for 15 min at 10,000 × g to eliminate bacterial debris. The concentration of total proteins in extract was determined by Pierce dosage by using bicinchoninic acid assay (Interchim, Montluçon, France). In parallel, the CAT concentration was determined by an enzyme-linked immunosorbent assay (ELISA) technique with anti-CAT digoxigenin-marked antibodies, followed by a colorimetric assay (CAT-ELISA Kit; Roche), as previously described (12). Each measurement was performed in triplicate. For each sample, CAT concentration was expressed relatively to the total protein concentration. Theses values were analyzed by the Student t test in order to determine P values for differences in expected versus actual values.
In vivo pathogenicity assays.
The pathogenicity assays were performed on the common cutworm Spodoptera littoralis as previously described (28). Briefly, 20 μl of exponentially growing bacteria diluted in PBS were injected into the hemolymph of 20 fifth-instar larvae of S. littoralis reared on an artificial diet. Insect larvae were then individually incubated at 23°C for up to 96 h, and the CFU of bacteria were determined by plating dilutions on LB agar. Insect death was monitored every 5 h. Three independent experiments for each treatment (bacterial doses or strains) were performed. Statistical analysis with SPSS version 11.0.1 (SPSS, Inc., Chicago, Ill.) was performed to construct a life table. Survival curves were compared by using Gehan's generalized Wilcoxon test. This test compares each survival time in each group with every survival time in other group.
A second construction was performed with gfpmut3 as a reporter gene by using pBBR1-KGFP (33). Cloning of the same region upstream of phlB as in pJT-PB-MAC was performed into the BamHI and KpnI sites of pBBR1-KGFP. About 3,000 CFU of the bacterial cells harboring these plasmids were injected in S. littoralis larvae as described above or, as a control, cultured in LB medium. Bacteria collected from LB medium or from insect hemolymph at 24 h postinoculation were examined for fluorescence under a Leica microscope.
Nucleotide sequence accession number.
The nucleotide sequence of P. luminescens hemolysin genes phlB and phlA were deposited in EMBL under accession no. AL662784.
RESULTS
phlBA sequence analysis.
The deduced amino acid sequence of two large open reading frames of 4,440 and 1,665 bp found in the P. luminescens TT01 genome showed 62 and 76% similarity, respectively, with the products responsible for hemolytic activity in Serratia marcescens, ShlA and ShlB (accession no. M22618). By analogy, we designated these two P. luminescens genes phlA and phlB. The deduced amino acid sequence of phlA and phlB also showed similarities (44 to 72% similarity to the all length of the protein sequence) with products of hemolysin genes from E. tarda (accession no. D89876), Proteus mirabilis (accession no. M30186), and H. ducreyi (accession no. U32175) and with putative products of hemolysin genes identified in the Yersinia pestis genome (accession no. AJ414158). All of these hemolysins were genetically organized as two adjacent genes, in the same orientation, as were phlB and phlA (Fig. 1A). In Serratia marcescens, Schiebel et al. showed that ShlB is essential for activation and secretion of ShlA (47).
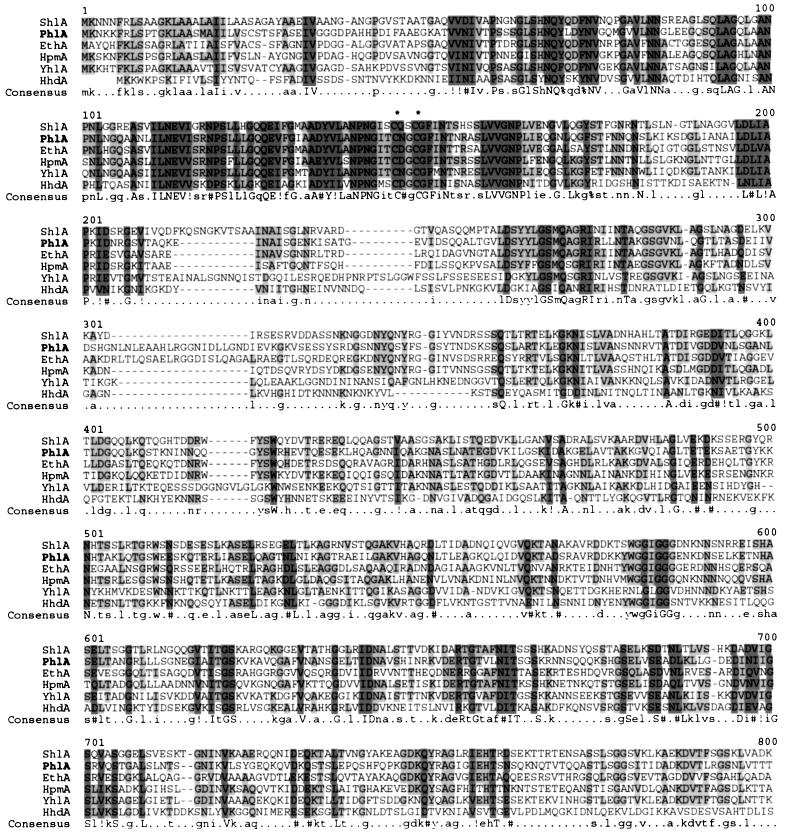
A novel family of protein secreted by the two-partner secretion (TPS) pathway was recently proposed to include HpmA (52), EthA (29), and HhdA (39) hemolysins from Proteus mirabilis, E. tarda, and H. ducreyi, respectively (31). However, this TpsA family not only includes hemolysins but also potential virulence proteins with other functions such as adhesins. A multiple alignment between PhlA, these four hemolysins, and a putative hemolysin from Y. pestis was performed by using Multalin program (Fig. 2) (15). This alignment revealed three groups of at least four amino acids each that were conserved between all of these hemolysins as follows: ILNEV (at positions 111 to 115), NPNG (positions 140 to 143), and LVVGNP (positions 159 to 164). Two cysteine residues found in PhlA showed a conserved position in the five other hemolysins of this family (Fig. 2). Multiple alignments between PhlB and the five other secretion or activation proteins showed that the positions of two cysteine residues and the C-terminal phenylalanine, which is characteristic of outer membrane proteins (50), were conserved (data not shown).
FIG. 2.
Multiple alignments between the N-terminal regions of the hemolysins of Serratia marcescens (ShlA), P. luminescens (PhlA), E. tarda (EthA), Proteus mirabilis (HpmA), and H. ducreyi (HhdA) and the putative hemolysin of Y. pestis (YhlA) as determined by using Multalin version 5.4.1 (15). The total numbers of residues are as follows: 1,608 for ShlA, 1,481 for PhlA, 1,594 for EthA, 1,577 for HpmA, 1,175 for HhdA, and 1,635 for YhlA. Cystein residues are marked by an asterisk. Consensus levels: high, 90% (dark gray shading); low, 50% (light gray shading). Consensus symbols: $, any of LM; !, any of IV; %, any of FY; #, any of NDQEBZ.
As shown on Fig. 1B, a putative −10 promoter region and a putative ribosome-binding site were found upstream of the start codon of phlB. In E. coli, iron regulation is achieved via the ferric uptake regulator (Fur) repressor protein, which binds to a 19-nucleotide consensus sequence (GATAATGATAATCATTATC) (16, 18). Overlapping Fur boxes are frequently found in the promoter regions of iron-regulated genes in E. coli (37). Four putative overlapping Fur boxes with homology with the E. coli consensus ranging from 9 to 12 of 19 were found in the 5′ upstream region of phlB (Fig. 1B). A putative rho-independent terminator was found 18 bp downstream of the stop codon of the phlA gene (data not shown).
Identification of the phlA-encoded function.
In Serratia marcescens, studies of C-terminally truncated ShlA polypeptides have indicated that this region is involved in the hemolytic activity (42). Because of the strong homologies between PhlA and ShlA, we decided to truncate 62% of the C-terminal region of PhlA. This mutation was performed after allelic exchange by insertion of an Ω interposon in phlA, leading to a truncated PhlA polypeptide lacking its 910 C-terminal amino acid residues. The resulting mutant, TT01c-phlA::Ω, was assayed for hemolytic activity. Comparison of TT01c and TT01c-phlA::Ω on a sheep or horse blood agar plate did not show any differences, since both strains displayed a perceptible halo of hemolysis with a size of <5 mm around colonies (data not shown). These data indicated that an additional hemolysin was produced on blood agar medium and that phlA was not involved in the hemolytic phenotype observed on such media.
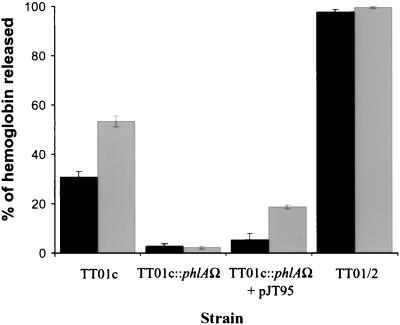
Using a liquid hemolytic assay, we found that exponentially growing TT01c bacteria produced hemolytic activity on horse erythrocytes. In contrast, no hemolysis production by TT01c-phlA::Ω strain could be detected under the same conditions (Fig. 3). After complementation of the phlA mutant with pJT95, hemolytic activity was partially restored (Fig. 3). Thus, this assay allowed us to identify the presence of PhlA hemolytic activity. Interestingly, this liquid hemolytic assay did not reveal any hemolytic activity in filter-sterilized bacterial supernatants of the strains tested (data not shown).
FIG. 3.
PhlA hemolytic activity as represented by the percentage of hemoglobin release from horse erythrocytes. Bacterial strains were grown in LB broth supplemented with 100 μM FeSO4 (▪) or iron depleted by the addition of 300 μM 2,2′-dipyridyl (░⃞) to an optical density at 540 nm (OD540) of 1.0 prior to the hemolytic assay (see Materials and Methods). The data represent the mean values ± the standard error of the mean for triplicate determinations from one of four similar experiments.
All attempts to clone the entire phlBA locus in E. coli failed with the medium-copy plasmid pBBR1MCS as a vector, but it was possible with the low-copy vector, pRK404. This may indicate a toxicity of the phlBA locus in E. coli, as was also observed for the ethBA locus, which could only be cloned on a low-copy number vector (29). Surprisingly, pJT95 containing the phlBA locus did not confer a hemolytic phenotype when cloned in E. coli either on a blood agar plate nor in a liquid hemolytic assay.
Increase of hemolytic activity in low-iron conditions.
The effects of iron deprivation were assayed on P. luminescens hemolytic activity. After addition of dipyridyl, a nonutilizable iron chelator, the hemolytic activity of P. luminescens was increased compared to that of iron-supplemented cells (Fig. 3). As expected, TT01c-phlA::Ω displayed no hemolytic activity in either iron-rich or iron-depleted media (Fig. 3). Surprisingly, the phase II variant TT01/2, which usually has many reduced exoenzymatic activities or is deficient in these activities compared to the wild-type phase I (5), displayed a hemolytic activity at least twice that of the wild type in iron-rich medium. In iron-depleted medium, TT01/2 hemolytic activity was not increased (Fig. 3).
Transcriptional analysis of phlBA.
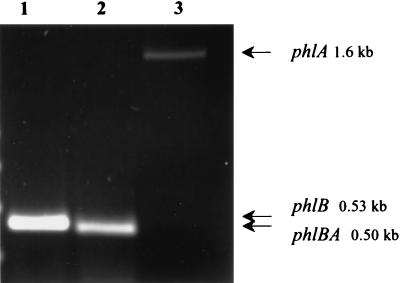
RT-PCR analysis revealed the amplification of fragments of expected sizes corresponding to mRNA of phlB (Fig. 4, lane 1) and phlA (Fig. 4, lane 3) when P. luminescens phase I cells were grown in low-iron conditions, whereas no such transcript could be observed in cells growing in standard medium (data not shown). In contrast, an intensive band was observed on an agarose gel after RT-PCR amplification of TT01/2 total RNA cells grown in standard medium (data not shown). These data indicate that the level of transcription of phlB and phlA is higher in TT01/2 than in phase I cells under these growth conditions.
FIG. 4.
RT-PCR detection of phlB, phlA, and a region overlapping the phlB and phlA open reading frames with P. luminescens TT01c RNA grown in iron-depleted LB broth. The following primers were used: lane 1, BF and BR (nucleotide positions in phlBA sequences 380 and 915, respectively); lane 2, 2F and 2R (positions 1471 and 1967); and lane 3, AF and AR (positions 3027 and 4685, see Materials and Methods).
When a couple of primers for which the amplified fragment overlapped phlB and phlA were used, a fragment of the expected size was observed after RT-PCR amplification (Fig. 4, lane 2), indicating that phlB and phlA were cotranscribed. Therefore, transcriptional analysis with the CAT reporter gene was only performed on the 5′-upstream region of phlBA.
Studies with cat transcriptional fusions indicated transcription of phlBA compared to the negative control (Table 2). In low-iron conditions, when hemolytic activity is induced, transcription of phlBA was significantly induced (Table 2). Interestingly, the CAT production by a phase II variant containing phlBA transcriptional fusions was at least twofold increased compared to those measured in TT01c in the same conditions. We confirmed that the phase II variant harboring pBBR1MAC2 (negative control) showed no significant transcription of the reporter gene (Table 2). These results were correlated to the measurements of the hemolytic activity previously described (Fig. 3).
TABLE 2.
Transcriptional activity of 5′ upstream region of phlBA operon as measured by expression of CAT reporter gene
| Phase: strain | Mean CAT concn (pg/μg of protein)a |
|
|---|---|---|
| With iron | Without iron | |
| Phase I (negative control): TT01c(pBBR1MAC2) | 8.7 ± 3.4b | 5.9 ± 0.1b |
| Phase I: TT01c(pJT-PB-MAC) | 289.9 ± 15.8c | 566.2 ± 26c |
| Phase II (negative control): TT01/2(pBBR1MAC2) | 21.3 ± 1.2d | ND |
| Phase II: TT01/2(pJT-PB-MAC) | 661.8 ± 2.4d | ND |
Means ± standard deviations of three measurements are presented. Bacteria were grown in LB medium either supplemented with FeSO4 (100 μM) or iron depleted with 2,2′-dipyridyl (300 μM) until reaching an OD540 of 1.0. ND, not determined.
Differences between data are not significant (P > 0.05).
Differences between data are significant (P < 0.01).
Differences between data are highly significant (P < 0.001).
Virulence of phlA mutant for S. littoralis.
In order to evaluate whether phlA is involved in the pathogenicity of P. luminescens for susceptible insect larvae, two doses (10 to 100 and 250 to 500 CFU) of the wild-type strain and TT01c-phlA::Ω were injected into S. littoralis larvae. We monitored insect mortality for 3 days postinjection (data not shown). After injection of fewer than 30 CFU into the S. littoralis hemolymph, all larvae were dead within 48 h, indicating that both strains are highly virulent. When the survival curves for two doses of injected bacteria were significantly different (P < 0.001), survival analysis with grouped injection levels of bacteria indicated no significant difference (P > 0.05) between the wild type and the phlA mutant. For instance, 50% of larvae were dead after 31.5 and 32 h with injected doses per insect of 430 CFU of TT01c and 475 CFU of TT01c-phlA::Ω, respectively. Thus, it can be concluded that TT01c-phlA::Ω displays behavior similar to that of the wild type during pathology in S. littoralis.
Septicemia was observed 24 h after injection of both strains in insect larvae. Transcriptional fusions with gfpmut3 reporter gene revealed a positive signal in bacteria collected from S. littoralis hemolymph at 24 h after the injections (ca. 6 h before insect death), whereas no induction was observed in the negative control (data not shown). Similar signals were also observed for bacteria grown in LB medium (data not shown).
DISCUSSION
Characterization of phlBA hemolysin locus from P. luminescens.
The phlBA locus of P. luminescens was identified on the basis of its sequence similarities with the Serratia marcescens hemolysin locus. In Serratia marcescens, ShlB that belongs to the TpsB protein family has been shown to be essential for secretion and activation of the ShlA hemolysin (47). HpmB and HpmA in Proteus mirabilis (52), EthB and EthA in E. tarda (29), and HhdB and HhdA of H. ducreyi (39), which all belong to the TpsB and TpsA protein families, have the same functions (31). Furthermore, putative hemolysin genes were also identified in the Y. pestis genome. In view of the strong similarities with the hemolysins (Fig. 2) and the secretion or activation proteins of these bacteria, the conserved positions of the cysteine residues, and an identical genetic organization, it might therefore be concluded that PhlB and PhlA most probably have the same functions in P. luminescens.
Several groups of at least four amino acids were conserved between all of these hemolysins (Fig. 2). In particular, the NPNG and the ANPN consensus found in PhlA were reported to be important for activation and secretion of ShlA (48). These conserved regions are located in the first 200 residues, the region involved in ShlA secretion, whereas the C-terminal region is responsible for hemolytic activity. This N-terminal region also showed similarity with nonhemolytic members of the TpsA family, such as FHA hemagglutinin of Bordetella pertussis (36), and the haem:haemopexin-binding protein of Haemophilus influenzae (14).
Because of the strong amino acid sequence conservation of these hemolysins, including both secretion/activation protein (protein B) and the hemolysin (protein A), PhlB and PhlA should belong to the TpsB and TpsA protein families, respectively. However, with regard to the relatively low conservation in the corresponding nucleotide sequences of these family members, the hypothesis of a recent horizontal transfer may be excluded. Alternatively, we can predict the existence of a common ancestral gene that might have evolved only slowly because of strong selective pressures, indicating an important role of this hemolysin family for the maintenance of the bacteria in their respective ecological niches.
In an investigation on the distribution of Serratia hemolysin genes among hemolytic bacteria, all Serratia marcescens strains tested, including two strains isolated from insects, have been found to contain sequences homologous to shlB and shlA (43). In P. luminescens W14, the genomic sample sequence presented similarities to shlA and shlB (23). We also detected some homologous segments between phlBA and several clones from this genomic sample sequence by using tblastX in the genome survey sequences database of GenBank (23). The present study shows the functionality of PhlA in P. luminescens subsp. laumondii TT01c. Moreover, homologous genes were recently found in the genome of Y. pestis, which persists in the flea gut for long periods. These aspects of life cycle of Yersinia in insects bears a striking resemblance to one stage of the interaction between Photorhabdus and its nematode hosts when the bacterial symbionts are within the midgut of infective juvenile larvae. Because of the occurrence of these genes in insect-parasite combinations, insect pathogenic bacteria, or nematode symbionts, we may therefore hypothesize that TpsA hemolysins could be involved in mutualistic or detrimental relationships between the invertebrate hosts and bacteria.
The transcription of phlBA operon is iron regulated.
For many pathogenic bacteria, iron is often a rate-limiting growth factor in their host. Production of cytolysins might be a mean to release iron present in eucaryotic cells (35). In the insect hemocoel, the free-iron concentration is probably low, as indicated by the isolation of transferrins in the hemolymph of several orders of insects, including Lepidopteran (3). Measurements of P. luminescens hemolysis in iron-supplemented or iron-depleted media showed an induction of PhlA activity in low-iron conditions (Fig. 3). These results have been confirmed by transcriptional analysis of 5′ upstream region of phlBA (Table 2). We also found four putative overlapping Fur boxes upstream of phlB, with identities of 9 to 12 nucleotides with the E. coli Fur box (Fig. 1B). Apparently, the E. coli Fur protein tolerates considerable variability in operator sequences (41), as illustrated by the Fur-regulated E. coli genes exbD and fhuE, which have a conservation of only 9 and 10 nucleotides with the Fur box consensus sequence (20, 45, 46). Furthermore, the presence of predicted sequences homologous to the E. coli Fur protein have been identified in P. luminescens TT01 genome (Duchaud E., unpublished data), and in P. luminescens W14 genomic sample sequence (23). The iron regulation of PhlA is in accordance with the previously described iron regulation of ShlA and EthA hemolysins from Serratia and Edwardsiella (29, 41).
The hemolytic activity of the phase II variant was not affected by iron rates in growth media (Fig. 3), and it was at least twofold higher than that of the wild type (phase I). RT-PCR analysis (data not shown) and studies of phlB fusions with cat reporter gene (Table 2) indicated a deregulation of phlBA transcription in this phase variant. The expression of other iron-regulated genes in this strain compared to the wild type is currently being studied.
In vivo role of PhlA hemolysin.
In a mouse challenge, intravenous injection of Proteus mirabilis or its corresponding hpmA mutant was performed. The results show a sixfold increase in the 50% lethal dose in the hpmA mutant compared to the wild type (51). Our insect pathology experiments revealed that the phlA mutant is still highly virulent. These data showed that PhlA is not a major virulence factor when P. luminescens is injected into S. littoralis hemolymph.
However, transcriptional studies with gfpmut3 as a reporter gene showed a positive signal for bacteria grown in vivo in insect hemolymph. In a previous work, we assayed insect hemocyte cytotoxicity in bacterial supernatants only and concluded that no cytolysin could be detected in P. luminescens supernatant (11). Although TPS hemolysins are secreted by their specific secretion/activation protein, the hemolytic activity detected in natural bacterial producer supernatant is usually absent or very low. Furthermore, pulse-labeling experiments have determined that ShlA has a half-life of only 3 min at 37°C (47). Our experiments indicate that bacteria and erythrocytes need to be coincubated to detect the hemolytic activity caused by PhlA. With regard to the present results, we can hypothesize a possible cytolytic activity of PhlA against insect hemocytes when bacteria are present in the hemolymph.
The transcriptional expression of phlBA promoter in hemolymph before insect death might indicate a unexpected role of PhlA hemolysin in this ecological niche. Once the septicemia is well established, the insect cadaver tissues are bioconverted into a source of nutrients for growth of both nematode and bacteria. PhlA hemolysin probably causes some insect cell disruption, which might play a role in nematode and bacteria multiplication.
Acknowledgments
We thank Steve Forst and Stephan Köhler for providing some plasmids used in this study and also Dorothée Murat and Sylvie Pagès for their assistance. We acknowledge Stephan Köhler for help in revising the manuscript.
J.B. was funded by a MENRT grant (no. 98-5-11869).
REFERENCES
- 1.Akhurst, R. J., R. G. Mourant, L. Baud, and N. E. Boemare. 1996. Phenotypic and DNA relatedness between nematode symbionts and clinical strains of the genus Photorhabdus (Enterobacteriaceae). Int. J. Syst. Bacteriol. 46:1034-1041. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartfeld, N. S., and J. H. Law. 1990. Isolation and molecular cloning of transferrin from the tobacco hornworm, Manduca sexta: sequence similarity to the vertebrate transferrins. J. Biol. Chem. 265:21684-21691. [PubMed] [Google Scholar]
- 4.Blackburn, M., E. Golubeva, D. Bowen, and R. H. ffrench-Constant. 1998. A novel insecticidal toxin from Photorhabdus luminescens, toxin complex a (Tca), and its histopathological effects on the midgut of Manduca sexta. Appl. Environ. Microbiol. 64:3036-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boemare, N. E., and R. J. Akhurst. 1988. Biochemical and physiological characterization of colony form variants in Xenorhabdus spp. (Enterobacteriaceae). J. Gen. Microbiol. 134:1835-1845. [DOI] [PubMed] [Google Scholar]
- 6.Boemare, N. E., R. J. Akhurst, and R. G. Mourant. 1993. DNA relatedness between Xenorhabdus spp. (Enterobacteriaceae), symbiotic bacteria of entomopathogenic nematodes, and a proposal to transfer Xenorhabdus luminescens to a new genus, Photorhabdus gen. nov. Int. J. Syst. Bacteriol. 43:249-255. [Google Scholar]
- 7.Bowen, D., M. Blackburn, T. Rocheleau, C. Grutzmacher, and R. H. ffrench-Constant. 2000. Secreted proteases from Photorhabdus luminescens: separation of the extracellular proteases from the insecticidal Tc toxin complexes. Insect Biochem. Mol. Biol. 30:69-74. [DOI] [PubMed] [Google Scholar]
- 8.Bowen, D., T. A. Rocheleau, M. Blackburn, O. Andreev, E. Golubeva, R. Bhartia, and R. H. ffrench-Constant. 1998. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science 280:2129-2132. [DOI] [PubMed] [Google Scholar]
- 9.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 10.Braun, V., H. Gunther, B. Neuss, and C. Tautz. 1985. Hemolytic activity of Serratia marcescens. Arch. Microbiol. 141:371-376. [DOI] [PubMed] [Google Scholar]
- 11.Brillard, J., C. Ribeiro, N. Boemare, M. Brehélin, and A. Givaudan. 2001. Two distinct hemolytic activities in Xenorhabdus nematophila are active against immunocompetent insect cells. Appl. Environ. Microbiol. 67:2515-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell, J. W., and J. E. Cronan, Jr. 2001. Escherichia coli FadR positively regulates transcription of the fabB fatty acid biosynthetic gene. J. Bacteriol. 183:5982-5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke, D. J., and B. C. A. Dowds. 1995. Virulence mechanisms of Photorhabdus sp. strain K122 toward wax moth larvae. J. Invertebr. Pathol. 66:149-155. [Google Scholar]
- 14.Cope, L. D., S. E. Thomas, J. L. Latimer, C. A. Slaughter, U. Muller-Eberhard, and E. J. Hansen. 1994. The 100-kDa haem:haemopexin-binding protein of Haemophilus influenzae: structure and localization. Mol. Microbiol. 13:863-873. [DOI] [PubMed] [Google Scholar]
- 15.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crosa, J. H. 1997. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol. Mol. Biol. Rev. 61:319-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daborn, P. J., N. Waterfield, M. A. Blight, and R. H. ffrench-Constant. 2001. Measuring virulence factor expression by the pathogenic bacterium Photorhabdus luminescens in culture and during insect infection. J. Bacteriol. 183:5834-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lorenzo, V., S. Wee, M. Herrero, and J. B. Neilands. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 169:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ditta, G., T. Schmidhauser, E. Yakobson, P. Lu, X. W. Liang, D. R. Finlay, D. Guiney, and D. R. Helinski. 1985. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid 13:149-153. [DOI] [PubMed] [Google Scholar]
- 20.Eick-Helmerich, K., and V. Braun. 1989. Import of biopolymers into Escherichia coli: nucleotide sequences of the exbB and exbD genes are homologous to those of the tolQ and tolR genes, respectively. J. Bacteriol. 171:5117-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farmer, J. J., III, G. V. Pierce, G. O. Poinar, Jr., P. A. D. Grimont, G. P. Carter, E. Ageron, R. J. Akhurst, F. W. Hickman-Brenner, J. H. Jorgensen, K. L. Wilson, and J. A. Smith. 1989. Xenorhabdus luminescens (DNA hybridization group 5) from human clinical specimens. J. Clin. Microbiol. 27:1594-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 23.ffrench-Constant, R. H., N. Waterfield, V. Burland, N. T. Perna, P. J. Daborn, D. Bowen, and F. R. Blattner. 2000. A genomic sample sequence of the entomopathogenic bacterium Photorhabdus luminescens W14: potential implications for virulence. Appl. Environ. Microbiol. 66:3310-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer-Le Saux, M., V. Viallard, B. Brunel, P. Normand, and N. E. Boemare. 1999. Polyphasic classification of the genus Photorhabdus and proposal of new taxa: P. luminescens subsp. luminescens subsp. nov., P. luminescens subsp. akhurstii subsp. nov., P. luminescens subsp. laumondii subsp. nov., P. temperata sp. nov., P. temperata subsp. temperata subsp. nov., and P. asymbiotica sp. nov. Int. J. Syst. Bacteriol. 49:1645-1656. [DOI] [PubMed] [Google Scholar]
- 26.Forst, S., B. Dowds, N. Boemare, and E. Stackebrandt. 1997. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu. Rev. Microbiol. 51:47-72. [DOI] [PubMed] [Google Scholar]
- 27.Frangeul, L., K. E. Nelson, C. Buchrieser, A. Danchin, P. Glaser, and F. Kunst. 1999. Cloning and assembly strategies in microbial genome projects. Microbiology 145:2625-2634. [DOI] [PubMed] [Google Scholar]
- 28.Givaudan, A., and A. Lanois. 2000. flhDC, the flagellar master operon of Xenorhabdus nematophilus: requirement for motility, lipolysis, extracellular hemolysis, and full virulence in insects. J. Bacteriol. 182:107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirono, I., N. Tange, and T. Aoki. 1997. Iron-regulated haemolysin gene from Edwardsiella tarda. Mol. Microbiol. 24:851-856. [DOI] [PubMed] [Google Scholar]
- 30.Isono, K., J. D. McIninch, and M. Borodovsky. 1994. Characteristic features of the nucleotide sequences of yeast mitochondrial ribosomal protein genes as analyzed by computer program GeneMark. DNA Res. 1:263-269. [DOI] [PubMed] [Google Scholar]
- 31.Jacob-Dubuisson, F., C. Locht, and R. Antoine. 2001. Two-partner secretion in gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol. Microbiol. 40:306-313. [DOI] [PubMed] [Google Scholar]
- 32.Kim, U. J., B. W. Birren, T. Slepak, V. Mancino, C. Boysen, H. L. Kang, M. I. Simon, and H. Shizuya. 1996. Construction and characterization of a human bacterial artificial chromosome library. Genomics 34:213-218. [DOI] [PubMed] [Google Scholar]
- 33.Kohler, S., S. Ouahrani-Bettache, M. Layssac, J. Teyssier, and J. P. Liautard. 1999. Constitutive and inducible expression of green fluorescent protein in Brucella suis. Infect. Immun. 67:6695-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop, Jr., and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 35.Litwin, C. M., and S. B. Calderwood. 1993. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Locht, C., P. Bertin, F. D. Menozzi, and G. Renauld. 1993. The filamentous haemagglutinin, a multifaceted adhesion produced by virulent Bordetella spp. Mol. Microbiol. 9:653-660. [DOI] [PubMed] [Google Scholar]
- 37.Newman, D. L., and J. A. Shapiro. 1999. Differential fiu-lacZ fusion regulation linked to Escherichia coli colony development. Mol. Microbiol. 33:18-32. [DOI] [PubMed] [Google Scholar]
- 38.Ouahrani-Bettache, S., F. Porte, J. Teyssier, J. P. Liautard, and S. Kohler. 1999. pBBR1-GFP: a broad-host-range vector for prokaryotic promoter studies. BioTechniques 26:620-622. [DOI] [PubMed] [Google Scholar]
- 39.Palmer, K. L., and R. S. Munson, Jr. 1995. Cloning and characterization of the genes encoding the hemolysin of Haemophilus ducreyi. Mol. Microbiol. 18:821-830. [DOI] [PubMed] [Google Scholar]
- 40.Peel, M. M., D. A. Alfredson, J. G. Gerrard, J. M. Davis, J. M. Robson, R. J. McDougall, B. L. Scullie, and R. J. Akhurst. 1999. Isolation, identification, and molecular characterization of strains of Photorhabdus luminescens from infected humans in Australia. J. Clin. Microbiol. 37:3647-3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poole, K., and V. Braun. 1988. Iron regulation of Serratia marcescens hemolysin gene expression. Infect. Immun. 56:2967-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poole, K., E. Schiebel, and V. Braun. 1988. Molecular characterization of the hemolysin determinant of Serratia marcescens. J. Bacteriol. 170:3177-3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruan, Y., and V. Braun. 1990. Hemolysin as a marker for Serratia. Arch. Microbiol. 154:221-225. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 45.Sauer, M., K. Hantke, and V. Braun. 1987. Ferric-coprogen receptor FhuE of Escherichia coli: processing and sequence common to all TonB-dependent outer membrane receptor proteins. J. Bacteriol. 169:2044-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sauer, M., K. Hantke, and V. Braun. 1990. Sequence of the fhuE outer-membrane receptor gene of Escherichia coli K-12 and properties of mutants. Mol. Microbiol. 4:427-437. [DOI] [PubMed] [Google Scholar]
- 47.Schiebel, E., H. Schwarz, and V. Braun. 1989. Subcellular location and unique secretion of the hemolysin of Serratia marcescens. J. Biol. Chem. 264:16311-16320. [PubMed] [Google Scholar]
- 48.Schonherr, R., R. Tsolis, T. Focareta, and V. Braun. 1993. Amino acid replacements in the Serratia marcescens haemolysin ShIA define sites involved in activation and secretion. Mol. Microbiol. 9:1229-1237. [DOI] [PubMed] [Google Scholar]
- 49.Simon, R. 1984. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-Mob transposon. Mol. Gen. Genet. 196:413-420. [DOI] [PubMed] [Google Scholar]
- 50.Struyve, M., M. Moons, and J. Tommassen. 1991. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 218:141-148. [DOI] [PubMed] [Google Scholar]
- 51.Swihart, K. G., and R. A. Welch. 1990. Cytotoxic activity of the Proteus hemolysin HpmA. Infect. Immun. 58:1861-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uphoff, T. S., and R. A. Welch. 1990. Nucleotide sequencing of the Proteus mirabilis calcium-independent hemolysin genes (hpmA and hpmB) reveals sequence similarity with the Serratia marcescens hemolysin genes (shlA and shlB). J. Bacteriol. 172:1206-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]