Abstract
Analysis of potential virulence factors of oral spirochetes focuses on surface and secreted proteins. The Treponema denticola chymotrypsin-like protease (CTLP) is implicated in degradation of host cell molecules and contributes to tissue invasion. The CTLP complex, composed of the 72-kDa PrtP protein and two auxiliary proteins with molecular masses of approximately 40 and 30 kDa, is also involved in localization and oligomerization of the T. denticola major surface protein (Msp). The larger auxiliary protein was reported to be encoded by an open reading frame (ORF2) directly upstream of prtP. The deduced 39-kDa translation product of ORF2 contains a sequence matching the N-terminal sequence determined from one of the CTLP complex proteins. No proteins with significant homology are known, nor was information available on the third protein of the complex. DNA sequence analysis showed that ORF2 extended an additional 852 bp upstream of the reported sequence. The complete gene, designated prcA, encodes a predicted N-terminally-acylated polypeptide of approximately 70 kDa. Isogenic mutants with mutations in prtP, prcA, and prcA-prtP all lacked CTLP protease activity. The prcA mutant lacked all three CTLP proteins. The prcA-prtP mutant produced only a C-terminally-truncated 62-kDa PrcA protein. The prtP mutant produced a full-length 70-kDa PrcA. Immunoblot analysis of recombinant PrcA constructs confirmed that PrcA is cleaved to yield the two smaller proteins of the CTLP complex, designated PrcA1 and PrcA2. These data indicate that PrtP is required for cleavage of PrcA and suggest that this cleavage may be required for formation or stability of outer membrane complexes.
Analysis of potential virulence factors of oral spirochetes focuses on surface and secreted proteins (reviewed in reference 8). The surface-expressed chymotrypsin-like protease (CTLP) is implicated in Treponema denticola cytotoxicity (6), degradation of fibronectin (5, 28) and host cell protease inhibitors (13), and detachment of cultured cells (28). By disrupting intercellular junctions (4), CTLP has been shown to mediate migration of T. denticola through model basement membranes (14) and increase permeability of a multilayer epithelial cell model (28).
Studies of isogenic T. denticola mutants indicated that a relationship exists between CTLP activity and expression of another potential virulence factor of this organism, the pore-forming major surface protein (Msp) (6). T. denticola MPE, a defined msp mutant expressing a C-terminally-truncated Msp monomer, produced no CTLP complex proteins or protease activity (10). Two previous studies reported construction of isogenic prtP mutants: strain K1, carrying an ermF/AM cassette inserted in prtP (18); and strain CKE, in which an ermF/AM cassette replaces a KpnI fragment that includes the 3′ end of prcA and the 5′ end of prtP (10). Both mutants exhibited defects in Msp production levels and oligomerization, in addition to the expected lack of PrtP protease activity. The connection between Msp expression and CTLP activity appears to be posttranscriptional (J. C. Fenno, unpublished results), suggesting that Msp and one or more proteins of the CTLP complex are required for proper localization or formation of native outer membrane complexes.
Native CTLP, a detergent-stable complex with an apparent molecular mass of 95 kDa, resolves to three polypeptides upon heating. The largest, the 72-kDa protein PrtP (dentilisin) encoded by prtP, exhibits homology with the Bacillus subtilis serine protease subtilisin (19). The prtP gene and its activity are conserved among several species of oral treponemes (17). Isogenic prtP mutants lack CTLP activity (10, 18). The sizes of the two smaller proteins of the CTLP complex have been variously reported as 27 and 23 kDa (27), 39 and 32 kDa (22), and 43 and 38 kDa (19), resulting in some confusion as to the identity and composition of the protease complex.
An open reading frame (ORF) designated ORF2 directly upstream of prtP has been proposed to encode the larger of the two auxiliary proteins of the CTLP complex (19). One of the two N-terminal amino acid sequences that can be determined from native CTLP (19; D. Grenier, personal communication) matches a sequence in the deduced amino region of the ORF2 peptide (19). No proteins with homology to the deduced 39-kDa product of ORF2 are known. Although ORF2 is cotranscribed with prtP and insertional mutagenesis of ORF2 results in loss of PrtP activity (K. Ishihara, H. K. Kuramitsu, T. Miura, and K. Okuda, Abstr. 79th Meet. Int. Assoc. Dent. Res., abstr. 941, 2001), no studies have directly confirmed the identities of the two protease-associated proteins or characterized their activities. The present study identifies the complete sequence of the gene encoding a protease-associated protein and describes a novel posttranslational processing event mediated by PrtP.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
T. denticola ATCC 35405 and isogenic mutants (Table 1) were grown and maintained in NOS broth medium as previously described (16), with erythromycin (40 μg ml−1) added as appropriate. For allelic replacement, mutants were selected on NOS/GN plates (3) containing erythromycin (40 μg ml−1) as described previously (10, 21). Cultures were examined by phase-contrast microscopy for purity and typical strain morphology before use.
TABLE 1.
T. denticola strains used in this study
Escherichia coli strains JM109 (30) and TOP10 (Invitrogen, Carlsbad, Calif.) were used as hosts for routine subcloning and plasmid preparations. E. coli NovaBlue (Novagen) was used as a host strain for direct cloning of PCR fragments in plasmid vector pSTBlue-1 (Novagen). Plasmid pCTLP (10) carries a 4-kb ORF2-prtP fragment from T. denticola ATCC 35405. Plasmid pVA2198 (11) was used as the source of the ermF/AM gene cassette. For expression studies, DNA fragments were cloned in pET17b (Novagen) and introduced into E. coli BL21(DE3)/pLysS. Expression from the vector-encoded T7 promoter was induced as described previously (9). E. coli strains were grown in Luria-Bertani (LB) broth or agar medium supplemented with ampicillin (50 μg ml−1), kanamycin (50 μg ml−1), chloramphenicol (34 μg ml−1), or erythromycin (200 μg ml−1) as appropriate.
Chemicals.
Unless otherwise noted, chemicals were purchased at the highest available purity from Sigma Chemical Co. (St. Louis, Mo.) or Fisher Scientific (Chicago, Ill.).
Recombinant DNA methods.
Unless stated otherwise, standard methods described by Ausubel et al. (2) or Sambrook et al. (26) were followed. DNA fragments were eluted from agarose gels with the Gene Clean II kit (Bio101, La Jolla, Calif.). Genomic and plasmid DNAs were isolated with the Wizard Genomic DNA Purification kit and Wizard Plus SV Minipreps kit (Promega, Madison, Wis.), respectively. Oligonucleotide primers (Invitrogen, Carlsbad, Calif.) were designed by using the GeneFisher algorithm (12). For Southern blot analysis, HindIII-digested genomic DNAs separated on 0.7% agarose gel were transferred to nylon membranes (Immobilon-Ny; Millipore) and hybridized with biotin-labeled DNA probes, followed by incubation of the blots with streptavidin, biotinylated alkaline phosphatase, and chemiluminescence detection reagent (New England Biolabs) according to the manufacturer's instructions. Chemiluminescence was detected with a Fluor-S Multi-Imager (Bio-Rad).
Construction of plasmids for expression studies and allelic replacement mutagenesis.
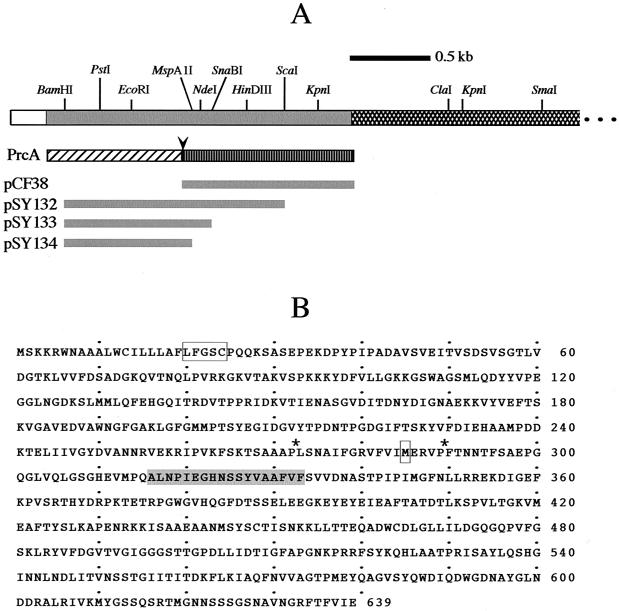
For expression studies, DNA fragments of interest were cloned in-frame with the T7 Gene 10 leader sequence in pET17b (Novagen). Oligonucleotide primers KX34 and KX35 (Table 2) complementary to the 5′ and 3′ ends of ORF2, with specific restriction sites added for cloning in pET17b, were used to amplify the predicted ORF2 coding region from T. denticola genomic DNA. The BamHI-XhoI-digested PCR product was gel purified and cloned in pET17b, yielding pCF38. Similarly, the PCR product amplified with CX252 and CX259 (Table 2) was digested to yield BamHI-ScaI, BamHI-SnaBI, or BamHI-MspA1I fragments, which were gel purified and cloned in pET17b, yielding pSY132, pSY133, and pSY134, respectively (Fig. 1A).
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Targeta | Sequenceb |
|---|---|---|
| KX34 | ORF2 5′ end (F) | 5′ dGGGGGATCCAATGGAAAGAGTTCCCTTT 3′ BamHI |
| KX35 | ORF2/prcA 3′ end (R) | 5′ dGGGCTCGAGTTATTCAATTACAAAGGTA 3′ XhoI |
| CX247 | ermF/AM 5′ end (F) | 5′ dGGCATATGCGATAGCTTCCGCTATTG3′ NdeI |
| CX249 | ermF/AM 3′ end (R) | 5′ dGGCATATGAGCTGTCAGTAGTATACC 3′ NdeI |
| CX251 | 3′ to prcA (R) | 5′ dCTTATTACCGATAACGGCAG 3′ |
| CX252 | Upstream of prcA (F) | 5′ dCTGCAATAAAGACAGGGAAG 3′ |
| CX259 | prcA 1233-1252 (R) | 5′ dAATGGGAATAGGCGTAGAAG 3′ |
| CX260 | prcA 10 bp from 5′ end (F) | 5′ dGAAAGATGGAACGCTG 3′ |
The orientation of the primer with respect to the gene of interest is shown (F, forward; R, reverse).
Restriction sites added for cloning are underlined. The ORF2 start codon is double underlined in KX34, as is the ORF2/prcA stop codon in KX35.
FIG. 1.
The prcA gene, its product, and regions expressed in E. coli. The map in panel A shows the relative locations of the prcA coding region (shaded) and the 5′ end of prtP (cross-hatched). Aligned below prcA is the translation product, including PrcA1 (diagonal hatching) and PrcA2 (vertical hatching). The putative cleavage site between PrcA1 and PrcA2 is indicated by an arrowhead. The shaded bars below show the relative locations of prcA DNA fragments cloned in E. coli expression vector pET17b. Panel B shows the deduced peptide sequence of PrcA. The spirochetal lipobox (L-F-G-S-C, residues 20 to 24) present in the hydrophobic N-terminal signal peptide is boxed. Potential cleavage sites for PrtP are indicated by asterisks. The N-terminal methionine formerly proposed for the ORF2 peptide (19) is boxed. The N-terminal amino acid sequence of PrcA2 (19; D. Grenier, personal communication) is shaded.
The ermF/AM cassette PCR amplified from pVA2198 with CX247 and CX249 (Table 2) was cloned in pSTBlue-1, yielding pSY120, which was used as the source for ermF/AM in constructing plasmids for allelic replacement mutagenesis.
For mutagenesis of prtP, a 6.1-kb ClaI-SmaI fragment of pCTLP was gel purified and ligated to the AccI-BstZ17I fragment of pSY120 containing the ermF/AM gene cassette. In the resulting plasmid, pSY130, ermF/AM replaces a 581-bp internal fragment of prtP.
For mutagenesis of prcA, the 4-kb EcoRI-BamHI fragment of pCTLP containing ORF2-prtP was first cloned in pSTBlue-1, yielding pSY119. A PCR product generated with CX260 and CX251 (Table 2) containing all but the first 10 bp of the 5′ end of prcA was amplified from T. denticola genomic DNA and cloned in pSTBlue-1, yielding pSY123. The 1-kb XhoI-NdeI fragment of pSY123 containing prcA DNA was isolated and ligated to the 7.5-kb XhoI-NdeI fragment of pSY119, yielding pSY125. pSY125 was linearized at the unique NdeI site in prcA and ligated to the NdeI-digested fragment of pSY120 containing ermF/AM, yielding pSY126.
DNA sequence analysis.
Templates for DNA sequencing included plasmid DNA and PCR products. Sequencing reactions were performed with ABI PRISM BigDye Terminator Cycle Sequencing kits with fluorescent-labeled dideoxynucleoside triphosphates (Applied Biosystems, Inc., Foster City, Calif.) and sequence-derived primers, according to the manufacturer's instructions. DNA sequences were resolved with an Applied Biosystems model 310 automated DNA sequencer. Both strands of the DNA sequence reported here were sequenced in their entirety. Analysis of DNA sequence data was performed with SeqEd 1.0 (Applied Biosystems, Inc.) and DNA Strider (Service de Biochimie, Department de Biologie, Institut de Recherche Fondamentale Commissariat a l'Energie Atomique, Saclay, France). The nonredundant SWISS-PROT, PIR, EMBL and GenBank databases were searched for homologous peptide and nucleotide sequences by using the BLAST (1) network service at the National Center for Biotechnology Information, Bethesda, Md. Protein sequence motifs were detected with PSORT and SignalP software (23, 24).
Allelic replacement mutagenesis.
Isogenic defined mutants were constructed by the method of Li et al. (21) by electroporation of T. denticola with the selectable ermF/AM gene cassette (11) cloned between fragments of the target sequence as described previously (10), except that linear DNA fragments to be introduced into T. denticola were UV irradiated at 25 mJ cm−2 as described by Picardeau et al. (25) prior to electroporation.
Preparation of antisera.
Polyclonal antisera to the purified native CTLP protein complex and to the recombinant ORF2 polypeptide were raised in New Zealand White rabbits as described previously by intramuscular injections with approximately 1 mg of purified protein in complete Freund's adjuvant (6, 9, 14). The titers of the antisera were determined by enzyme-linked immunosorbent assay, with alkaline phosphatase-conjugated goat anti-rabbit antibody (1:5,000; Life Technologies, Gaithersburg, Md.). A monoclonal antibody that recognizes the T7 gene 10 leader peptide encoded on pET17b (Novagen) was used as a positive control in expression studies.
Gel electrophoresis and immunoblotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western immunoblotting were done as described previously (9). E. coli cells were harvested by centrifugation at 10,000 × g (10 min, 4°C). The pellets were resuspended in 100 μl of sample buffer containing β-mercaptoethanol and 2 mM phenylmethylsulfonyl fluoride. Whole-cell extracts or the detergent phase of Triton X-114 extracts of T. denticola cells were prepared as described previously (6). Samples were heated at 100°C for 5 min prior to electrophoresis in 8 to 16% gradient gels. Proteins in gels were detected by Coomassie brilliant blue staining. Proteins blotted to nitrocellulose membranes were probed with rabbit polyclonal primary antibodies followed by horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG) or goat anti-mouse IgG (Pierce Chemical Co., Rockford, Ill.) as appropriate. Protein bands of interest were developed with SuperSignal West Pico chemiluminescent substrate (Pierce) and detected with a Fluor-S Multi-Imager (Bio-Rad). For reprobing, blots were incubated in stripping buffer (2% SDS, 62.5 mM Tris-HCl [pH 6.8], 10 mM β-mercaptoethanol; 60°C, 30 min) and washed as suggested by the SuperSignal West Pico kit protocol (Pierce).
Enzymatic activity assays.
Enzymatic activities of T. denticola parent and mutant strains were tested by hydrolysis of chromogenic substrates succinyl-l-alanyl-l-alanyl-l-prolyl-l-phenylalanine-p-nitroanilide (SAAPFNA) and N-α-benzoyl-l-arginine-p-nitroanilide (BApNA) as described previously (7). Four-day cultures were adjusted to an A600 of 0.25 in NOS broth and then diluted 1:8 in deionized water for assays. NOS broth diluted 1:8 in deionized water served as a negative control.
Nucleotide sequence accession number.
The nucleotide sequence of T. denticola prcA has been assigned GenBank accession no. AY069957.
RESULTS
Identification of the complete prcA gene.
Our previous studies using isogenic T. denticola mutant CKE, which carries a deletion of a KpnI fragment spanning the 3′ end of ORF2 and the 5′ end of prtP in the CTLP locus (Fig. 1), demonstrated involvement of the CTLP complex in expression and oligomerization of Msp (10). A concurrent study reported similar results for an isogenic prtP mutant (18). To construct other defined mutations in this locus, we designed oligonucleotide primers based on preliminary unannotated contigs of the T. denticola genome (http://www.tigr.org) sequences upstream of the previously reported prtP sequence (19). The preliminary genome sequence data showed that ORF2 upstream of prtP extended an additional 852 bases upstream of the published sequence, suggesting that the previously reported sequence was incomplete. We amplified this upstream region and ORF2 from T. denticola ATCC 35405 genomic DNA using oligonucleotide primers CX251 and CX252 and determined its sequence. The DNA sequence obtained was identical to that of the preliminary genome sequence contig (data not shown). The newly identified complete gene, designated prcA (for protease complex associated) comprises an ORF of 1,917 nucleotides that could encode a 639-residue polypeptide of approximately 70 kDa (data not shown; GenBank accession no. AY069957), compared with the 39-kDa deduced product of ORF2. The deduced PrcA peptide contains a hydrophobic N-terminal region that could serve as a membrane-targeting or translocation signal. The N-terminal region includes a potential recognition site (L-F-G-S-C, residues 20 to 24 [boxed in Fig. 1]) for spirochetal signal peptidase II (15). Searches of GenBank protein sequences and translated contigs of available microbial genomes revealed no significant homologues of PrcA in organisms other than oral treponemes. Based on cleavage of chromogenic substrates by the purified protease complex (19, 22, 27), residues 272 and 289 (marked with an asterisk in Fig. 1) are the only potential cleavage sites for PrtP activity. The previously reported deduced peptide product of ORF2 begins at residue 285 (boxed in Fig. 1). The N-terminal sequence determined from one of the native CTLP complex proteins (19; D. Grenier, personal communication) is present in PrcA (residues 316 to 333 [shaded in Fig. 1]).
Construction of isogenic mutants in prcA and prtP.
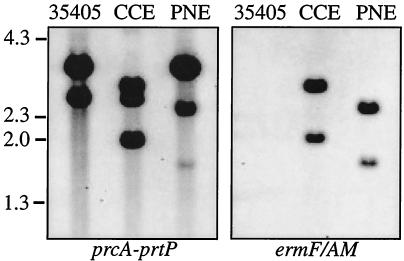
Isogenic mutants in prcA and prtP were constructed by allelic replacement mutagenesis. Plasmids pSY126 and pSY130, containing disrupted prcA and prtP, respectively, were digested with PvuII to separate the vector from insert fragments. T. denticola cells were electroporated with linear DNA from pSY126 or pSY130 and selected for erythromycin resistance. Erythromycin-resistant colonies were isolated, and the structures of the mutant constructs in strains CCE (prtP mutant) and PNE (prcA mutant) were confirmed by Southern blot analysis. HindIII-digested genomic DNAs of T. denticola strains 35405 (parent), CCE, and PNE, separated by agarose gel electrophoresis and transferred to nylon membranes, were probed with the prcA-prtP insert fragment of pCTLP or the ermF/AM cassette from pSY120. As shown in Fig. 2, the ermF/AM probe hybridized with two bands in strains CCE and PNE (ermF/AM contains a single HindIII site) and did not hybridize with 35405 DNA. The prcA-prtP probe, which contains a single HindIII site, hybridized with all three strains and showed the expected band pattern in each strain. It was noted that the 1.7-kb hybridizing signal of the prcA-prtP probe in strain PNE was weak, because fewer than 300 bp of this fragment are homologous with the probe.
FIG. 2.
Southern blot confirming construction of prcA and prtP mutants. HindIII-digested genomic DNAs of T. denticola 35405 (parent), CCE (prtP mutant), and PNE (prcA mutant) were probed with prcA-prtP or ermF/AM as shown. Both prcA and ermF contain single HindIII sites.
Enzymatic activity of parent and mutant strains.
T. denticola 35405 exhibited strong hydrolysis of SAAPFNA, a chromogenic substrate recognized by the CTLP protease complex, while the SAAPFNA activities of CCE (prtP mutant), PNE (prcA mutant), CKE (prcA-prtP mutant), and MPE (CTLP-defective msp mutant) (10) were below detectable levels (data not shown). In contrast, there were no significant differences between strains in hydrolysis of BApNA (data not shown), a chromogenic substrate recognized by the OpdB peptidase (7).
Protein expression and processing of PrcA in parent and mutant strains.
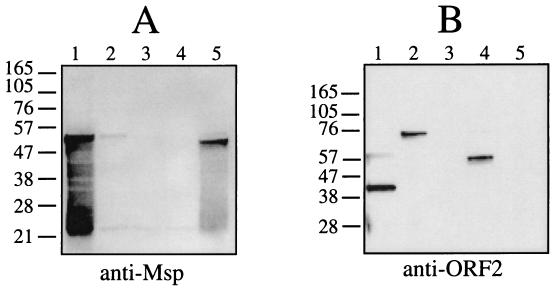
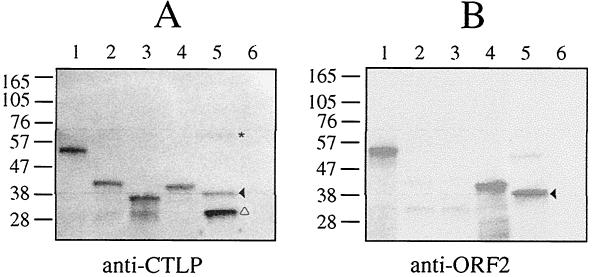
Expression of PrcA and Msp was assayed in parent and mutant strains. As shown in Fig. 3A, anti-Msp antibodies recognized the large amount of Msp and breakdown products produced in the parent strain, compared with the much smaller amount of Msp produced in the various mutants. Msp was at most barely detectable in strains CCE (prtP mutant, lane 2), PNE (prcA mutant, lane 3) and CKE (prcA-prtP mutant, lane 4). Compared with the parent strain, T. denticola MPE (lane 5) produced a lesser amount of a C-terminally-truncated Msp, as previously reported (8).When probed with anti-ORF2 antiserum, no reactive proteins were detected in PNE (prcA mutant, lane 3) or MPE (CTLP-defective msp mutant, lane 5), while a band of 39 kDa corresponding to the size of the larger of the two auxiliary proteins of the protease complex was recognized in 35405. Interestingly, the 39-kDa band was absent in both CCE and CKE, and a higher-molecular-mass band was present in each. The reactive band in CCE migrated at 70 kDa, while in CKE, the reactive band was at 62 kDa. These bands correspond in size to the predicted translation products of prcA in these strains: full-length protein in CCE and truncated protein in CKE. To determine whether the N-terminal region of PrcA was produced in the prcA mutant PNE, the parent and mutant strains were probed with antibodies raised against the native CTLP complex. Immunoreactive bands at 72 kDa (PrtP) and 39 and 32 kDa (CTLP auxiliary proteins) were detected in the parent strain (Fig. 4A, lane 5), while no reactive bands were detected in the mutant PNE (Fig. 4A, lane 6). This indicates that the prcA mRNA is translated as a 70-kDa polypeptide that is then cleaved to the approximately 39-kDa mature protein PrcA2 (Fig. 1) in strains expressing PrtP protease activity. The stability of the PrcA translation product in mutant strains appeared to be dependent on the particular strain construct.
FIG. 3.
Western immunoassays showing expression of Msp and PrcA in T. denticola parent and mutant strains. Panel A shows T. denticola strains probed with antiserum raised against recombinant Msp. Panel B shows the same blot probed with antiserum raised against recombinant PrcA2 (ORF2). Lanes: 1, 35405 (parent); 2, CCE (prtP mutant); 3, PNE (prcA mutant); 4, CKE (prcA-prtP mutant); 5, MPE (msp mutant).
FIG. 4.
Western immunoassays showing PrcA expression in E. coli strains carrying recombinant PrcA constructs and in the T. denticola parent and prcA mutant. Panel A was probed with antiserum raised against native CTLP complex. The three peptides of the CTLP complex are indicated as follows: asterisk, PrtP; open triangle, PrcA1; and solid triangle, PrcA2. Panel B shows the same blot probed with antiserum raised against recombinant PrcA2 (ORF2). Lanes: 1, E. coli(pSY132); 2, E. coli(pSY133); 3, E. coli(pSY134); 4, E. coli(pCF38); 5, T. denticola 35405; 6, T. denticola PNE (prcA mutant).
Expression of PrcA in E. coli.
To determine whether PrcA might encode both of the two auxiliary proteins of the CTLP complex, PrcA expression in E. coli was studied. Attempts to clone the full-length prcA gene were unsuccessful, suggesting that expression of the hydrophobic N terminus was toxic in E. coli. Several fragments of prcA were expressed as fusion proteins with the T7 gene 10 leader sequence under the control of a T7 RNA polymerase promoter in pET17b (Fig. 1). Fragments of prcA, beginning at the BamHI site near the 5′ end of the gene (Fig. 1) and ending at recognition sites for ScaI (pSY132), SnaBI (pSY133), or MspA1I (pSY134) were cloned and expressed. The fusion proteins expressed from these plasmids include PrcA residues 36 to 497 (pSY132, 52.3 kDa), 36 to 350 (pSY133, 41 kDa) and 36 to 307 (pSY134, 36 kDa). DNA encoding the predicted 39-kDa ORF2 product (19), which corresponds to residues 285 to 639 of PrcA, was amplified from genomic DNA and similarly cloned in pET17b, yielding pCF38, which encodes a T7-PrcA fusion protein of 41 kDa. The recombinant protein expressed from pCF38 was excised from an SDS-PAGE gel and used to generate polyclonal rabbit antiserum. In Western immunoassays, these antibodies recognized recombinant proteins of the expected sizes expressed from pSY132 and pCF38, respectively (Fig. 4B, lanes 1 and 4). Anti-ORF2 sera did not recognize the recombinant proteins expressed from pSY133 and pSY134, demonstrating that, if the PrcA polypeptide was cleaved to yield the two observed smaller proteins of the CTLP complex, these antibodies would recognize only the protein derived from the C-terminal portion of PrcA. As a positive control, a duplicate blot was probed with a monoclonal antibody directed against the T7 gene 10 leader peptide. All four recombinant proteins were recognized by this antibody (data not shown). When probed with antibodies raised against the native CTLP complex (Fig. 4A), recombinant T7-PrcA proteins of the predicted sizes were recognized in all four E. coli strains. Anti-CTLP antibodies recognize the three proteins of the CTLP complex in T. denticola 35405 that are absent in the prcA mutant PNE (Fig. 4A, lanes 5 and 6). These data indicate that the amino-terminal region of PrcA comprises the 32-kDa PrcA1, the third protein of the CTLP complex.
DISCUSSION
Native expression of two potential virulence determinants of T. denticola, the CTLP protease complex and the Msp protein, appears to be related. We previously reported that, in addition to the expected loss of CTLP protease activity, disruption of the prtP locus results in a greatly decreased ability of T. denticola to produce the native oligomeric form of the major surface protein, Msp (10). We also found an apparently related phenomenon in a defined T. denticola msp mutant that was unable to oligomerize Msp due to a mutation in the C terminus of the Msp peptide and also lacked all detectable CTLP proteins and protease activity (10). The two smaller proteins that form the native CTLP complex with the 72-kDa PrtP protease had no known functions, and in the case of the one protein whose gene had been reported, no known homologues (19). For this reason, we chose to further characterize these proteins and their potential role(s) in assembly or stability of outer membrane complexes in this spirochete. The present study characterized expression of PrcA, the protease complex-associated polypeptide, and the PrtP-dependent processing of PrcA that results in the two native protease complex-associated proteins PrcA1 and PrcA2.
The prcA gene identified in this study includes the sequence previously reported as ORF2 directly upstream of the gene encoding the PrtP protease component of the CTLP complex (19). ORF2 was first proposed to encode one of the two protease complex-associated proteins, based on its location adjacent to prtP, the size of its deduced protein product, and the presence within the deduced protein of an amino acid sequence identical to the N-terminal sequence one of the native CTLP complex proteins. The absence of an identifiable signal sequence on the deduced ORF2 product that could direct it to a secretory pathway was not previously addressed and led us to further examine DNA sequences upstream of ORF2. Our results showed that prcA, including ORF2 and 852 bp upstream, encodes a protein of 70 kDa with a hydrophobic N-terminal region that could serve as a membrane-targeting or translocation signal. The N-terminal region includes a potential recognition site (L-F-G-S-C, residues 20 to 24) for spirochetal signal peptidase II (15). The prediction that PrcA is acylated is consistent with the observed segregation of the entire CTLP complex to the detergent phase of Triton X-114 extracts (6).
Identification of the prcA gene as encoding a polypeptide of 70 kDa raised further issues: (i) whether this protein is in fact part of the protease complex; and (ii) if PrcA is CTLP-associated, how it might be processed to the native molecular mass. Characterization of PrcA expression in E. coli and in T. denticola parent and isogenic mutant strains demonstrated that the PrcA polypeptide is the source of both of the two smaller proteins that, together with PrtP, form the native CTLP protease complex. In addition, cleavage of PrcA to yield PrcA1 and PrcA2 appeared to be due to PrtP protease activity, since T. denticola strains CCE and CKE (both mutated in prtP) lacked PrtP activity, and neither strain cleaved the PrcA polypeptide to a native form. The 70-kDa PrcA in CCE and the 62-kDa PrcA in CKE correspond to the predicted PrcA translation products of these strains. It is likely that cleavage of PrcA to PrcA1 and PrcA2 occurs after export across the cytoplasmic membrane. Both the deduced peptide sequence and the determined N-terminal sequence of PrtP suggest that PrtP is processed to an active form subsequent to cleavage of its N-terminal signal peptide and secretion across the cytoplasmic membrane (19) and thus is unlikely be proteolytically active in the cytoplasm.
Several other members of the subtilisin group of bacterial proteases require an associated chaperone-like protein for proper translocation and maturation (20). The PrtM protein of Lactococcus lactis, a typical member of this group, is a 33-kDa putative lipoprotein required for proper maturation of the L. lactis PrtP cell envelope-associated subtilisin homologue (29). The prtM gene is located upstream of prtP in L. lactis and is transcribed from the same promoter region, although in the opposite direction. While there is no identifiable homology between L. lactis PrtM and T. denticola PrcA, our results support the hypothesis that PrcA functions similarly as a chaperone-like molecule in the T. denticola CTLP complex.
Several areas remain for future molecular characterization of this intriguing protease complex. Most importantly, the nature of the processing event requires more study. There are two potential recognition sites for PrtP protease activity in PrcA, either of which would yield proteins of the observed sizes of PrcA1 and PrcA2. A unique Pro-Phe sequence (residues 289 to 290) contains the reporter site of SAAPFNA, the chromogenic substrate used to characterize CTLP enzyme activity (19, 22, 27). The unique sequence Pro-Leu (residues 268 to 272) is also a possible cleavage site (19). Studies of cleavage of native PrcA are likely to be problematic until methods are developed to generate defined point mutations in T. denticola. No method of purifying proteolytically active PrtP separate from PrcA1 and PrcA2 has ever been reported. The extremely close association between PrtP and PrcA1 and PrcA2 suggests that this association may be required for protease activity or localization of the complex. Similarly, while we have been unable to directly demonstrate cleavage of exogenous PrcA by PrtP present in the protease complex (data not shown), it is also likely that such cleavage may be dependent on specific association and binding between PrtP and PrcA as part of the maturation process of the protease complex.
While it was not possible to directly determine the role of PrcA1 and PrcA2 in Msp expression because T. denticola isogenic mutants deficient in either or both prcA and prtP had similar Msp phenotypes and lacked CTLP activity, these strains will be useful in planned studies examining the biological role of the CTLP protease complex in the interactions between T. denticola and host cells. The present study characterized expression and processing of a putative chaperone molecule required for proper expression and activity of T. denticola outer membrane-associated proteins that are implicated in periodontal disease cytopathology.
Acknowledgments
This work was supported by Public Health Service grant DE13565 from the National Institute of Dental and Craniofacial Research (J.C.F.). Preliminary studies for this work were supported by the Medical Research Council of Canada (B.C.M.). Partial support for S.Y.L. was provided by Kangnung National University, Korea (2000).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology, vol. 1. Wiley-Interscience, New York, N.Y.
- 3.Chan, E. C. S., A. DeCiccio, R. McLaughlin, A. Klitorinos, and R. Siboo. 1997. An inexpensive solid medium for obtaining colony-forming units of oral spirochetes. Oral Microbiol. Immunol. 12:372-376. [DOI] [PubMed] [Google Scholar]
- 4.Ellen, R. P., K. S. Ko, C. M. Lo, D. A. Grove, and K. Ishihara. 2000. Insertional inactivation of the prtP gene of Treponema denticola confirms dentilisin's disruption of epithelial junctions. J. Mol. Microbiol. Biotechnol. 2:581-586. [PubMed] [Google Scholar]
- 5.Ellen, R. P., M. Song, and C. A. G. McCulloch. 1994. Degradation of endogenous plasma membrane fibronectin concomitant with Treponema denticola 35405 adhesion to gingival fibroblasts. Infect. Immun. 62:3033-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenno, J. C., P. M. Hannam, W. K. Leung, M. Tamura, V.-J. Uitto, and B. C. McBride. 1998. Cytopathic effects of the major surface protein (Msp) and the chymotrypsinlike protease (CTLP) of Treponema denticola. Infect. Immun. 66:1869-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenno, J. C., S. Y. Lee, C. H. Bayer, and Y. Ning. 2001. The opdB locus encodes the trypsin-like peptidase activity of Treponema denticola. Infect. Immun. 69:6193-6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fenno, J. C., and B. C. McBride. 1998. Virulence factors of oral treponemes. Anaerobe 4:1-17. [DOI] [PubMed] [Google Scholar]
- 9.Fenno, J. C., K.-H. Müller, and B. C. McBride. 1996. Sequence analysis, expression and binding activity of recombinant major outer sheath protein (Msp) of Treponema denticola. J. Bacteriol. 178:2489-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenno, J. C., G. W. K. Wong, P. M. Hannam, and B. C. McBride. 1998. Mutagenesis of outer membrane virulence determinants of the oral spirochete Treponema denticola. FEMS Microbiol. Lett. 163:209-215. [DOI] [PubMed] [Google Scholar]
- 11.Fletcher, H. M., H. A. Schenkein, R. M. Morgan, K. A. Bailey, C. R. Berry, and F. L. Macrina. 1995. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect. Immun. 63:1521-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giegerich, R., F. Meyer, and C. Schleiermacher. 1996. GeneFisher—software support for the detection of postulated genes. Proc. Int. Conf. Intell. Syst. Mol. Biol. 4:68-77. [PubMed] [Google Scholar]
- 13.Grenier, D. 1996. Degradation of host protease inhibitors and activation of plasminogen by proteolytic enzymes from Porphyromonas gingivalis and Treponema denticola. Microbiology 142:955-961. [DOI] [PubMed] [Google Scholar]
- 14.Grenier, D., V.-J. Uitto, and B. C. McBride. 1990. Cellular location of a Treponema denticola chymotrypsinlike protease and importance of the protease in migration through the basement membrane. Infect. Immun. 58:347-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haake, D. A. 2000. Spirochaetal lipoproteins and pathogenesis. Microbiology 146:1491-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haapasalo, M., U. Singh, B. C. McBride, and V.-J. Uitto. 1991. Sulfhydryl-dependent attachment of Treponema denticola to laminin and other proteins. Infect. Immun. 59:4230-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heuner, K., I. Bergmann, K. Heckenbach, and U. B. Gobel. 2001. Proteolytic activity among various oral Treponema species and cloning of a prtP-like gene of Treponema socranskii subsp. socranskii(1). FEMS Microbiol. Lett. 201:169-176. [DOI] [PubMed] [Google Scholar]
- 18.Ishihara, K., H. K. Kuramitsu, T. Miura, and K. Okuda. 1998. Dentilisin activity affects the organization of the outer sheath of Treponema denticola. J. Bacteriol. 180:3837-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishihara, K., T. Miura, H. K. Kuramitsu, and K. Okuda. 1996. Characterization of the Treponema denticola prtP gene encoding a prolyl-phenylalanine-specific protease (dentilisin). Infect. Immun. 64:5178-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunji, E. R., I. Mierau, A. Hagting, B. Poolman, and W. N. Konings. 1996. The proteolytic systems of lactic acid bacteria. Antonie Leeuwenhoek 70:187-221. [DOI] [PubMed] [Google Scholar]
- 21.Li, H., J. Ruby, N. Charon, and H. Kuramitsu. 1996. Gene inactivation in the oral spirochete Treponema denticola: construction of an flgE mutant. J. Bacteriol. 178:3664-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mäkinen, P. L., K. K. Mäkinen, and S. A. Syed. 1995. Role of the chymotrypsin-like membrane-associated proteinase from Treponema denticola ATCC 35405 in inactivation of bioactive peptides. Infect. Immun. 63:3567-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakai, K., and M. Kanehisa. 1991. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins 11:95-110. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 25.Picardeau, M., A. Brenot, and I. Saint Girons. 2001. First evidence for gene replacement in Leptospira spp. Inactivation of L. biflexa flaB results in non-motile mutants deficient in endoflagella. Mol. Microbiol. 40:189-199. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Uitto, V.-J., D. Grenier, E. C. S. Chan, and B. C. McBride. 1988. Isolation of a chymotrypsinlike enzyme from Treponema denticola. Infect. Immun. 56:2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uitto, V.-J., Y.-M. Pan, W. K. Leung, H. Larjava, R. P. Ellen, B. B. Finlay, and B. C. McBride. 1995. Cytopathic effects of Treponema denticola chymotrypsin-like proteinase on migrating and stratified epithelial cells. Infect. Immun. 63:3401-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vos, P., M. van Asseldonk, F. van Jeveren, R. Siezen, G. Simons, and W. M. de Vos. 1989. A maturation protein is essential for production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J. Bacteriol. 171:2795-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]