Abstract
A genome-independent microarray and new statistical techniques were used to genotype Bacillus strains and quantitatively compare DNA fingerprints with the known taxonomy of the genus. A synthetic DNA standard was used to understand process level variability and lead to recommended standard operating procedures for microbial forensics and clinical diagnostics.
Discriminating between closely related strains of microorganisms is obviously required for identifying biological agents or pathogens. However, providing actionable, quantifiable, and diagnostic information to physicians or policy makers requires a level of certainty and statistical confidence that go beyond descriptive methodologies used for current microbial epidemiological studies (1, 6, 13). The need for high-resolution genotyping is, in part, dependent upon the genome diversity of the species in question. Bacillus species and strains, for example, can be very difficult to identify or resolve taxonomically with conventional techniques (14, 17), and full-genome sequencing was ultimately employed (18) to identify the strain of Bacillus anthracis recovered from the 2001 B. anthracis mail release (9). However, full-genome sequencing is neither practical nor cost-effective for routine public health and epidemiology applications.
Because diagnostic nucleic acid signatures are not and may not be known a priori for all organisms of interest, gel-based DNA fingerprinting techniques continue to dominate microbial epidemiology studies (see, e.g., references 12, 16, and 20). However, it is well recognized that current genotyping methods frequently do not discriminate between isolates. Gel-to-gel positional variations in internal standards and the test sample is particularly troubling, for example, because it necessarily leads to increased bin sizes and decreased resolving power in cross-gel comparisons (see, e.g., reference 21). The positional variations in gels, however, also begs the following questions: what are the objective criteria for including or excluding data from a gel-based DNA fingerprint and how does one generate error bars and statistical confidence to test the hypothesis of profile equivalence?
DNA microarrays provide physically fixed data features, are readily amenable to replication, and provide an alternative technology base for developing quantitative DNA fingerprinting methods. We are particularly interested in developing a simple, low-cost, diagnostic genotyping product and method for microbial epidemiology, while retaining sufficient resolving power to discriminate between strains that may be indistinguishable by conventional techniques. Inherent in this objective is a need to develop standard operating protocols and normalization controls that will (ultimately) allow for quantifiable and objective comparisons across days, users, or laboratories.
Bacterial isolates used for this study are listed in Table 1. Bacillus near-neighbor isolates were grown in nutrient broth (Difco, Sparks, MD) at 29°C and 450 rpm for ∼48 h. American Type Culture Collection (ATCC) isolates (e.g., outliers) were purchased as genomic DNA preparations from the vendor. B. anthracis isolates were cultivated, and genomic DNA was isolated under appropriate biosafety level 3 controls as described in reference 11. Nucleic acid integrity from all genomic DNA preparations was analyzed by gel electrophoresis on 0.8% single-comb E-gels (Invitrogen, Carlsbad, CA). DNA concentrations were determined in solution by UV absorbance (UV/visual-light spectrophotometer Lambda Bio 10; Perkin Elmer, Boston, MA) and in-gel by ethidium bromide staining of chromosomal DNA. Only intact genomic DNA of >10 kbp in length was utilized for subsequent PCR and microarray analysis.
TABLE 1.
Bacterial isolates used in this study
| Bacteria | Sourcea |
|---|---|
| Bacillus cereus | ATCC 4342 |
| Bacillus cereus strains D17 and FRI-13 | LANL |
| Bacillus cereus strains 3A and FRI-41 | LANL |
| Bacillus cereus strains S2-8 and FRI-42 | LANL |
| Bacillus cereus strains F1-15 and FRI-43 | LANL |
| Bacillus thuringiensis strain 97-27 | LANL |
| Bacillus thuringiensis strain HD-571 | LANL |
| Bacillus thuringiensis strain Al Hakum | LANL |
| Bacillus anthracis strains A0392 and K2165 | BYU |
| Bacillus anthracis strains A0362 and K8091 | BYU |
| Salmonella enterica serovar Typhimurium strain LT2 | ATCC 700720D |
| Campylobacter jejuni strain NCTC 11168 | ATCC 700819D |
| Yersinia enterocolitica | ATCC 9610D |
LANL, Los Alamos National Laboratory, Los Alamos, NM; BYU, Brigham Young University, Provo, UT. ATCC numbers with a D suffix were purchased as genomic DNA preparations.
A set of repetitive extragenic palindromic (REP) consensus PCR primers (22) were used to sample the bacterial genomes and generate amplified fragments for hybridization and analysis on the oligonucleotide microarray; it should be recognized that nondegenerate and/or alternative repetitive DNA primer sequences can likewise be used to generate microarray fingerprints. PCR amplification and microarray hybridization conditions were essentially as described in reference 25 but utilized 100 ng bacterial DNA, 1× PCR buffer, 2.5 mM Mg2+, 200 μM each deoxynucleoside triphosphate, 2.5 U Taq polymerase, and 0.6 μM each Cy3-labeled REP primer per PCR. PCR amplification was confirmed by analyzing 5-μl aliquots of the amplification reaction mixture on a 2% agarose single-comb E-gel. The remaining (labeled) amplification products were hybridized directly to microarrays without further manipulation.
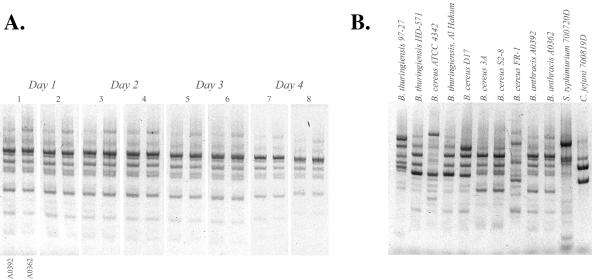
Figure 1A shows that the REP-PCR is quite reproducible by conventional microbiological standards, with the same (qualitative) level of PCR reproducibility observed for all other isolates in the study (not shown). Thus, variations in microarray fingerprints (below) are not due to PCR bias or error during the sample-processing steps. From this simple gel analysis, however, it is readily apparent that the two B. anthracis isolates are indistinguishable based on a conventional REP-PCR test. In the same way, the REP-PCR gels could not differentiate between B. thuringiensis strains HD-571 and Al Hakum and between B. cereus strains 3A and S2-8 (Fig. 1B). The positional variation in gel bands, background smears, and related gel artifacts underscore the qualitative nature of gel-based genotype comparisons.
FIG. 1.
Reproducibility of B. anthracis REP-PCR. (A) Agarose gel electrophoresis of REP-PCR products amplified with genomic DNA from two Bacillus anthracis isolates, shown in sequential order (A0392 followed by A0362 for all 4 days). One hundred nanograms of each genomic DNA template was amplified in replicate tubes each day for four separate days, as described in the text, and 10% of the PCR amplification products were analyzed on a 2% agarose gel containing ethidium bromide. Gel images were stretched and adjusted in Adobe Photoshop 7.0 to account for different electrophoresis results and exposure times in the creation of this image. While the fingerprints are qualitatively reproducible, the two B. anthracis isolates cannot be differentiated (but see results in Fig. 3 and 4). Similar patterns of PCR reproducibility were obtained for all other isolates used in this study (not shown). (B) REP-PCR fingerprints for 12 of the 13 isolates used in this study.
Microarray capture probes (nonamers) were generated by random computer selection based on the sequence of the Escherichia coli K-12 genome (GenBank accession number U00096), with 190 probes derived from a previous study (25) and 200 probes selected de novo for this study in order to extend the range of probe G+C % content and the frequency of occurrence relative to that of the K-12 genome. Nonamer capture probes were synthesized in-house, purified by isopropanol precipitation, reconstituted in Milli-Q water, and quantified by UV absorption. A control set of Cy3-labeled 9-mer oligonucleotides (Table 2) (Mix-10 control) with perfect complementarity to 10 of the nonamer capture probes were also synthesized (Sigma-Genosys, The Woodlands, TX) to measure the effects of printing, hybridization, replication, and normalization strategies on the resulting DNA fingerprints.
TABLE 2.
Mix-10 control targets
| Matched probe | Sequence (5′-3′) |
|---|---|
| 56 | Cy3-TTACAGCGC |
| 66 | Cy3-AGTCTGGAT |
| 119 | Cy3-GTTGGGTTG |
| 140 | Cy3-ACCCGCATT |
| 181 | Cy3-TGGAAACGA |
| 205 | Cy3-TTTTGTGAA |
| 225 | Cy3-TCGACGGAT |
| 284 | Cy3-GCATCTGCA |
| 322 | Cy3-TCAGCATCT |
| 366 | Cy3-TATGCTCCG |
Microarrays were manufactured on ready-to-go epoxy silane slides (Erie Scientific Company, Portsmouth, N.H.) as described previously (25), using nonamer probes diluted to 0.1 to 0.2 mM in 150 mM sodium phosphate buffer, pH 8.5, containing 0.01% sodium dodecyl sulfate. In addition to the nonamer capture probes, the microarray contained a Cy3-labeled quality control probe (5′-Cy3-TTGTGGTGGTGGTGTGGTGG-3′; Sigma-Genosys, The Woodlands, TX) that served as positional reference and spotting quality points, and a negative control buffer blank to test for nonspecific interactions and residual fluorescence on the microarray surface. Slides were printed in batches of 20 each with four arrays per slide, and each batch was tested for spot quality and reproducibility by staining several slides (from the beginning, middle, and end of the print run) with SYBR green II (Molecular Probes, Eugene, OR), a fluorophore with specific affinity for single-stranded DNA (2). If SYBR green slides showed missing spots, the entire print lot (20 slides) was discarded and prepared anew. SYBR green quality control slides were not used for hybridization experiments.
The target DNA for the 12 hybridizations for each bacterial isolate was derived from six independent amplification reactions, split evenly between two arrays. REP-PCR fragments from three different organisms were hybridized to three arrays on the same slide according to a balanced incomplete block design, where each slide is treated as a block (24) so that paired strains were directly compared on the same slide exactly twice. The fourth array on each slide was hybridized with the Mix-10 standard targets, all diluted in an equimolar ratio to a final concentration of 1.53 nM (each) in the hybridization solution. Each bacterial species or strain was hybridized to 12 replicate arrays, and paired strains were directly compared on the same slide exactly twice, with no triple repeated. Hybridization and washing proceeded as described elsewhere (25).
Microarray images were acquired on a custom-built, temperature-controlled fluorescence microscope operating at room temperature. Briefly, slides were illuminated with a 100-W mercury lamp through a D525/50 bandpass filter and Cy3 emissions were collected through a 590DF35 filter. Microarrays were illuminated for 20 s, and images were captured through a custom lens (LINOS Photonics, Inc., Milford, MA) as img files with a 12-bit SenSys charge-coupled-device camera (Photometrics, Tucson, AZ) at a resolution of 1,536 by 1,024 pixels. Image analysis was performed using the freely available Automated Microarray Image Analysis Toolbox for Matlab (23; http://www.pnl.gov/statistics/amia). Spot identification routines are implemented using a seeded-region-growing method adapted from Hojjatoleslami and Kittler (10). To ensure accurate spot finding against a potentially distorted printing grid, an analyst manually identified several spots and reran the automated algorithm until all spots in the alignment were accurately identified. For each spot, the average pixel intensity value and the average (local) background intensity value were exported to a spreadsheet for statistical analysis as described in detail elsewhere (24). In brief, we calculated the log(mean spot pixel intensity) minus the log(mean background pixel intensity) for each probe over all replicate arrays (n = 12). The only explicit normalization performed in the analysis is to center low-end probe histogram modes for each array. Summary statistics for each probe were computed using a mixed-effects linear analysis of variance (ANOVA) model that parallels the incomplete block design. F-statistics from the ANOVA calculations were used to identify discriminating probes significant at α = 0.01. For the discriminating probes, the cell means parameterization of the linear model was used to obtain estimates of relative hybridization, which were grouped using a finite mixture procedure for grouping treatment means. The number or proportion of microarray probes and their relative hybridization intensities therefore provide a quantitative measure of difference between the isolates that can be tested for significant differences with any number of multivariate statistical procedures.
It is clear from the literature that microarray results can be highly varied (see, e.g., reference 15). The Mix-10 control targets were therefore used to understand underlying microarray variability, independent of the genetic variation between organisms or method level variability associated with nucleic acid extraction and/or PCR amplification prior to microarray hybridization. For the microarray design reported here, each array consisted of 400 spots (391 probes with nine controls) printed with a 4-pin print head (i.e., 100 spots per pin, wherein 1 pin defines a subarray). To determine if calculated probe intensities varied between pins during microarray manufacture, the average hybridization intensities in each subarray (over all 100 probe spots) were normalized to mean 0 and standard deviation 1 and compared for each array using ANOVA. Among the 52 Mix-10 control arrays, at least 1 subarray was statistically distinct from the other subarrays in approximately 31% of the slides (at α = 0.05). Similarly, we utilized a linear ANOVA to analyze the hybridization intensity of each of the perfectly matched Mix-10 capture probes across days (or print lots). Table 3 shows that for all but one of the Mix-10 probes and control targets (probe/target 119), there is a significant print day effect at α = 0.05. Hence, there are clearly significant pin and day effects during the manufacture of printed arrays, effects that must be managed in a standard operating protocol through biological replication across print lots (and pins).
TABLE 3.
Average log signal/background values and standard deviations for perfectly matched Mix-10 control probes and targets based on print lot (or day)
| Probe | Mean log signal/background ± SD
|
F statistic | P valuea | |||
|---|---|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | Day 4 | |||
| 56 | 3.9 ± 0.9 | 4.6 ± 0.3 | 2.7 ± 0.5 | 4.1 ± 0.2 | 30.79 | 2.0 × 10−11 |
| 66 | 4.1 ± 0.3 | 3.8 ± 0.3 | 3.8 ± 0.5 | 3.4 ± 0.5 | 5.11 | 0.0037 |
| 119 | 3.6 ± 0.5 | 3.4 ± 0.4 | 3.8 ± 0.4 | 3.6 ± 0.3 | 2.36 | 0.0824 |
| 140 | 3.4 ± 0.4 | 2.7 ± 0.4 | 1.6 ± 0.9 | 2.3 ± 0.6 | 19.42 | 1.8 × 10−8 |
| 181 | 4.0 ± 0.4 | 3.7 ± 0.4 | 3.9 ± 0.4 | 3.4 ± 0.2 | 8.01 | 0.00019 |
| 205 | 4.3 ± 0.9 | 5.4 ± 0.5 | 5.7 ± 0.4 | 4.7 ± 0.5 | 11.63 | 1.3 × 10−6 |
| 225 | 2.2 ± 0.9 | 2.2 ± 0.9 | 2.4 ± 0.7 | 3.0 ± 0.6 | 3.14 | 0.0335 |
| 284 | 4.7 ± 0.7 | 4.2 ± 0.7 | 4.2 ± 0.4 | 3.2 ± 0.3 | 16.15 | 1.8 × 10−7 |
| 322 | 2.4 ± 0.6 | 1.8 ± 0.3 | 2.1 ± 0.5 | 3.7 ± 0.6 | 37.90 | 6.4 × 10−13 |
| 366 | 1.7 ± 0.9 | 1.3 ± 0.4 | 1.4 ± 0.3 | 3.5 ± 0.2 | 45.88 | 2.2 × 10−14 |
F statistics and P values were determined by one-way ANOVA, as described in the text.
All of the Mix-10 perfect matches generated hybridization signals significantly above background, but the hybridization intensity for the Mix-10 probes was not uniform (Table 3), even though the synthetic standards were all applied at equimolar concentration to each of the control arrays (e.g., see the differing response of probe 205 from that of 366). Surprisingly (and unexpectedly), we also observed 56 consistent and reproducible false-positive signals for mismatched capture probes and the Mix-10 standard (66 total positive probes, including the perfect matches). However, regression analysis showed that there is no correlation between the number of mismatched (or matched) nucleotides and signal intensity for the Mix-10 standard (not shown). In fact, several mismatched probes (false positives) generated greater average signal intensities than perfectly matched probes. Hence, we cannot necessarily equate positive hybridization on the nonamer array with sequence identity in the nucleic acid targets, using the conventional sense or definition of hybridization specificity and probe design (4, 7, 8, 19). A similar conclusion was recently reached by Belosludtsev et al. (3) for a high-density array of 12- and 13-mer oligonucleotide probes. The extent to which false-positive hybridization contributes to method level variability during the analysis of REP-PCR amplification products cannot be known a priori. What becomes more important, then, is the reproducibility of the microarray pattern through time and space and our ability to utilize a positive control (such as the Mix-10 standard) as part of a cross-slide normalization strategy in the face of manufacturing variability and false-positive hybridization.
Scatter plots were generated to compare actual hybridization intensity for each positive probe on each array with the median hybridization intensity for each probe over all replicate arrays (n = 12 for bacterial isolates; n = 52 for the Mix-10 standard). Both Fig. 2 and Table 4 show that the Mix-10 standard and REP-PCR hybridizations for bacterial isolates are equally varied and reproducible, even in the face of unpredictable cross-hybridization and false positives. The average R2 for the Mix-10 data are somewhat lower than those for the bacterial isolates, most likely due to the few number of positive capture probes (66 reproducibly positive signals) relative to the number from a typical bacterial REP-PCR hybridization pattern (>200 reproducible signals). As such, the Mix-10 itself may not be an ideal or perfectly representative standard as presently configured, but it nonetheless accurately reflects hybridization behavior and microarray probe responses to bacterial REP-PCR products. Hence, the fingerprinting method described here is making conservative, unbiased conclusions relative to underlying biological differences between isolates, rather than displaying differences due to measurement noise or error. From these data (Fig. 2; Table 4), we are cautiously optimistic about using a Mix-10 (or similar) synthetic standard as part of a quality control and normalization procedure and recommending 12 replicate hybridizations as an upper boundary on any standard experimental procedure. The extent to which the total number of replicates can be reduced while still providing statistically significant and quantifiable DNA fingerprints will be determined in future studies with larger collections of isolates.
FIG. 2.
Fingerprinting array method level reproducibility. Scatter plots for hybridization (A) and method level (B-D) reproducibility. Panel A shows results for a randomly selected Mix-10 array relative to the median intensity for 66 probes (52 replicate arrays), reflecting the behavior and reproducibility of the microarray itself during hybridization. The R2 value is for the single array and regression line shown in the panel. Average R2 values are in Table 4. Panels B-D show results for B. thuringiensis 97-27, B. anthracis A0392, and Campylobacter jejuni 11168 (12 replicate arrays), respectively, which are inclusive of PCR amplification and labeling steps. Results were similar for all other isolates, as shown in Table 4.
TABLE 4.
Average R2 values for bacterial REP-PCR products and the Mix-10 standard
| Isolate(s)a | Avg R2 | SD |
|---|---|---|
| Bacillus cereus ATCC 4342 | 0.91 | 0.03 |
| Bacillus cereus strains D17 and FRI-13 | 0.90 | 0.04 |
| Bacillus cereus strains 3A and FRI-41 | 0.91 | 0.04 |
| Bacillus cereus strains S2-8 and FRI-42 | 0.90 | 0.02 |
| Bacillus cereus strains F1-15 and FRI-43 | 0.90 | 0.04 |
| Bacillus thuringiensis strain 97-27 | 0.89 | 0.05 |
| Bacillus thuringiensis strain HD-571 | 0.89 | 0.04 |
| Bacillus thuringiensis strain Al Hakum | 0.90 | 0.05 |
| Bacillus anthracis strain A0392 | 0.90 | 0.04 |
| Bacillus anthracis strain A0362 | 0.91 | 0.04 |
| Salmonella enterica serovar Typhimurium LT2 | 0.92 | 0.03 |
| Campylobacter jejuni NCTC 11168 | 0.88 | 0.05 |
| Yersinia enterocolitica | 0.91 | 0.05 |
| Mix-10 standard targetb | 0.85 | 0.07 |
Bacterial isolates included 391 probe spots and 12 replicate arrays.
The Mix-10 standard included 66 probe spots and 52 replicate arrays.
Probe intensity values were compared across the 13 isolates using a linear mixed-effects (ANOVA) model that parallels the incomplete block design (a slight variation of the model used in reference 24). At α = 0.01, 212 of the 391 probes (54%) are differentially detected and/or hybridized across the 13 isolates. That is, for the 212 probes, the intensity differences between isolates are significantly greater than the intensity differences between replicate hybridizations for the same isolate. Discriminatory fingerprints were constructed from the ANOVA output using a specially designed finite mixture procedure described in reference 24, which provides a discrete fingerprint (analogous to gel-based fingerprints) in which isolates are assigned to a small number of probe level groups according to relative intensity. Figure 3 shows the discriminatory fingerprints for the 13 isolates, where the probes have been reordered (clustered) according to their similarity across isolates, and bands are shaded according to relative hybridization intensity at each discriminating probe.
FIG. 3.
Simultaneous clustering of probes and isolates. Only the 212 discriminatory probes are plotted here. Probes are clustered and reordered according to their correlation across isolates. Jaccard's distance was used to create the dendrogram.
The number of fingerprint differences between each isolate pair is displayed in Fig. 4. By this measure, Yersinia enterocolitica is separated from the two B. anthracis strains by 110 and 108 differences, respectively, while the two B. anthracis strains are differentiated by 10 discriminating probes (with sequences [5′ to 3′] of CAGCTAATG, TGCAGATGC, CGTCAACTT, CAACACTCG, CCAGCGATA, TGCAGAAGC, TGCCATGAG, TCACGGTAG, TTTACTGAC, and GTTGAGTTG). A more rigorous statistical test that controls the false-discovery rate for the large number of comparisons made also found differences between all pairs of isolates based on their normalized probe intensity values (24). What is evident from Fig. 3 and 4, then, is that all isolates are statistically and quantifiably distinguishable (at α = 0.01) one from the other, even amid the microarray manufacturing and hybridization variability described above and despite the fact that three isolate pairs could not be differentiated based on a typical gel electrophoresis pattern (Fig. 1B).
FIG. 4.
Numbers of fingerprint differences between pairs of isolates. There are 212 discriminatory probes (out of 391) for the set of 13 isolates tested in this study and analyzed with the balanced incomplete block statistical design. Cells in the image plot are color coded according to the number of discriminating probes, from fewest (black) to most (white). The actual number of discriminating probes between isolate pairs is shown within the color-coded cells. B thur, Bacillus thuringiensis.
While it was not necessary for us to normalize the REP-PCR results across slides with a balanced incomplete block study design, it will be practically impossible to organize microarray hybridizations and analyses as complete data sets (e.g., reference 5 or the balanced experimental design described herein) for each and every isolate that is tested across days, users, and laboratories. Routine clinical applications—as opposed to the designed experiment described here—will likely require a synthetic standard (such as Mix-10), but further study is needed to evaluate synthetic standards for this purpose. For the time being, we envision using the reference target on each and every test array and generating M-versus-A plots similar to the cyclic loess or quantile normalization procedures described by Bolstad et al. (5). In this case, M represents the difference in log intensity values and A represents the average of the log intensity values for the reference target applied to two separate arrays. Therefore, an M-versus-A plot for normalized data should show a point cloud scattered about the M = 0 axis. Such a normalization strategy makes sense in routine implementation, because the data described here (Mix-10 standard and test isolates) suggest that linear normalization is appropriate (Fig. 2; Table 4).
It can certainly be argued that multiple PCR amplifications and 12 replicate arrays per isolate are impractical for routine forensic or diagnostic purposes, but any (present) judgment should be balanced against the quality and statistical rigor of the resulting information (microbial fingerprint). That is, a natural inclination or assumption underlying the development of universal genotyping microarrays is that more probes are “better” and that large data sets are a suitable substitute or proxy for method level reproducibility (e.g., see arguments and rationale in reference 3). For a complete data set or experimental design, these assumptions may hold and generate a robust microarray pattern for qualitative comparison (depending upon how one analytically defines “robust”). On the other hand, we argue that data are not equivalent to information. Hence, for the diagnostic problems of library construction, quantitative comparisons against libraries and reference databases, and dealing with the practicality of routine analyses, we argue that method level reproducibility and information quality are more important than data volume. In this context, a question worth asking is whether high-density arrays are even necessary or helpful for the end user. What we have shown here is that a very simple (391 probe) nonamer array, combined with appropriate (method level) replication and quantitative statistical techniques, can easily and reproducibly differentiate between strains of Bacillus anthracis and near neighbors, a group of organisms that are notoriously monomorphic and difficult to differentiate by many classical molecular taxonomy techniques.
Acknowledgments
This work was supported by grants from the U.S. Department of Homeland Security and National Institutes of Health (NIH). Argonne National Laboratory is operated for the U.S. DOE by the University of Chicago under contract W-31-109-ENG-38. Pacific Northwest National Laboratory is operated for the U.S. DOE by Battelle Memorial Institute under contract DE-AC06-76RLO 1830.
We thank Anne Gemmell for assistance in the cultivation and extraction of nucleic acids from bacterial isolates.
REFERENCES
- 1.Alper, J. 2003. Standardized systems needed to detect and track microbial biocrimes. ASM News 69:379-383. [Google Scholar]
- 2.Battaglia, C., G. Salani, C. Consolandi, I. R. Bernardi, and G. De Bellis. 2000. Analysis of DNA microarrays by non-destructive fluorescent staining using SYBR green II. BioTechniques 29:78-81. [DOI] [PubMed] [Google Scholar]
- 3.Belosludtsev, Y. Y., D. Bowerman, R. Weil, N. Marthandan, R. Balog, K. Luebke, J. Lawson, S. A. Johnston, C. R. Lyons, K. O'Brien, H. R. Garner, and T. F. Powdrill. 2004. Organism identification using a genome sequence-independent universal microarray probe set. BioTechniques 37:654-660. [DOI] [PubMed] [Google Scholar]
- 4.Bhanot, G., Y. Louzoun, J. Zhu, and C. DeLisi. 2003. The importance of thermodynamic equilibrium for high throughput gene expression arrays. Biophys. J. 84:124-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolstad, B. M., R. A. Irizarry, M. Astrand, and T. P. Speed. 2003. A comparison of normalization methods of high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185-193. [DOI] [PubMed] [Google Scholar]
- 6.Budowle, B., S. E. Schutzer, A. Einseln, L. C. Kelley, A. C. Walsh, J. A. L. Smith, B. L. Marrone, J. Robertson, and J. Campos. 2003. Building microbial forensics as a response to bioterrorism. Science 301:1852-1853. [DOI] [PubMed] [Google Scholar]
- 7.Forman, J. E., I. D. Walton, D. Stern, R. P. Rava, and M. O. Trulson. 1998. Thermodynamics of duplex formation and mismatch discrimination on photolithographically synthesized oligonucleotide arrays. ACS Symp. Ser. 682:206-228. [Google Scholar]
- 8.Gotoh, M., Y. Hasegawa, Y. Shinohara, M. Shimizu, and M. Tosu. 1995. A new approach to determine the effect of mismatches on kinetic parameters in DNA hybridization using an optical biosensor. DNA Res. 2:285-293. [DOI] [PubMed] [Google Scholar]
- 9.Higgins, J. A., M. Cooper, L. Schroeder-Tucker, S. Black, D. Miler, J. S. Karns, E. Manthey, R. Breeze, and M. L. Perdue. 2003. A field investigation of Bacillus anthracis contamination of U.S. Department of Agriculture and other Washington, D.C., buildings during the anthrax attack of October 2001. Appl. Environ. Microbiol. 69:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hojjatoleslami, S. A., and J. Kittler. 1998. Region growing: a new approach. IEEE Trans. Image Process. 7:1079-1084. [DOI] [PubMed] [Google Scholar]
- 11.Jackson, P. J., E. A. Walthers, A. S. Kalif, K. L. Richmond, D. M. Adair, K. K. Hill, C. R. Kuske, G. L. Andersen, K. H. Wilson, M. E. Hugh-Jones, and P. Keim. 1997. Characterization of the variable-number tandem repeats in vrrA from different Bacillus anthracis isolates. Appl. Environ. Microbiol. 63:1400-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansson, A., J. Farlow, P. Larsson, M. Dukerich, E. Chambers, M. Bystrom, J. Fox, M. Chu, M. Forsman, A. Sjostedt, and P. Keim. 2004. Worldwide genetic relationships among Francisella tularensis isolates determined by multiple-locus variable-number tandem repeat analysis. J. Bacteriol. 186:5808-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keim, P. 2003. Microbial forensics: a scientific assessment. American Academy of Microbiology, Washington, D.C.
- 14.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, M.-L. T., F. C. Kuo, G. A. Whitmore, and J. Sklar. 2000. Importance of replication in microarray gene expression studies: statistical methods and evidence from repetitive cDNA hybridizations. Proc. Natl. Acad. Sci. USA 97:9834-9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leng, X., D. A. Mosier, and R. D. Oberst. 1996. Differentiation of Cryptosporidium parvum, C. muris, and C. baileyi by PCR-RFLP analysis of the 18s rRNA gene. Vet. Parasitol. 62:1-7. [DOI] [PubMed] [Google Scholar]
- 17.Radnedge, L., P. G. Agron, K. K. Hill, P. J. Jackson, L. O. Ticknor, P. Keim, and G. L. Andersen. 2003. Genome differences that distinguish Bacillus anthracis from Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 69:2755-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. Jiang, E. Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296:2028-2033. [DOI] [PubMed] [Google Scholar]
- 19.Rouillard, J.-M., M. Zuker, and E. Gulari. 2003. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 31:3057-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swaminathan, B., T. J. Barrett, S. B. Hunter, R. V. Tauxe, and C. P. T. Force. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ticknor, L. O., A.-B. Kolstø, K. K. Hill, P. Keim, M. T. Laker, M. Tonks, and P. J. Jackson. 2001. Fluorescent amplified fragment length polymorphism analysis of Norwegian Bacillus cereus and Bacillus thuringiensis soil isolates. Appl. Environ. Microbiol. 67:4863-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White, A. M., D. S. Daly, A. R. Willse, M. Protic, and D. P. Chandler. 2005. Automated microarray image analysis toolbox for MATLAB. Bioinformatics 21:3578-3579. [DOI] [PubMed] [Google Scholar]
- 24.Willse, A., D. P. Chandler, A. White, M. Protic, D. S. Daly, and S. Wunschel. 2005. Comparing bacterial DNA microarray fingerprints. Stat. Appl. Genet. Mol. Biol. 4:Article 19. [Online.] [DOI] [PubMed]
- 25.Willse, A., T. M. Straub, S. C. Wunschel, J. A. Small, D. R. Call, D. S. Daly, and D. P. Chandler. 2004. Quantitative oligonucleotide microarray fingerprinting of Salmonella enterica isolates. Nucleic Acids Res. 32:1848-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]