Abstract
Pseudomonas cellulosa is a highly efficient xylan-degrading bacterium. Genes encoding five xylanases, and several accessory enzymes, which remove the various side chains that decorate the xylan backbone, have been isolated from the pseudomonad and characterized. The xylanase genes consist of xyn10A, xyn10B, xyn10C, xyn10D, and xyn11A, which encode Xyn10A, Xyn10B, Xyn10C, Xyn10D, and Xyn11A, respectively. In this study a sixth xylanase gene, xyn11B, was isolated which encodes a 357-residue modular enzyme, designated Xyn11B, comprising a glycoside hydrolase family 11 catalytic domain appended to a C-terminal X-14 module, a homologue of which binds to xylan. Localization studies showed that the two xylanases with glycoside hydrolase family (GH) 11 catalytic modules, Xyn11A and Xyn11B, are secreted into the culture medium, whereas Xyn10C is membrane bound. xyn10C, xyn10D, xyn11A, and xyn11B were all abundantly expressed when the bacterium was cultured on xylan or β-glucan but not on medium containing mannan, whereas glucose repressed transcription of these genes. Although all of the xylanase genes were induced by the same polysaccharides, temporal regulation of xyn11A and xyn11B was apparent on xylan-containing media. Transcription of xyn11A occurred earlier than transcription of xyn11B, which is consistent with the predicted mode of action of the encoded enzymes. Xyn11A, but not Xyn11B, exhibits xylan esterase activity, and the removal of acetate side chains is required for xylanases to hydrolyze the xylan backbone. A transposon mutant of P. cellulosa in which xyn11A and xyn11B were inactive displayed greatly reduced extracellular but normal cell-associated xylanase activity, and its growth rate on medium containing xylan was indistinguishable from wild-type P. cellulosa. Based on the data presented here, we propose a model for xylan degradation by P. cellulosa in which the GH11 enzymes convert decorated xylans into substituted xylooligosaccharides, which are then hydrolyzed to their constituent sugars by the combined action of cell-associated GH10 xylanases and side chain-cleaving enzymes.
The recycling of photosynthetically fixed carbon through the action of microbial plant cell wall hydrolases is a key biological process. Xylan is one of the most abundant plant structural polysaccharides. This heterogeneous polymer consists of a backbone of β1,4-linked xylopyranose residues which are decorated with acetyl, arabinofuranosyl, and 4-methyl-O-glucuronyl side chains (6). The backbone xylose polymer is hydrolyzed by endo-β1,4-xylanases (xylanase), whereas the side chains are removed by the action of arabinofuranosidases, α-glucuronidases, and acetyl xylan esterases (6). Based on amino acid sequence similarity, xylanases have been grouped into glycoside hydrolase families (GH) 10 and 11 (http://afmb.cnrs-mrs.fr/∼cazy/CAZY/index.html [18]). GH 10 xylanases expressed by fungi and Streptomyces lividans display more relaxed specificity than GH 11 enzymes from the same microorganisms (7, 8), although it is unclear whether this general view of hemicellulase specificity can be extended to gram-negative prokaryotes. Xylanases, in common with other plant cell wall hydrolases, are encoded by large multigene families (13, 16, 22, 25), although the evolutionary pressures leading to this complex array of genes are not readily apparent.
The majority of plant cell wall hydrolases are not constitutively expressed but are induced by plant structural polysaccharides (9, 20, 30). Several of these enzymes are induced only by their target substrate, while others are synthesized in response to a range of plant structural polysaccharides (9, 26, 33). The general model for induction of microbial plant cell wall-degradative enzymes involves the low-level constitutive expression of a “sensor” enzyme, which generates small oligo- or disaccharides from plant cell wall material that enter the microorganism and elicit the high-level synthesis of plant cell wall hydrolases (26, 30, 33). This model has been demonstrated for Trichoderma reesei xylanases; xyn1 is only induced by xylan or its degradative products, whereas xyn2 displays low-level constitutive expression and its synthesis is activated by both xylan and cellulose (33). Similarly, it has been proposed that the xylanase encoded by Streptomyces cyaneus was the constitutively expressed sensor protein that activated the synthesis of the xylan-degrading system in this bacterium (5). Currently, regulatory proteins that control xylanase gene expression have only been identified in filamentous fungi. In Aspergillus niger XlnR was shown to be a general transcriptional activator of the xylanolytic system, binding to a consensus heptanucleotide sequence that is also present in the promoters of Penicillium chrysogenum xylanase genes (32). Recently, a novel Trichoderma transcriptional activator, ACEII, was identified that induced expression of the majority of cellulases and the xylanase encoded by xyn2 (2).
Although there have been numerous studies on the regulation of xylanase gene expression in fungi, the regulatory mechanisms that induce the synthesis of the corresponding bacterial enzymes and the cellular location of these glycoside hydrolases are poorly understood. In this study we investigated the expression patterns and the cellular location of the xylanases produced by the bacterium Pseudomonas cellulosa. P. cellulosa is an efficient xylan-degrading bacterium (15), which was previously shown to contain genes encoding five xylanases in addition to several side chain-cleaving enzymes (4, 11). The bacterium displays xylanase activity when cultured on xylan or cellulose (13, 14, 16, 17, 22, 25); however, it is unclear whether specific subsets of xylanase genes are synthesized in response to different polysaccharides. The cellular location of these enzymes in P. cellulosa, apart from Xyn10A (which is extracellular, i.e., secreted into the culture medium when the bacterium is grown on Luria broth [LB] supplemented with xylan [17]), has also not been elucidated, and it is unknown whether the bacterium synthesizes a sensor xylanase. In this report we identify a new P. cellulosa GH11 xylanase, Xyn11B, and demonstrate that all of the xylanases from this pseudomonad are induced by the same polysaccharides, although there was evidence for differential temporal expression of the genes encoding the GH11 enzymes. Although one of the highly expressed GH10 enzymes, Xyn10C, is located on the cell membrane, the two GH11 enzymes appear to be the major xylanases in the extracellular milieu. The pattern of xylanase expression, and their cellular location, is discussed in relation to the capacity of the bacterium to use xylan as a carbon and energy source.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The Escherichia coli strains used in this study were BL21(DE3)/pLysS (Novagen) and XL1-Blue (Stratagene). The bacteriophage employed comprised a genomic library of P. cellulosa (NCIMB 10462) constructed previously in λZAPII (Stratagene [25]). The plasmid vectors used were pCR-Blunt (Invitrogen), pET16b (Novagen), and pRG960sd (31). All E. coli strains containing recombinant plasmids were cultured in LB supplemented with 50 μg of ampicillin or kanamycin/ml, as needed, at 37°C unless otherwise stated. E. coli cells used to propagate bacteriophage were grown on LB supplemented with 10 mM MgSO4 and 0.2% (wt/vol) maltose and were plated out on NZYM top agar (0.7%). To screen the genomic library, ca. 40,000 recombinant phage were plated at a density of three plaques per cm2. After incubation at 37°C for 24 h, the phage was subjected to plaque hybridization with the pseudomonad insert of pKE5, which comprises the 5′ region of xyn11A, as the probe. To synthesize Xyn11B, E. coli XL1-Blue harboring pKE2 was cultured in LB containing ampicillin and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 16 h. To grow P. cellulosa, a rifampin-resistant mutant of the bacterium was cultured in either LB or minimal medium, which consists of M9 salts and the appropriate carbon source, either monosaccharide or polysaccharide, at a final concentration of 0.2%. The pseudomonad was grown in liquid culture at 37°C with high aeration (200 rpm) for up to 36 h in media comprising <10% (vol/vol) of the incubator vessel (conical flask) or on solid medium at 28°C for up to 2 weeks. All media used to culture the pseudomonad were supplemented with 50 μg of rifampin/ml, whereas streptomycin and spectinomycin (both at 50 μg/ml) were added to cultures of P. cellulosa harboring derivatives of the promoter probe vector pRG960sd.
Recombinant DNA techniques.
Standard recombinant DNA techniques such as Southern hybridization, plaque hybridization, bacterial transformation, agarose gel electrophoresis, plasmid DNA preparation, restriction digestion, and ligation were as described previously (27). The nucleotide sequence of DNA was determined by using an ABI Prism Ready Reaction DyeDeoxy terminator cycle sequencing kit. The sequencing reactions were electrophoresed, and the fluorescent signals emitted by the DNA molecules were detected by using an Applied Biosystems 377A sequencing system. The complete sequence of the DNA (both strands) was determined by using a series of custom-made primers to initiate DNA synthesis. The compiled DNA sequence was analyzed by using the DNAsis computer program.
Transfer of DNA into P. cellulosa.
Recombinants of the promoter probe vector pRG960sd were transferred into P. cellulosa either by filter mating or by electroporation. Filter mating was carried out similarly to the method previously described (3), except that triparental filter mating was carried out with a third E. coli strain containing the helper plasmid pRK2013 (12). The electroporation method was carried out as follows. P. cellulosa was cultured in 400 ml of LB to mid-exponential phase, and the bacterial cells were then harvested by centrifugation at 6,000 × g for 10 min at 4°C. The cell pellet was washed three times with 400 ml of 5% (vol/vol) glycerol and then resuspended in 1 ml of the glycerol solution. The cells were divided into 50-μl aliquots and stored at −70°C. Electroporation was carried out by pipetting 50 μl of cells into Gene Pulser cuvettes (0.1-cm gap), and the electrical current was provided by a Gene Pulser (Bio-Rad) set at 1.6 kV cm−1, with a resistance of 100 Ω and a pulse time of 4 ms. The bacterial cells were then diluted with 250 μl of LB and incubated with aeration for 1 h at 37°C before being plated out on media supplemented with streptomycin and spectinomycin (both at 50 μg/ml). To introduce the transposon Tn10 into the P. cellulosa genome, E. coli S17-1-λpir harboring pLOFKm (contains a mini-Tn10 element [19]) and the pseudomonad were cultured overnight in LB supplemented with appropriate antibiotics. The cells were pelleted, washed, and then resuspended in phosphate-buffered saline. The cell suspensions were then mixed in a P. cellulosa/E. coli ratio of 5:1, and a 6-ml portion was filter mated as described previously (3), except that the filters were incubated overnight on Luria agar containing 1 mM IPTG to induce transposase expression, and transconjugants were selected on medium containing 50 μg of rifampin and kanamycin/ml.
RNA quantification.
RNA was isolated from cultures of P. cellulosa grown at 30°C at different stages of the growth phase as described previously (3). Aliquots of RNA (5 μg) were electrophoresed on 1.2% (wt/vol) agarose-formaldehyde denaturing gels (27) and transferred to Hybond N membranes (Amersham-Pharmacia) by using 300 mM sodium citrate buffer (pH 7.2) containing 3 M NaCl, and the nucleic acid was immobilized by UV cross-linking. The filters were then probed with radiolabeled DNA encoding the various xylanase genes. Probe bound to the transcripts was determined by electronic autoradiography (phosphorimager) by using a Packard Instant Imager. To confirm that the signals were in the linear range of detection, samples which gave the highest and lowest intensity signals were subjected to Northern hybridization with 2.5-, 5-, 10-, and 20-μg RNA samples, and the intensity of the signal of the bound probe was determined. In all cases there was a linear correlation between signal intensity and the amount of RNA electrophoresed. The probes used to detect transcripts derived from xyn10A (16), xyn10B (22), xyn10C (13), xyn10D (25), xyn11A, and xyn11B (Fig. 1, pKE5) encoded the catalytic domains of the respective enzymes, except the xyn10D probe, which encoded the full-length xylanase. To distinguish between xyn11A and xyn11B, probes encoding the xylan-binding modules of Xyn11A (pKE3) and Xyn11B (pKE4) were used.
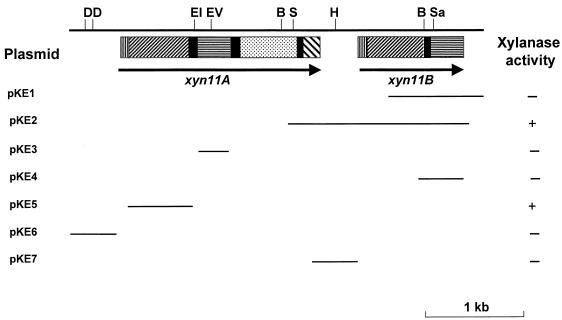
FIG. 1.
Physical map of xyn11A/xyn11B locus. The positions of the cleavage sites for the following restriction enzymes are as follows: BglII (B), DraI (D), EcoRI (EI), EcoRV (EV), HindIII (H), SmaI (S), and SacII (Sa). The solid arrows show the extent and orientation of the genes encoding Xyn11A (xyn11A) and Xyn11B (xyn11B). The lines denote the region of pseudomonad DNA present in the plasmids, which were constructed as described in Materials and Methods. The capacity of the plasmids to encode a functional xylanase was determined by plating E. coli containing these DNA molecules on either Blue-Xylan or Azo-wheat arabinoxylan. Clear haloes surrounding the colonies are indicative of xylanase activity (+); no halo indicates the enzyme is absent (−). The modules in the schematic of Xyn11A and Xyn11B are as follows: signal peptide (▥), catalytic domain (▨), linker sequence (▪), family 4 carbohydrate esterase module (░⃞), xylan-binding CBM (▤), and family 10 CBM (▧).
To measure transcripts by real-time PCR, 1 μg of total P. cellulosa RNA, prepared as described above, was used as a template to synthesize cDNA by using random primers and the Superscript preamplification cDNA synthesis kit (Gibco-BRL). The cDNA was then subjected to PCR with 200 μM concentrations of each transcript-specific primer, 3 mM MgCl2, and the Light Cycler-DNA SYBR Green 1 Master Mix (Roche Diagnostics). The PCRs were carried out in a Roche Light Cycler instrument with the following temperature parameters: 95°C for 20 min, followed by 50 cycles of 95°C for 15 s, 60°C for 5 s, and 72°C for 10 s. The fluorometric intensity of SYBR Green 1, a specific dye for double-stranded DNA, was measured at the end of each elongation. Fluorescent signals caused by primer dimers and nonspecific background were evaluated by melting-curve analysis and agarose gel electrophoresis, as recommended by the manufacturer. The sizes of the amplified DNA were as follows: xyn10A, 380 bp; xyn10B, 320 bp; xyn11A, 320 bp; xyn11B, 238 bp; xyn10D, 385 bp; and xyn10C, 900 bp.
Construction of recombinant plasmids.
To clone the 447-bp (−463 to −16) and 445-bp (−461 to −17) regions immediately upstream of P. cellulosa, xyn11A and xyn11B, respectively, were amplified by PCR. The PCRs were performed with 2 U of the thermostable DNA polymerase VentR (New England Biolabs). The reactions contained 2 mM MgSO4, 1× Thermopol reaction buffer (New England Biolabs), 0.5 μM concentrations of primers, and 60 ng of target DNA in a final volume of 100 μl. The reactions were subject to 25 cycles at the following temperatures: 95°C for 1 min, 55°C for 1 min, and 72°C for 2 min. The amplified DNAs were inserted into pCR-Blunt, the sequences were verified, and the nucleic acids were then excised by restricting with BamHI and PspAI (an isoschizomer of SmaI) and cloned into similarly digested pRG960sd to generate pKE6 and pKE7, which contain the promoter regions of xyn11A and xyn11B, respectively.
To clone the 5′ region of xyn11B, primers were designed which hybridized to the region of xyn11A that encoded the family 4 carbohydrate esterase domain of Xyn11A and the region of xyn11B encoding the C-terminal sequence of the xylan-binding carbohydrate-binding module (CBM) of Xyn11B. The primers were used to amplify the target sequence from P. cellulosa genomic DNA, and the 1,728-bp product was cloned into pCR-Blunt to generate pKE2. To clone DNA encoding the xylan-binding CBM, the appropriate region of xyn11B was amplified by PCR. The product was cloned into pCR-Blunt to generate pKE4, and the resultant plasmid was used in Northern and Southern analyses of P. cellulosa RNA and DNA, respectively. To clone DNA encoding the catalytic domain of Xyn11A, the appropriate region of xyn11A was amplified by PCR and the product was restricted with NdeI and XhoI and cloned into similarly digested pET16b to generate pKE5.
To clone the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene (gpd) from P. cellulosa, the primary structures of the enzyme from 50 prokaryotic and eukaryotic microorganisms were compared. The data revealed a highly conserved sequence at the N and C termini of these proteins, from which primers were designed based on the nucleotide sequence of gpd from Pseudomonas aeruginosa (M74256). These primers were used in a PCR which generated a 934-bp product that was cloned into the pCR-Blunt and sequenced. Analysis of the sequence revealed that the DNA encoded GAPDH. The amplified DNA was used as a probe in Northern hybridization of P. cellulosa RNA as described above.
All of the primers used in the various PCRs described above are available on request.
Assays.
Protein was measured by the dye-binding method of Sedmak and Grossberg (28) with bovine serum albumin as the standard. Xylanase activity was determined by using a standard reducing sugar assay with dinitrosalicylic acid (24). The reporter enzyme β-glucuronidase was assayed as follows. A 500-μl reaction mixture of 50 mM sodium phosphate buffer (pH 7.0), 1 mM 4-methylumbelliferyl-β-d-glucuronide, 1 mg of bovine serum albumin/ml, and an appropriate dilution of the bacterial cell extract was incubated at 37°C for 10 min and was terminated by the addition of 2.5 ml of 50 mM glycine-NaOH buffer (pH 10.4). The release of the fluorescent reaction product 4-methylumbelliferone was determined with a RF-1501 spectrofluorophotometer (Shimadzu) by using excitation and emission wavelengths of 365 and 460 nm, respectively.
Fractionation of P. cellulosa and Western analysis.
Stationary cells of P. cellulosa were harvested by centrifugation at 6,000 × g for 10 min from a 400-ml culture comprising minimal medium in which wheat arabinoxylan was the carbon source. The cell pellet was then resuspended in 10 ml of 50 mM sodium phosphate-12 mM citric acid buffer (pH 6.5) and sonicated. The cell membranes were then pelleted by centrifugation at 100,000 × g at 4°C for 1 h, and the supernatant, consisting of the cytoplasm and the periplasm (referred to here as the cell extract) was retained for further use. The purity of the different fractions was assessed by measuring the activity of the marker enzymes malate dehydrogenase (cell extract) and NADH oxidoreductase (membrane fraction) as described previously (13). Western analysis of the various fractions was carried out as described previously (13) with antisera raised against the catalytic domains of Xyn11A, Xyn10C, and Xyn10D.
Nucleotide sequence accession number.
The nucleotide sequence for xyn11B has been deposited in the GenBank database under accession no. AY065640.
RESULTS
Identification of a novel P. cellulosa xylanase gene xyn11B.
In previous studies five genes encoding xylanases were isolated from P. cellulosa. These genes were designated xyn10A (formerly xynA), xyn10B (formerly xynB), xyn11A (formerly xynE), xyn10C (formerly xynF), and xyn10D (formerly xynG), respectively (13, 16, 22, 25). To investigate the regulation of xylanase expression in P. cellulosa, it is important that all of the xylanase genes are identified in this bacterium. Exhaustive screening of 106 clones of a λZAPII library of P. cellulosa failed to identify any xylanase-expressing clones not previously characterized. Southern hybridization with xyn11A as a probe, however, revealed the presence of a sixth xylanase gene, defined as xyn11B. The observation that the probe bound to single BamHI, BglII, DraI, and PstI restriction fragments may point to linkage between xyn11A and xyn11B, although this interpretation of the data must be viewed with some caution (Fig. 2). Screening of the genomic library with xyn11A led to the isolation of a clone (pKE1) that contained an incomplete open reading frame (ORF), designated xyn11B, which exhibited sequence similarity to the 3′ region of xyn11A. To clone the missing sequence of the ORF, primers that bound to the 3′ region of xyn11A and xyn11B (primers were directed against the 3′ region of xyn11B, which was not conserved in xyn11A), respectively, were used in a PCR of P. cellulosa genomic DNA. The resultant DNA product was cloned into pCR-Blunt to generate pKE2. The sequence of the pseudomonad insert in pKE2 showed that the plasmid contained the missing 5′ region of xyn11B. The physical map of the xyn11A/xyn11B locus is shown in Fig. 1.
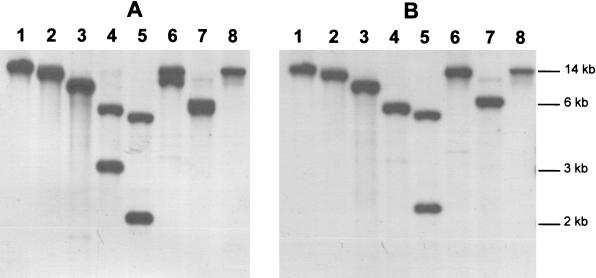
FIG. 2.
Southern hybridization of the xyn11A/xyn11B locus of the P. cellulosa genome. P. cellulosa genomic DNA was digested with BamHI (lane 1), BglII (lane 2), DraI (lane 3), EcoRI (lane 4), EcoRV (lane 5), HindIII (lane 6), SmaI (lane 7), and PstI (lane 8). The restricted DNA was electrophoresed, blotted onto nylon membranes, and then probed with the pseudomonad inserts in pKE5 (encodes the catalytic domain of Xyn11A) (A) and pKE4 (encodes the xylan-binding CBM of Xyn11B) (B).
Translation of the xyn11B ORF (present in GenBank under the accession no. AY065640) showed that the gene encoded a protein, designated Xyn11B, of Mr 37,889. Comparison of the primary structure of Xyn11B with protein databases indicated that the enzyme had a modular structure. The N-terminal 27 amino acids conform to a typical signal peptide; the 9 N-terminal hydrophilic residues contain several basic amino acids and are followed by a stretch of 18 small hydrophobic residues that are capable of forming an α-helix, which is followed by a potential signal peptidase I cleavage site. The putative signal peptide is followed by 210 amino acids that exhibit 94% sequence identity with the catalytic module of Xyn11A (encoded by xyn11A [25]), demonstrating that this region comprises a catalytic module belonging to GH11. Inspection of the catalytic module of Xyn11A showed that Glu116 and Glu213 were the key catalytic residues of the xylanase since they correspond to Glu78 and Glu172 of the Bacillus circulans GH 11 enzyme, which are the catalytic nucleophile and catalytic acids, respectively (21). The GH11 catalytic module is separated by a 20-residue linker sequence rich in serines and glycines from a 110-amino-acid module that displays 57% sequence identity with residues 254 to 362 of Xyn11A. Recent studies have shown that this region constitutes a xylan-binding CBM (H. Xie et al., unpublished data). Thus, Xyn11B is highly homologous to the N-terminal region of Xyn11A, which comprises the xylanase GH11 catalytic module and the xylan-binding CBM, but it lacks the family 4 carbohydrate esterase and the family 10 CBM that are present at the C terminus of Xyn11A (25). The xyn11B ORF is preceded by a typical ribosome-binding sequence, GGAGA, 9 bp upstream of the translational start codon. The sequence immediately downstream of the ORF of xyn11B is GC-rich and contains a region of dyad symmetry that was followed and preceded by a series of thymine and adenine nucleotides, respectively. This region is likely to act as a typical rho-independent transcriptional terminator sequence when either the sense or the antisense strand is transcribed. Upstream of the ORF is a 34-nucleotide sequence that exhibits almost perfect dyad symmetry, although it is not preceded by a succession of thymines, and thus the functional significance of this region is currently unknown.
To determine whether the putative catalytic module of Xyn11B is functional, DNA encoding this region (pKE2; Fig. 1) was transformed into E. coli. The recombinant bacterium, when plated onto LB containing IPTG and either either Blue-Xylan (Sigma Chemical Co.) or Azo-wheat arabinoxylan (Megazyme International Ltd.), displayed xylanase activity as evidenced by the appearance of clear haloes surrounding the bacterial colonies in a blue background.
Localization of xylanase activity.
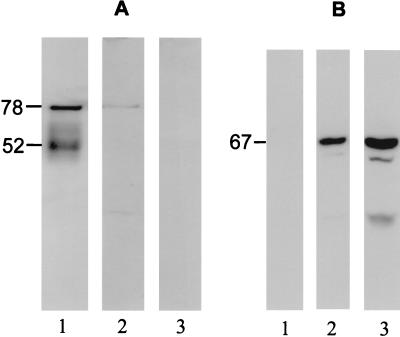
To determine the location of the xylanase activity expressed by P. cellulosa, the bacterium was cultured on xylan and the cells, in mid-log phase, were fractionated, and the xylanase activity in each fraction was assessed. The data, presented in Table 1, showed that ca. 65% of xylanase activity was present in the culture supernatant and that the remainder was cell associated. It would appear that the majority of cell-associated xylanase activity was on the outer membrane since xylan degradation by whole cells was similar to that by sonicated cell extracts, in which the cell membranes had been disrupted releasing the periplasmic and cytosolic proteins (data not shown). Antisera raised against the xylanases were then used to probe the cellular location of the different enzymes. The data, presented in Fig. 3, showed that Xyn10C was predominantly associated with the cell membrane, although the protein could also be detected in the cell extract, whereas the GH11 xylanases appeared to be present in the culture supernatant; Xyn10D gave no obvious signal on Western analysis (data not shown). The size of Xyn10C, determined by Western analysis, was very similar to its deduced molecular size, indicating a lack of posttranslational modification.
TABLE 1.
Xylanase expression in P. cellulosaa
| Growth medium | Xylanase activity (U/ml of culture) |
|
|---|---|---|
| Culture supernatant | Cell associated | |
| Oat spelt xylan | 0.73 ± 0.07 | 0.42 ± 0.11 |
| Glucuronoxylan | 0.65 ± 0.07 | 0.39 ± 0.02 |
| Rye arabinoxylan | 0.81 ± 0.10 | 0.48 ± 0.08 |
| Wheat arabinoxylan | 0.91 ± 0.11 | 0.52 ± 0.1 |
| β-Glucan | 0.21 ± 0.02 | 0.13 ± 0.03 |
| Carbo galactomannan | <0.01b | <0.01 |
| Pectin | <0.01 | <0.01 |
| LB | <0.01 | <0.01 |
| LB and oat spelt xylan | 0.21 ± 0.04 | 0.13 ± 0.06 |
| Oat spelt xylan/glucose | <0.01 | <0.01 |
P. cellulosa was grown to mid-log phase, and xylanase activity in the culture medium and that was cell-associated was determined. Values are given as means ± the SD where indicated.
The assay can detect 10−2 U of xylanase activity/ml.
FIG. 3.
Western analysis of P. cellulosa xylanases. P. cellulosa cultured on wheat arabinoxylan for 20 h was fractionated into culture media (lanes 1), cell extract comprising periplasm and cytoplasm (lanes 2), and the membrane fraction (lanes 3). The fractions were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis with a 10% (wt/vol) polyacrylamide gel and subjected to Western analysis with antibodies raised against the catalytic module of Xyn11A (A) and full-length Xyn10C (B).
Expression of xylanase activity in different media.
P. cellulosa was grown on a variety of different media, and the expression of xylanase activity was monitored with time. The data, presented in Table 1, showed that xylan induced the expression of xylanase activity in both the culture supernatant and the cell fraction. Although cellulosic substrates also induced xylanase activity, the level of expression was lower than cells cultured on xylan. No xylanase activity was detected when the bacterium was cultured on LB, mannan, or pectic polysaccharides. When xylan media were supplemented with LB, the level of xylanase expression was greatly reduced, although the inclusion of mannan or pectin in xylan media did not repress the synthesis of these enzymes. No xylanase activity was detected in xylan medium supplemented with glucose until the stationary phase was reached. Assaying the culture supernatant for glucose showed that xylanase activity only appeared when all of the hexasaccharide had been metabolized (data not shown). These data show xylanase activity is not constitutively expressed but is induced by xylan and, to a lesser extent, by β-glucan; however, β-mannans or pectins do not influence the expression of these enzymes.
Regulation of xylanase gene transcripts.
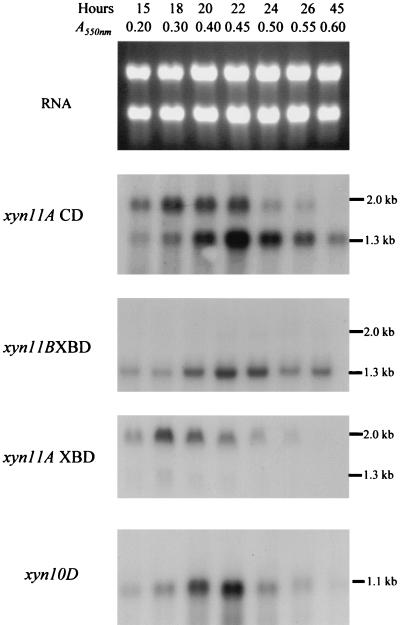
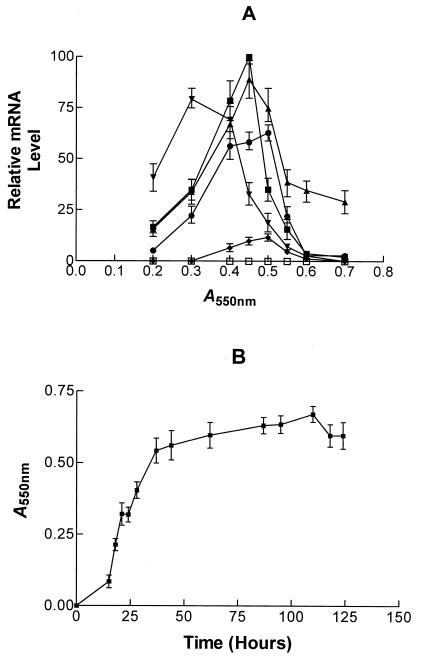
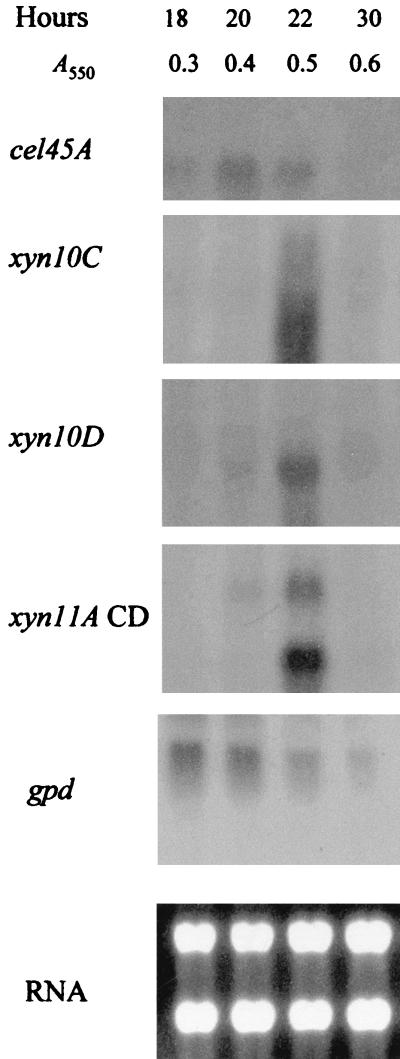
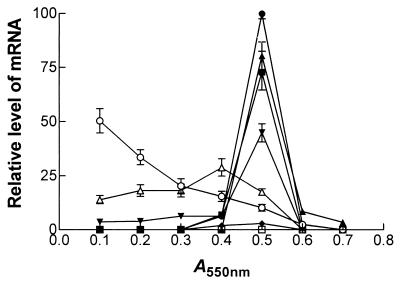
The expression of the individual xylanase genes was determined by Northern hybridization. The results obtained from P. cellulosa cultured on wheat arabinoxylan are presented in Fig. 4. These data are qualitatively similar to the transcript profile of the xylanase genes when P. cellulosa was grown on oat spelt xylan, rye arabinoxylan, and glucuronoxylan (data not shown). The sizes of mRNA and of the respective ORFs (in parentheses) for the various genes were as follows: xyn10A, 1.8 kb (1.83 kb); xyn10C, 1.8 (1.82 kb); xyn10D, 1.2 kb (1.13 kb); xyn11A, 2.0 kb (1.98 kb); and xyn11B, 1.3 kb (1.1 kb). Thus, the lengths of the transcripts were consistent with the sizes of the ORFs encoded by the respective genes. It should be noted that when the region of xyn11A encoding the catalytic domain was used in Northern hybridizations, transcripts of 2.0 and 1.1 kb were detected, a finding consistent with the size of xyn11A and xyn11B mRNA. Probes that consisted of the regions of xyn11A and xyn11B that encoded the xylan-binding CBMs of the respective enzymes hybridized to 2.0- and 1.1-kb mRNA species, respectively, showing that these probes were specific for transcripts derived from their target genes (Fig. 4). The data revealed that the highest levels of transcripts were from the xyn10D and xyn11B genes, although xyn11A and xyn10C were also efficiently transcribed (Fig. 5). Low levels of mRNA derived from xyn10A were observed, and no expression of xyn10B could be detected. In general, the xylanase genes were transcribed in mid-log phase when cultured on xylan, although only xyn11B expression persisted into stationary phase. The one very clear exception to this pattern was xyn11A mRNA, which appeared in early log phase, and its level had greatly decreased when the other xylanase transcripts were synthesized (Fig. 5). On β-glucan, xylanase mRNA molecules appeared in late log phase, shortly after the cellulase transcripts had reached their respective highest levels (Fig. 6 and 7), and the accumulations of these mRNAs were lower than on xylan. No xylanase transcripts were detected by Northern analysis on LB or on media containing glucose and xylan or when either mannan or pectin were the sole carbon sources (data not shown). To determine whether the lack of transcripts on mannan, LB, or pectin was due to the relatively low sensitivity of Northern hybridization, the RNA preparations were subjected to real-time PCR with primers that were specific for the six xylanase genes. The results were very similar to the Northern hybridization data. Transcripts encoding five of the xylanases were present when the bacterium was cultured on xylan-containing or β-glucan-containing media, but these mRNA species were not detected when the pseudomonad was grown on media containing mannan, pectin, or LB (data not shown). This methodology did not detect xyn10B on any of the media evaluated. To validate that the lack of xylanase transcripts on various media was not due to mRNA degradation, these RNA preparations were probed for the presence of transcripts derived from the housekeeping gene gpd. The data (examples of which are displayed in Fig. 6 and 7) showed that mRNA encoded by gpd could easily be detected, and in all RNA samples the two major rRNA species were clearly visible.
FIG. 4.
Northern hybridization of P. cellulosa RNA with xylanase-specific probes. An example of Northern hybridization of P. cellulosa RNA, derived from the bacterium cultured on wheat arabinoxylan minimal medium, is shown. The blots were probed with the region of xyn11A encoding the catalytic domain (xyn11A CD) and xylan-binding CBM (xyn11A XBD) of Xyn11A, the region of xyn11B that encodes the xylan binding CBM of Xyn11B (xyn11B XBD), and full-length xyn10D. The time and optical density of the cultures from which the RNA was extracted are shown, as is the RNA electrophoresed on a 1.2% agarose gel and stained with ethidium bromide (RNA).
FIG. 5.
Relative level of xylanase transcripts in P. cellulosa cultured on wheat arabinoxylan. (A) At various times RNA was extracted and subjected to Northern hybridization with probes that recognized mRNA derived from xyn10A (♦), xyn10B (□), xyn10C (•), xyn10D (▪), xyn11A (▾), and xyn11B (▴). The signal was quantified by using a phosphorimager. (B) Growth curve of the bacterium.
FIG. 6.
Northern hybridization of P. cellulosa RNA with xylanase- and cellulase-specific probes. An example of Northern hybridization of P. cellulosa RNA, derived from the bacterium cultured on β-glucan minimal medium, is shown. The blots were probed with the region of xyn11A that encodes the catalytic domain (xyn11A CD), the region of cel45A encoding the catalytic domain of Cel45A (cel45A), the gpd probe (gpd), and full-length xyn10C (xyn10C) and xyn10D (xyn10D). The time and optical density of the cultures from which the RNA was extracted are shown, as is the RNA electrophoresed on a 1.5% agarose gel and stained with ethidium bromide (RNA).
FIG. 7.
Relative levels of xylanase transcripts in P. cellulosa cultured on β-glucan. P. cellulosa was cultured on barley β-glucan, and at various times RNA was extracted and subjected to Northern hybridization with probes that recognized mRNA derived from cel45A (▵; encodes a cellulase formerly called endoglucanase B [14]), xyn10A (♦), xyn10B (□), xyn10C (•), xyn10D (▪), xyn11A (▾), xyn11B (▴), and gpd (○). The signal was quantified by using a phosphorimager.
To further probe the regulation of xylanase gene expression, the 450-bp untranslated 5′ regions of xyn11A and xyn11B were inserted immediately upstream of the β-glucuronidase gene (uidA) present in the promoter probe vector pRG960sd, which replicates in a range of gram-negative bacteria (31). The recombinant plasmids, pKE6 and pKE7, which contain the putative xyn11A and xyn11B promoters, respectively, were introduced into P. cellulosa by using either conjugation, with a helper plasmid, or electroporation, demonstrating for the first time that it is possible to transform the gram-negative bacterium. The copy number of the plasmid in the pseudomonad was ca. 1. The level of β-glucuronidase in wild-type P. cellulosa or in the bacterium containing pRG960sd was negligible; however, the pseudomonad containing either pKE6 or pKE7 expressed β-glucuronidase activity when cultured in the presence of xylan (Table 2). The addition of glucose to the xylan-containing medium prevented expression of β-glucuronidase in mid-log-phase cultures (Table 2); however, when the hexose sugar had been fully metabolized in stationary phase β-glucuronidase activity was apparent (data not shown). Negligible levels of the marker enzyme were produced when P. cellulosa containing either pKE6 or pKE7 was cultured on LB. These data show that the promoter regions of both xyn11A and xyn11B have been successfully cloned and thus demonstrate that the two genes are not transcribed from a single operon but rather contain discrete regulatory sequences.
TABLE 2.
β-Glucuronidase activity of recombinant P. cellulosaa
| Medium | Mean β-glucuronidase activity (nmol/mg protein/min) ± SD |
|||
|---|---|---|---|---|
| Wild type | pRG960sd | pKE6 | pKE7 | |
| Wheat arabinoxylan | <0.001b | 0.46 ± 0.04 | 70 ± 7.9 | 55 ± 5.6 |
| LB | <0.001 | 0.35 ± 0.13 | 0.01 ± 0.001 | 0.02 ± 0.01 |
| Wheat arabinoxylan- glucose | <0.001 | 0.33 ± 0.06 | <0.001 | 0.01 ± 0.002 |
Wild-type P. cellulosa or the bacterium containing pRG960sd, pKE6, and pKE7 was grown to mid-log phase, the cells were pelleted and disrupted by sonication, and β-glucuronidase activity was determined as described in Materials and Methods.
The assay can detect 1 pmol of product/mg of protein/min.
Transposon mutagenesis of P. cellulosa.
In a recent report we showed Tn5 can be conjugated into P. cellulosa and can be used to disrupt gene function. The integration of this mobile element into the pseudomonad genome was not random, with 90% of insertions occurring at a single locus (3). Thus, we evaluated the potential of an alternative transposon, a “mini” form of Tn10 (19), to randomly mutagenise the P. cellulosa genome. The mini-Tn10 was present on a mobilizable plasmid, and the transposase gene was outside the inverted repeats of the transposon element, ensuring stable integration of the transposon into the target genome (19). By using conjugation, >30,000 kanamycin-resistant transconjugants were obtained, and the site of Tn10 integration was examined in 10 of these bacterial strains by Southern hybridization. The transposon had integrated into a different site in each of the bacteria, and only one copy of the mobile element was detected in the genome of nine of the pseudomonads analyzed (data not shown). It would appear, therefore, that the Tn10 minitransposon is a much more useful genetic tool than Tn5 in disrupting gene function in P. cellulosa.
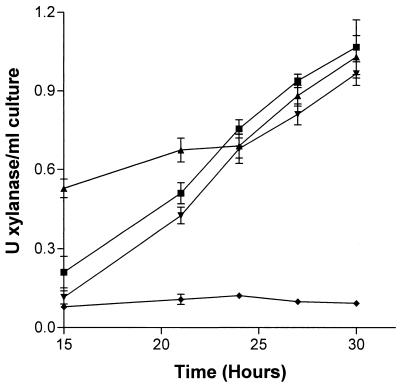
The P. cellulosa transposon library was screened for altered xylanase activity to investigate further the regulation of these enzymes and the functional importance of the various isoforms. The data revealed two transconjugants that displayed low levels of xylanase activity, as judged by clear haloes when grown on media containing Azo-wheat arabinoxylan. Quantitative assays (Fig. 8) showed that, although there was a substantial reduction in extracellular xylanase activity, the hydrolytic capacity of the cell-bound form of the enzyme was similar to that of the wild-type bacterium. Surprisingly, the growth rate of the transconjugants on media containing xylan as the sole carbon source was similar to that of the wild-type bacterium (data not shown). Southern hybridization showed that in these transconjugants the minitransposon had integrated into the xyn11A/xyn11B locus, mediating a deletion event encompassing both genes, which was confirmed by the absence of xyn11A and xyn11B transcripts when the transconjugants were cultured on xylan (data not shown). Thus, the reduction in the extracellular xylanase activity was due to the absence of both Xyn11A and Xyn11B.
FIG. 8.
Xylanase activity in wild-type and mutant P. cellulosa. Wild-type (▴ and ▪) P. cellulosa and a mutant of the bacterium lacking xyn11A and xyn11B (♦ and ▾) were cultured on minimal media containing xylan, and xylanase activity in the culture media (▴ and ♦) and cell-associated xylanase activity (▪ and ▾) were determined.
DISCUSSION
The main objective of this study was to examine the expression of the xylanase genes of P. cellulosa. The data showed that, whereas xyn10C, xyn10D, xyn11A, and xyn11B were efficiently expressed, only low levels of xyn10A transcripts were detected. The ratio of the different xylanase transcripts remained similar irrespective of the culture media, with none of the genes apparently expressed in the absence of xylan or cellulose. These data are in contrast to the xylanase system in T. reesei. In this fungus the two major xylanase genes are expressed in response to different nutrients, with xyn2 displaying significant constitutive expression that is induced by both xylan and cellulose, whereas xyn1 is only transcribed when media contain xylan (33). In the general model for induction of xylanase expression a sensor enzyme is constitutely expressed (e.g., T. reesei xyn2 [33]) which hydrolyzes xylan into oligosaccharides that enter the bacterium and activate the expression of all of the xylanase genes. It is possible that one or more of the repertoire of P. cellulosa xylanases are expressed at extremely low levels in the absence of xylan (i.e., <10 copies per cell, which is the minimum number of transcripts the real-time PCR method can detect) and thus function as the sensor enzyme. Further studies are clearly required to determine the mechanism by which xylan induces expression of the P. cellulosa xylanases.
Although the temporal expression of four of the xylanase genes was broadly similar, with maximum levels of transcripts appearing in mid-log phase, xyn11A, interestingly, was transcribed in early exponential phase. The expression pattern of Xyn11A may reflect the higher proportion of extracellular xylanase activity in the culture media, compared to the cell-associated forms of these enzymes, during the early component of the growth phase. Another interesting feature of the expression pattern of xyn11B is that transcripts of the gene persist into the late exponential and early stationary phases, at which point no other xylanase mRNAs were detected. These data may indicate that xyn11B mRNA exhibits an unusually high level of stability. The sequential synthesis of the two family 11 enzymes is consistent with their biochemical properties. Both xylanases are secreted into the extracellular culture medium, suggesting that they play a key role in plant cell wall degradation. Furthermore, they both contain xylan- and, in the case of Xyn11A, cellulose-binding CBMs, indicating that these enzymes target the plant cell wall as their primary substrate. Xyn11A also contains a family 4 xylan acetyl esterase and hydrolyzes acetylated xylan, whereas Xyn11B displays no activity against this decorated xylan. Since ca. 70% of xylose residues in plant cell wall xylans are decorated with acetate groups, the removal of these side chains is an essential prerequisite to xylan hydrolysis. Thus, we propose that the early expression of Xyn11A rapidly removes the acetate groups, making the backbone polysaccharide accessible to xylanase attack, and thus the second major extracellular xylanase, Xyn11B, is expressed at a later stage of the growth phase.
The importance of Xyn11A and Xyn11B in xylan degradation is illustrated by the observation that transposon mutants of P. cellulosa with greatly reduced xylanase activity lacked both of these enzymes but expressed the three family 10 xylanases at levels similar to that of the wild-type bacterium. This could reflect the nature of the screen which selected for bacteria with small haloes on Blue-Xylan. Since the secreted enzymes make the largest contribution to halo formation, deletions in xyn11A and xyn11B are likely to be isolated from the screen. A rather surprising feature of the mutants is that their growth rates were similar to that of the wild-type bacterium on media in which xylan was the sole carbon source. This suggests that the amount of extracellular xylanase activity is not a limiting factor in the growth of P. cellulosa on xylan. It would appear, therefore, that either the hydrolysis of xylooligosaccharides to xylose or the transport of these molecules is the rate-limiting step in the utilization of poorly substituted xylans. In contrast, deletion of any of the three xylanase genes of Streptomyces lividans reduced the growth rate of the bacterium, suggesting that in this prokaryote the rate of xylan degradation is the rate-limiting factor (1). It is likely, however, that the loss of the esterase activity of Xyn11A in the P. cellulosa mutant would limit its capacity to utilize “natural” highly acetylated xylans. Indeed, the importance of arabinose removal has already been shown to be a limiting factor in the utilization of arabinoxylans as a carbon source by the bacterium (3).
In contrast to the other xylanase genes, no expression of xyn10B was detected. It is interesting that xyn10B is 120 bp downstream of abf62A (formerly xynC), a gene encoding a xylan-specific arabinofuranosidase, which is also not expressed in P. cellulosa cultured on liquid media containing xylan, arabinoxylan, or glucuronoxylan (3). These data are somewhat puzzling since these genes encode functional enzymes when expressed in E. coli, indicating that they are likely to play a role in hemicellulose degradation in P. cellulosa. It is interesting that the enzymes encoded by these genes are the only xylan-degrading glycoside hydrolases to contain family 2a CBMs, which mediate tight and irreversible binding to crystalline cellulose (23). Thus, the primary function of these enzymes could be to attack xylans that are in intimate contact with cellulose, and thus it would be rational for these proteins to be expressed when P. cellulosa is presented with complex plant cell walls containing xylan-cellulose composites. This view is consistent with a previous study which showed that an Aspergillus xylanase was not expressed in monocultures of the fungus but was produced when the organism was inoculated into compost (10), a highly complex environment. Unfortunately P. cellulosa grew very poorly on plant cell walls or insoluble cellulose-xylan composites, precluding the analysis of xyn10B expression when the bacterium is presented with complex insoluble substrates.
The transposon mutant lacking xyn11A and xyn11B expressed similar levels of the other xylanases to wild-type P. cellulosa. This finding is in sharp contrast to those of a previous study on the Streptomyces lividans xylanase genes. The bacterium produced three major xylanases, and expression of two of the genes was intimately dependent on the expression of the other (1). The data presented here do not exclude such an interrelationship between the expression of the P. cellulosa xylanases, but such a phenomenon does not occur between the family 10 and 11 enzymes.
In P. cellulosa the two family 11 xylanases are extracellular, whereas Xyn10C is membrane bound, a finding consistent with its extensive linker sequence between the signal peptide and the catalytic module (25). It was not possible, however, to determine the location of Xyn10D; it is a highly unstable enzyme (data not shown) and was thus probably degraded during processing of the samples for Western analysis. The highly homologous enzyme Xyn10A (which exhibits 96% identity with P. cellulosa Xyn10D) from the related bacterium Cellvibrio mixtus subsp. cellulosa is periplasmic (13), and thus it is likely that Xyn10D is also cell associated. These data are in contrast with the generally held view that aerobic microorganisms secrete plant cell wall hydrolases into the extracellular medium, whereas anaerobic prokaryotes assemble the corresponding enzymes into large multienzyme complexes that are associated with the bacterial cell surface (29). The selective pressure that led to the different locations of the xylanases is an important issue. It is possible that the function of the membrane-bound xylanases may be related to the cell-associated location of the enzymes that remove the arabinose and glucurono decorations. The action of the arabinofuranosidases and α-glucuronosidases generates linear xylose polymers proximal to the bacterium. The cleavage of these polymers to xylose and small oligosaccharides by cell-associated xylanases would enable the products of hemicellulose degradation to be utilized primarily by P. cellulosa rather than by competing bacteria which inhabit these complex ecosystems. Support for this model is provided by the study of Shulami et al. (30) on the glucuronic acid gene cluster in Bacillus stearothermophilus, which showed that the bacterium synthesized an extracellular xylanase, similar to Xyn10C, that generates short decorated xylooligosaccharides that enter the bacterium and are further degraded to monosaccharides by a battery of intracellular hemicellulases.
Conclusions.
We have shown here that the synthesis of the different xylanases in P. cellulosa is induced by the same polysaccharides, although there are differences in the temporal expression of the family 11 enzymes. The differences in the temporal expression of Xyn11A and Xyn11B are consistent with their mode of action, with the former enzyme capable of removing acetyl side chains, as well as attacking the xylan backbone, whereas the latter enzyme only cleaves β1,4-xylosyl linkages. The spatial localization of the different xylanases indicates that the various isoforms of these enzymes play quite different roles in the xylan degradation process. The extracellular enzymes mediate initial xylan breakdown, while the cell-associated hemicellulases hydrolyze linear xylooligosaccharides in which the side chains have been removed by membrane-bound glycoside hydrolases. Transposon mutagenesis studies support the view that Xyn11A and Xyn11B play an important role in xylan degradation, although the hydrolysis of the polysaccharide does not appear to be the rate-limiting step in the utilization of xylan as a growth substrate. Finally, this study has generated genetics tools, a random transposon mutagenesis system, and a promoter probe vector in which the reporter gene is regulated by xylan promoters that will be invaluable in the further analysis of the mechanisms regulating the synthesis of the xylan degradative enzymes of P. cellulosa.
REFERENCES
- 1.Arhin, F. F., F. Shareck, D. Kluepfel, and R. Morosoli. 1994. Effects of disruption of xylanase-encoding genes on the xylanolytic system of Streptomyces lividans. J. Bacteriol. 176:4924-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aro, N., A. Saloheimo, M. Ilmen, and M. Penttila. 2001. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 276:24309-24314. [DOI] [PubMed] [Google Scholar]
- 3.Beylot, M. H., K. Emami, V. A. McKie, H. J. Gilbert, and G. Pell. 2001. Pseudomonas cellulosa expresses a single membrane-bound glycoside hydrolase family 51 arabinofuranosidase. Biochem. J. 358:599-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beylot, M. H., V. A. McKie, A. G. Voragen, C. H. Doeswijk-Voragen, and H. J. Gilbert. 2001. The Pseudomonas cellulosa glycoside hydrolase family 51 arabinofuranosidase exhibits wide substrate specificity. Biochem. J. 358:607-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biely, P. 1993. Biological aspects of the production of microbial hemicellulases, p. 20-31. In M. P. Coughlan and G. P. Hazlewood (ed.), Hemicellulose and hemicellulases. Portland Press, London, England.
- 6.Biely, P. 1985. Microbial xylanolytic systems. Trends Biotechnol. 3:286-290. [Google Scholar]
- 7.Biely, P., D. Kluepfel, R. Morosoli, and F. Shareck. 1993. Mode of action of three endo-β-1,4-xylanases of Streptomyces lividans. Biochim. Biophys. Acta 1162:246-254. [DOI] [PubMed] [Google Scholar]
- 8.Biely, P., M. Vrsanska, M. Tenkanen, and D. Kluepfel. 1997. Endo-β-1,4-xylanase families: differences in catalytic properties. J. Biotechnol. 57:151-166. [DOI] [PubMed] [Google Scholar]
- 9.de Graaff, L. H., H. C. van den Broeck, A. J. van Ooijen, and J. Visser. 1994. Regulation of the xylanase-encoding xlnA gene of Aspergillus tubigensis. Mol. Microbiol. 12:479-490. [DOI] [PubMed] [Google Scholar]
- 10.De Groot, P. W., D. E. Basten, A. Sonnenberg, L. J. van Griensven, J. Visser, and P. J. Schaap. 1998. An endo-1,4-β-xylanase-encoding gene from Agaricus bisporus is regulated by compost-specific factors. J. Mol. Biol. 277:273-284. [DOI] [PubMed] [Google Scholar]
- 11.Ferreira, L. M. A., T. M. Wood, G. Williamson, C. Faulds, G. P. Hazlewood, G. W. Black, and H. J. Gilbert. 1993. A modular esterase from Pseudomonas fluorescens subsp. cellulosa contains a non-catalytic cellulose-binding domain. Biochem. J. 294:349-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontes, C. M., H. J. Gilbert, G. P. Hazlewood, J. H. Clarke, J. A. Prates, V. A. McKie, T. Nagy, T. H. Fernandes, and L. M. Ferreira. 2000. A novel Cellvibrio mixtus family 10 xylanase that is both intracellular and expressed under non-inducing conditions. Microbiology 146:1959-1967. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert, H. J., J. Hall, G. P. Hazlewood, and L. M. Ferreira. 1990. The N-terminal region of an endoglucanase from Pseudomonas fluorescens subspecies cellulosa constitutes a cellulose-binding domain that is distinct from the catalytic centre. Mol. Microbiol. 4:759-767. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert, H. J., D. A. Sullivan, G. Jenkins, L. E. Kellett, N. P. Minton, and J. Hall. 1988. Molecular cloning of multiple xylanase genes from Pseudomonas fluorescens subsp. cellulosa. J. Gen. Microbiol. 134:3239-3247. [DOI] [PubMed] [Google Scholar]
- 16.Hall, J., G. P. Hazlewood, N. S. Huskisson, A. J. Durrant, and H. J. Gilbert. 1989. Conserved serine-rich sequences in xylanase and cellulase from Pseudomonas fluorescens subspecies cellulosa: internal signal sequence and unusual protein processing. Mol. Microbiol. 3:1211-1219. [DOI] [PubMed] [Google Scholar]
- 17.Hazlewood, G. P., J. I. Laurie, L. M. Ferreira, and H. J. Gilbert. 1992. Pseudomonas fluorescens subsp. cellulosa: an alternative model for bacterial cellulase. J. Appl. Bacteriol. 72:244-251. [DOI] [PubMed] [Google Scholar]
- 18.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ilmen, M., A. Saloheimo, M. L. Onnela, and M. E. Penttila. 1997. Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 63:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joshi, M. D., G. Sidhu, J. E. Nielsen, G. D. Brayer, S. G. Withers, and L. P. McIntosh. 2001. Dissecting the electrostatic interactions and pH-dependent activity of a family 11 glycosidase. Biochemistry 40:10115-10139. [PubMed] [Google Scholar]
- 22.Kellett, L. E., D. M. Poole, L. M. Ferreira, A. J. Durrant, G. P. Hazlewood, and H. J. Gilbert. 1990. Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem. J. 272:369-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLean, B. W., M. R. Bray, A. B. Boraston, N. R. Gilkes, C. A. Haynes, and D. G. Kilburn. 2000. Analysis of binding of the family 2a carbohydrate-binding module from Cellulomonas fimi xylanase 10A to cellulose: specificity and identification of functionally important amino acid residues. Protein Eng. 13:801-809. [DOI] [PubMed] [Google Scholar]
- 24.Miller, G. L. 1959. The use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426-428. [Google Scholar]
- 25.Millward-Sadler, S. J., K. Davidson, G. P. Hazlewood, G. W. Black, H. J. Gilbert, and J. H. Clarke. 1995. Novel cellulose-binding domains, NodB homologues and conserved modular architecture in xylanases from the aerobic soil bacteria Pseudomonas fluorescens subsp. cellulosa and Cellvibrio mixtus. Biochem. J. 312:39-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sachslehner, A., B. Nidetzky, K. D. Kulbe, and D. Haltrich. 1998. Induction of mannanase, xylanase, and endoglucanase in Sclerotium rolsii. Appl. Environ. Microbiol. 64:594-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Sedmak, J. J., and S. E. Grossberg. 1977. A rapid, sensitive, and versatile assay for protein using Coomassie brilliant blue G250. Anal. Biochem. 79:544-552. [DOI] [PubMed] [Google Scholar]
- 29.Shoham, Y., R. Lamed, and E. A. Bayer. 1999. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 7:275-281. [DOI] [PubMed] [Google Scholar]
- 30.Shulami, S., O. Gat, A. L. Sonenshein, and Y. Shoham. 1999. The glucuronic acid utilization gene cluster from Bacillus stearothermophilus T-6. J. Bacteriol. 181:3695-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van den Eede, G., R. Deblaere, K. Goethals, M. van Montagu, and M. Holsters. 1992. Broad host range and promoter selection vectors for bacteria that interact with plants. Mol. Plant-Microbe Interact. 5:228-234. [DOI] [PubMed] [Google Scholar]
- 32.van Peij, N. N., J. Visser, and L. H. de Graaff. 1998. Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol. Microbiol. 27:131-142. [DOI] [PubMed] [Google Scholar]
- 33.Zeilinger, S., R. L. Mach, M. Schindler, P. Herzog, and C. P. Kubicek. 1996. Different inducibility of expression of the two xylanase genes xyn1 and xyn2 in Trichoderma reesei. J. Biol. Chem. 271:25624-25629. [DOI] [PubMed] [Google Scholar]