Abstract
The three French overseas departments of the Americas are characterized both by insular (Guadeloupe and Martinique) and continental (French Guiana) settings with a tuberculosis case detection rate that varies from less than 10 per 100,000 per year in insular areas to an estimated incidence of more than 55 per 100,000 in French Guiana. Under a long-term genotyping program, more than three-fourths of all the Mycobacterium tuberculosis isolates (n = 744) received from the three settings were fingerprinted over a 10-year period (1994 to 2003) by spoligotyping and variable number of tandem DNA repeats (VNTRs) in order to understand the current trends in their detection rates, drug resistance, and groups and subpopulations at risk of contracting the disease and to pinpoint the circulating phylogeographical clades of the bacilli. The major difference in the study populations was the nationality of the patients, with a high percentage of immigrants from high-incidence neighboring countries in French Guiana and a low but increasing percentage in the French Caribbean. The rate of recent transmission was calculated to be 49.3% in French Guiana, compared to 27.2% and 16.9% in Guadeloupe and Martinique, respectively. At the phylogeographic level, 77.9% of the isolates studied belonged to four major clades (Haarlem, Latin-American and Mediterranean, T, and X) which are already reported from neighboring Caribbean islands in an international database and may underline potential interregional transmission events.
Since 1994, the tuberculosis unit of the Pasteur Institute of Guadeloupe has been serving as a regional reference laboratory for tuberculosis and mycobacterial diagnostics, identification, and drug resistance determination for the Caribbean (20). Under a long-term genotyping project, more than three-fourths of all the Mycobacterium tuberculosis isolates received from the three French overseas departments of the Americas (Guadeloupe, Martinique, and French Guiana) were systematically fingerprinted to realize an active survey of tuberculosis. The two sister islands of Guadeloupe and Martinique are situated south of Puerto Rico and are called the French West Indies (FWI), whereas French Guiana is situated in South America between Brazil and Surinam. Guadeloupe and Martinique, which cover respective areas of 1,705 and 1,100 km2, are highly urbanized islands with populations of 422,222 and 381,325 inhabitants (density of 255 and 353 inhabitants/km2), respectively, whereas French Guiana is a large continental area of 84,000 km2 with a population of 156,790 habitants (density of 2 inhabitants/km2, although 90% of the population is concentrated in the coastal region which represents 10% of the overall territory; http://www.insee.fr/en/home/home_page.asp). These three areas also differ strikingly by the proportion of foreign-born individuals, which varies from a low of about 1% of the population in Martinique to a high of 30% in French Guiana, whereas Guadeloupe has a foreign-born population of 5%, similar to the average for metropolitan France (http://www.lesechos.fr/regions/atlas/atlas_01_08_2004.htm). Most of the foreign-born individuals in these three overseas areas originate from neighboring countries where tuberculosis (TB) is highly endemic, such as Haiti, the Dominican Republic, Surinam, or Brazil; a previous study showed that both TB incidence as well as imported cases of TB were highest in French Guiana among all the French departments, including the metropolitan areas (13, 20). Consequently, it was decided to initiate a long-term systematic fingerprinting of Mycobacterium tuberculosis isolates in FWI and French Guiana regions so as to understand the current trends in their TB detection rates, drug resistance, and groups and subpopulations at risk of contracting the disease and to pinpoint the circulating phylogeographical clades of the bacilli in order to define epidemiological and demographical specificities that may be useful to better define the priorities in TB control strategy. This investigation, performed over a period of 10 years (1994 to 2003), permits the first detailed population-based study describing epidemiological characteristics of clustered versus nonclustered cases of TB as well as major circulating clades in this area of the Americas.
MATERIALS AND METHODS
Clinical isolates.
A total of 744 clinical isolates of M. tuberculosis were isolated from patients in Guadeloupe, Martinique, and French Guiana from January 1994 to December 2003. The cultures were performed using Löwenstein-Jensen slants at 37°C and duly identified as Mycobacterium tuberculosis complex using classical biochemical tests and the AccuProbe test (GenProbe Inc., San Diego, CA) at the Pasteur Institute of Guadeloupe, followed by drug susceptibility testing of isoniazid (INH), rifampin (RIF), ethambutol (EMB), streptomycin (SM), pyrazinamide (PZA), and ethionamide (ETH) by the 1% proportion method (13). The geographical repartition of the isolates was as follows: 242 isolates from Guadeloupe, 396 isolates from French Guiana, and 106 isolates from Martinique.
Molecular typing.
The bacterial DNA was prepared by a cetyl-trimethyl ammonium bromide method (29) and used for spoligotyping and variable number of tandem DNA repeat (VNTR) analysis. Spoligotyping was performed using a homemade membrane with 43 covalently bound oligonucleotides as described previously using primers designated DRa and DRb, with DRa biotinylated 5′ to amplify the whole DR region by PCR (16). As recommended, DNAs from M. tuberculosis type isolate H37Rv and M. bovis BCG, as well as sterile water, were used as controls. Detection of hybridizing DNA was done by using an enhanced chemiluminescence detection kit (ECL; Amersham, Buckhamshire, England), followed by exposure to X-ray film (Hyperfilm ECL; Amersham) according to the manufacturer's instructions. The X-ray film was developed using standard photochemicals after 2 h of exposure. A total of 585 isolates representing 78.6% of the isolates was arbitrarily selected for spoligotyping, and the results obtained were entered in a binary format as Excel (Microsoft, Cupertino, CA) spreadsheets and compared to the updated international spoligotyping database of the Pasteur Institute of Guadeloupe (12, 26; a SpolDB3 version is available online at http://www.pasteur-guadeloupe.fr/tb/spoldb3). At the time of the matching analysis, the updated spolDB4 version contained 39,295 patterns distributed into 1,939 shared types (a shared type, or ST, is defined as an identical spoligotype found in ≥2 individual patient isolates) and 3,370 orphan patterns consisting of entries occurring only once in the database from 122 countries (K. Brudey, unpublished data).
The isolates clustered by spoligotyping (n = 452) were further subtyped by VNTRs. This PCR-based method was performed as described previously (14) with slight modifications as follows. PCR was performed in a Perkin Elmer GeneAmp PCR system 9600 (Roche Diagnostic Systems, Basel, Switzerland). An initial denaturation of 12 min at 95°C was followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 1 min, and extension at 72°C for 2 min, with a final extension step at 72°C for 10 min. Thirty microliters of the reaction tubes was run on a 3% agarose gel (Gibco BRL, Grand Island, NY). Molecular weight standards (100-bp ladder; AP-Biotech) were run every seven lanes. The molecular weight determination of PCR fragments was performed using Taxotron software (Taxolab; PAD Grimont, Institut Pasteur, Paris, France) on images digitized using the Video-Copy system (Bioprobe, Montreuil, France). Once the length of the PCR fragments was precisely calculated, the number of copies for each exact tandem repeat (ETR) was deduced using an Excel spreadsheet and the data previously published and was documented as five-digit numbers representing allele profiles ETR-A to ETR-E (14).
Statistical analysis.
Data were analyzed by the EpiInfo 6.04 Fr.v program (Centers for Disease Control and Prevention, Atlanta, GA, and World Health Organization, Geneva, Switzerland), and observed frequencies were compared by means of two-by-two contingency tables using uncorrected Chi-squared tests. When a value was smaller than 5, Fisher's exact test was used. A P value of <0.05 was considered statistically significant.
RESULTS
Epidemiological data.
Among the isolates collected, the quality of data permitted statistical analysis concerning the demographic characteristics of the population affected by M. tuberculosis in Guadeloupe, Martinique, and French Guiana. These results are summarized in Table 1 for each area individually. The incidence of culture-positive TB cases varied from a low of about 3/100,000 in Martinique to 5.8/100,000 in Guadeloupe and as high as 25.3/100,000 in French Guiana. Nonetheless, these rates concerned only culture-positive cases and were obviously lower than the real incidence of the disease, which, based on notifications made to regional health authorities, were calculated to be 6/100,000 in Martinique, 7.6/100,000 in Guadeloupe, and 47.2/100,000 in French Guiana. The incidence of TB in Guadeloupe and Martinique is close to that in metropolitan France (3) but is eight times higher in French Guiana. The real incidence of TB in French Guiana is underestimated, as it was previously estimated to be more than 55/100,000 habitants (1).
TABLE 1.
Basic epidemiological features of strains retrieved from tuberculosis patients in French West Indies (Guadeloupe and Martinique) and French Guiana
| Epidemiological feature | Value for group
|
|||
|---|---|---|---|---|
| Guadeloupe | Martinique | FWI | French Guiana | |
| Total no. of strains | 242 | 106 | 348 | 396 |
| Sex ratio | 2.1 | 1.0 | 1.6 | 1.8 |
| Age known (no. of patients) | 151 | 74 | 225 | 370 |
| No. of patients in age groupa: | ||||
| 0-14 yr | 2 (1.3) | 2 (2.7) | 4 (1.8) | 13 (3.5) |
| 15-39 yr | 65 (43.1) | 15 (20.3) | 80 (35.6)b | 209 (56.5)b |
| 40-64 yr | 53 (35.1) | 26 (35.1) | 79 (35.1) | 118 (31.9) |
| >65 yr | 31 (20.5) | 31 (41.9) | 62 (27.6)b | 30 (8.1)b |
| Origin known (no. of patients) | 122 | NAc | 268 | |
| No. of foreign patientsd | 52 (42.6) | NA | 185 (69.0) | |
| No. of patients frome: | ||||
| BRA | 57 (30.8) | |||
| DOM | 6 (11.5) | |||
| GUY | 16 (8.6) | |||
| HTI | 39 (75.0) | 91 (49.2) | ||
| SUR | 9 (4.9) | |||
| Other countries | 7 (13.5) | 12 (6.5) | ||
| Drug susceptibilityf | ||||
| Any resistance | 33 (13.6) | 12 (11.3) | 45 (12.9) | 62 (15.7) |
| MDRf | 3 (1.2) | 0 (0.0) | 3 (0.9) | 10 (2.5) |
Values in parentheses are percentages of the total number of patients for whom age was known.
P < 0.05.
NA, not available.
Values in parentheses are percentages of the total number of patients for whom origin was known.
Values in parentheses are percentages of the total number of foreign patients. Abbreviations: BRA, Brazil; DOM, the Dominican Republic; GUY, Guyana; HTI, Haiti; SUR, Surinam.
Values are numbers of patients. Values in parentheses are percentages. MDR, multidrug resistance (resistance to isoniazide and rifampin).
As shown in Table 1 for culture-positive cases, the male-to-female ratio in the study population during the 10 years was estimated as 1.0 in Martinique, 1.8 in French Guiana, and 2.1 in Guadeloupe. Some demographic data were not available for Martinique, as the corresponding laboratories did not complete the questionnaire adequately. Nonetheless, the available data as well as previous results (20) suggested that the two sister islands (Guadeloupe and Martinique) presented very similar and homogeneous populations and could be taken as a whole; these were consequently treated together as FWI and were compared to the data from French Guiana. The distribution of cases was different among different age groups. The age group of 15 to 39 years was the most important and represented 35.6% and 56.5%, respectively, of the isolates in FWI and French Guiana (P < 0.05), followed by the age group 40 to 64 years, with 35.1% and 31.9% of the isolates, respectively. The age group 65 and older was significantly more represented in FWI than in French Guiana (27.6% versus 8.1% of the isolates; P < 0.05). The age group 0 to 14 years corresponded to 1.8% of the isolates in FWI and 3.5% in French Guiana. The results of human immunodeficiency (HIV) serology were not known for all cases; nonetheless, the available results underlined a TB-HIV coinfection in 19% of the patients in Martinique, 25% in French Guiana, and 28% in Guadeloupe; these values are significantly higher than those found in French metropolitan departments (20).
Drug susceptibility results showed nearly the same percentage of resistant isolates to at least one antibiotic in FWI and French Guiana (12.9% and 15.7%, respectively) but with different proportions of multidrug-resistant TB (MDR-TB; 0.9% in FWI and 2.5% in French Guiana; Table 1). The origin of the patients (Table 1) being different in the two settings was considered. The origin of patients from Martinique was not available. A major proportion of the patients in Guadeloupe was made up of French nationals (57.4%), compared to only 31% in French Guiana. Compared to Guadeloupe, where the foreign-born patients were essentially from Haiti and the Dominican Republic, the nationality of patients in French Guiana was more diverse. Nearly half of the foreign-born patients originated from Haiti (49.2%), followed by others that share a common frontier with French Guiana, i.e., Brazil (30.8%), Guyana (8.6%), and Surinam (4.9%). The diversity of nationalities found in French Guiana can easily be explained by the continental aspect of this French overseas department sharing a common border with high-TB-incidence countries (13).
Molecular typing results.
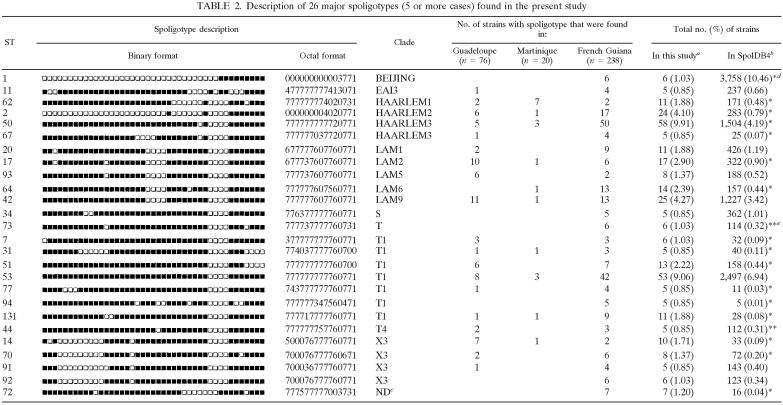
The spoligotyping was performed as a first-line screening method on 585 isolates followed by VNTR performed on all the 452 clustered isolates, and the results obtained are detailed in Table S1 of the supplemental material. Briefly, spoligotyping generated 182 distinct patterns among 585 typed isolates. The spoligotype ST designations were attributed by comparing the patterns obtained with those included in the world spoligotype database, SpolDB4. A total of 452 isolates, or 77.3% of the typed isolates, was grouped in 49 clusters (2 to 58 isolates per cluster). Among the remaining 133, or 22.7%, of the unclustered isolates, 61 or 10.4% were not yet reported to the SpolDB4 database and represented the true orphan isolates. The other 72, or 12.3%, of the unclustered isolates, though unique in our 10-year study, were already reported in SpolDB4, which allowed the attribution of an ST number to these. Among the 49 clusters, 23 included two to four isolates and were defined as minor spoligotypes. Major spoligotypes were defined as STs that contained five or more isolates each (n = 26; Table 2), consisting of 73.9% of clustered isolates. Analysis of the frequency of major spoligotypes with SpolDB4 allowed a differentiation between ubiquitous types present in all the continents (ST 1, 11, 20, 34, 42, 44, 50, 51, 53, 64, and 73) and types that were specific to the Americas (ST 14, 67, and 77). Several types were endemic in Guadeloupe (ST 13, 15, and 103), in French Guiana (ST 66, 76, 94, 1084, 1340, and 1526), and in Martinique (ST 936 and 1086). Some interesting variations were found among the three French overseas departments regarding the predominant spoligotypes. Three top STs from French Guiana represented 29.1% of all isolates (ST 50, n = 50; ST 53, n = 42; ST 2, n = 17), compared to 22% of the isolates in Martinique (ST 62, n = 7; ST 50, n = 3; ST 53, n = 3) and 19.2% of the isolates in Guadeloupe (ST 42, n = 11; ST 17, n = 10; ST 53, n = 8).
TABLE 2.
Description of 26 major spoligotypes (5 or more cases) found in the present study
Percentage in this study compared to total isolates by spongotyping (n = 585).
Percentage compared to total number of shared type strains in SpolDB4.0 (n = 35,925).
ND, not yet defined in SpolDB4.0.
*, P < 0.001.
**, P < 0.05.
Phylogeographic analysis.
As explained in a previous study (4), all the isolates with the exception of true orphans in the database were attributed a clade designation based on minor and major visual rules for spoligotyping-based classification (12, 22). As summarized in Tables 2 and 3, this permitted us to designate 11 clades: Africanum; Beijing; Bovis; Central Asia [CAS]; East-African Indian [EAI]; Haarlem, Latin-American, and Mediterranean [LAM]; Manu; S; X; and an ill-defined T clade. Among the 26 major STs present in our study, 17 were significantly more prevalent in our setting than in the SpolDB4, whereas a single ST (ST 1, Beijing type) was significantly less present than in the international database (Table 2). Our study permitted us to define five major M. tuberculosis clades (Tables 2 and 3). Isolates of the T family predominated in our study (30.1% of all isolates; 27.8% in Guadeloupe, 28.8% in Martinique, and 31.2% in French Guiana). The second major clade was the Haarlem family (21.7% of all isolates; 19.2% in Guadeloupe, 21.1% in French Guiana, and 32.2% in Martinique), followed by the LAM family (19.7% of all isolates; 11.9% in Martinique, 18.7% in French Guiana, and 25.2% in Guadeloupe), X family (6.5% of all isolates), and EAI family (4.8% of all isolates). The other families (Africanum, Beijing, Bovis, CAS, Manu, and S) seemed minor in our setting. It may be underlined that the Beijing clade (ST 1) that is among the most predominant clade in the world today (30) was not found in FWI; however, six or 1.6% of the isolates from French Guiana were Beijing type (Table 3). Lastly, some evolutionary hypotheses could be made based on the distribution of the major types found, e.g., within the X3 clade, ST 14, ST 70, and ST 91 may all have derived from ST 92 by the loss of single spacers (respectively spacers 2, 39, and 13).
TABLE 3.
Distribution of major M. tuberculosis families among shared types (STs) encountered in the French West Indies (Guadeloupe and Martinique) and French Guiana
| Family | No. (%) of strains
|
||||
|---|---|---|---|---|---|
| Total | Guadeloupe | Martinique | FWI | French Guiana | |
| Africanum | 3 (0.5) | 2 (1.3) | 0 | 2 (1.0) | 1 (0.3) |
| Beijing | 6 (1.0) | 0 | 0 | 0 | 6 (1.6) |
| Bovis | 1 (0.2) | 0 | 0 | 0 | 1 (0.3) |
| CAS | 1 (0.2) | 1 (0.7) | 0 | 1 (0.5) | 0 |
| EAI | 28 (4.8) | 4 (2.6) | 0 | 4 (1.9) | 24 (6.4) |
| Haarlem | 127 (21.7) | 29 (19.2) | 19 (32.2) | 48 (22.9) | 79 (21.1) |
| LAM | 115 (19.7) | 38 (25.2) | 7 (11.9) | 45 (21.4) | 70 (18.7) |
| Manu | 2 (0.3) | 0 | 0 | 0 | 2 (0.5) |
| S | 9 (1.5) | 2 (1.3) | 1 (1.7) | 3 (1.4) | 6 (1.6) |
| T | 176 (30.1) | 42 (27.8) | 17 (28.8) | 59 (28.1) | 117 (31.2) |
| X | 38 (6.5) | 14 (9.3) | 2 (3.4) | 16 (7.6) | 22 (5.8) |
| Undefineda | 79 (13.5) | 19 (12.6) | 13 (22.0) | 32 (15.2) | 47 (12.5) |
| Total | 585 | 151 | 59 | 210 | 375 |
Strains with a signature that is as-yet undefined in SpolDB4 and orphan strains.
Clusters analysis.
Out of the 585 isolates typed by spoligotyping, 452 clustered isolates were subtyped using VNTRs. A combined analysis of spoligotyping and VNTRs grouped 345 isolates in 68 clusters, ranging from 2 to 39 isolates per cluster (Table 4 and supplemental material available online). Different characteristics, such as the sex, the resistance to antibiotics, and the nationality, were compared between isolates with unique spoligotyping patterns, isolates clustered by spoligotyping alone, and those clustered by spoligotyping plus VNTR (Table 4). The sex ratio was nearly similar between isolates with unique patterns and clustered isolates. The proportion of patients in the different age groups for total isolates was also similar in the three categories: the age group 15 to 39 years corresponded to half of the isolates, followed by the age groups 40 to 64 years and 65 years and older. Nonetheless, the values, once split between the three settings, showed that the age group 15 to 39 years was highly predominant in French Guiana, with nearly 60% of clustered cases, followed by Guadeloupe (43.4%), and significantly less predominant in Martinique (13.3%). This distribution was inversed for higher age groups, with most of the clustered cases occurring among 65 years and older in Martinique (40%), followed by Guadeloupe (15.1%) and as low as 7.1% in French Guiana. However, it should be underlined that the degree of clustering is not affected by the age of the patient, as in most of the cases the percentage of clustering by age in a given setting simply reflected the total number of cases in that particular age group (Tables 1 and 4). On the other hand, there was a real difference in the overall degree of clustering between the three settings studied, ranging from 27% in Martinique to 65% in French Guiana (Table 4).
TABLE 4.
Molecular typing results for unclustered and clustered strains in this study and distribution of clusters by spoligotyping and VNTR analysis in each setting
| Epidemiological feature | Isolates with unique patterns | Isolates clustered by spoligotyping | Value for group
|
||||
|---|---|---|---|---|---|---|---|
| Isolates clustered by spoligotyping and VNTR
| |||||||
| All | Guadeloupe | Martinique | French West Indies | French Guiana | |||
| Total no. (%) of clustered strains | 452 (77.3) | 345 (59.0) | 57 (37.7) | 16 (27.1) | 73 (34.8) | 242 (64.5) | |
| No. of clusters | 49 | 68 | 16 | 6 | 22 | 57 | |
| Total no. of strains | 133 | 585 | 585 | 151 | 59 | 210 | 375 |
| Transmission rate (%) | 68.9 | 47.4 | 27.2 | 16.9 | 24.3 | 49.3 | |
| Sex ratio | 1.7 | 1.7 | 1.8 | 1.7 | 1 | 1.5 | 1.9 |
| Age known (no. of patients) | 101 | 412 | 318 | 53 | 15 | 68 | 224 |
| No. (%) of patients in age group: | |||||||
| 0-14 yr | 3 (3.0) | 13 (3.2) | 10 (3.1) | 0 (0) | 1 (6.7) | 1 (1.5) | 7 (3.2) |
| 15-39 yr | 52 (51.5) | 211 (51.2) | 169 (53.2) | 23 (43.4) | 2 (13.3) | 25 (36.8) | 134 (59.8) |
| 40-64 yr | 31 (30.7) | 136 (33.0) | 106 (33.3) | 22 (41.5) | 6 (40.0) | 28 (41.2) | 67 (29.9) |
| >65 | 15 (14.8) | 52 (12.6) | 33 (10.4) | 8 (15.1) | 6 (40.0) | 14 (20.6)a | 16 (7.1)a |
| Origin known (no. of patients) | 71 | 290 | 227 | 44 | NAb | NDc | 165 |
| No. (%) of foreign-born patients | 49 (69.0) | 177 (61.0) | 139 (61.2) | 20 (45.5) | NA | ND | 108 (65.5) |
| No. (%) of patients fromd: | |||||||
| BRA | 17 (34.7) | 39 (22.0) | 29 (20.9) | 28 (25.9) | |||
| DOM | 6 (3.4) | 2 (1.4) | |||||
| GUY | 2 (4.1) | 15 (8.5) | 14 (10.1) | 13 (12.0) | |||
| HTI | 25 (51.0) | 95 (53.7) | 78 (56.1) | 16 (80.0) | 54 (50.0) | ||
| SUR | 2 (4.1) | 7 (3.9) | 7 (5.0) | 7 (6.5) | |||
| Other countries | 3 (6.1) | 17 (8.5) | 9 (6.5) | 4 (20.0) | 6 (5.6) | ||
| Drug susceptibilitye | |||||||
| Any resistance | 19 (14.3) | 61 (13.5) | 43 (12.5) | 5 (8.8) | 1 (6.3) | 6 (8.2) | 32 (13.2) |
| MDR | 1 (0.8) | 11 (2.4) | 9 (2.6) | 2 (3.5) | 0 (0.0) | 2 (2.7) | 8 (3.3) |
P < 0.005.
NA, not available.
ND, not done.
Abbreviations: BRA, Brazil; DOM, the Dominican Republic; GUY, Guyana; HTI, Haiti; SUR, Surinam.
Values are numbers of patients. Values in parentheses are percentages. MDR, Multidrug resistance (resistance to isoniazide and rifampin).
Interestingly, although the proportion of patients of different origins did not vary significantly among clustered and unclustered isolates (Table 4), final analysis based on individual settings showed that while a major source of foreign-born clustered TB cases was nearly exclusively limited to Haiti in Guadeloupe (80%), the origin was much more diverse in French Guiana (Haiti, 50%; Brazil, 25.9%; Guyana, 12%; Surinam, 6.5%; see the supplemental material). This diversity observed for clustered cases in French Guiana unambiguously reflects the overall diversity of sources of imported TB cases in this setting as opposed to Guadeloupe, where one out of four foreign-born patients was from Haiti. Regarding drug resistance, some clusters shared isolates with similar resistance patterns, e.g., cluster with resistance to SM and PZA and to INH alone or in association with SM. Regarding ethnicity, patients identified among a few clusters in French Guiana were either exclusively Haitians or exclusively Brazilians. Some clusters presented more evident links than ethnicity or drug resistance alone, e.g., a cluster with three patients originating from Haiti, and all were HIV positive (cluster B1 in the supplemental material). A relatively large cluster, B2, contained 19 patients distributed among all three departments studied, with nearly half of the cases found among the foreign born. Interestingly, it also contained a high proportion of INH-resistant isolates (n = 4); among these, one was also resistant to SM, one to ETH, and two to RIF plus ETH. The latter two were isolated from HIV-positive patients residing in French Guiana. On the same lines, another large cluster, I, shared 11 isolates with nine patients from French Guiana; five were females, and among these, four were Haitian-born and HIV positive and shared isolates that were drug resistant. All four isolates were RIF resistant, and among these, one was also resistant to SM, two to INH, and one to INH plus SM. Lastly, very few clusters were defined among cases diagnosed in FWI, which essentially involved patients of old age in Martinique and foreign-born patients (essentially from Haiti, followed by the Dominican Republic) in Guadeloupe.
Regarding drug resistance, the percentages of isolates resistant to any drug among clustered and unclustered isolates were similar (Table 4), but the percentage of MDR-TB (combined resistance to INH and RIF) was higher in isolates clustered by spoligotyping and VNTR (2.6%) than in isolates with unique patterns (0.8%). In conclusion, most of the clusters with evident demographic links were from French Guiana, underlining the high impact of imported cases of the disease in this department and its further intra- and intercommunity transmission within both French Guiana and FWI.
DISCUSSION
Contrary to metropolitan France, the French departments of the Americas are characterized by a relatively younger age of active population as well as precariousness and an important immigration from countries with high TB incidence and high AIDS burden that has been previously implicated both in the spread of the TB disease and TB-HIV coinfection (3, 13, 20). All three settings studied share an important immigration from Haiti, which, with an estimated incidence of 200 new cases per 100,000 inhabitants per year (7), have a heightened risk of developing TB compared to the native population. Indeed, if the TB among the foreign born is not considered, the incidence of “culture-positive” TB cases in Guadeloupe and French Guiana will be 4.6/100,000 and 13.7/100,000, respectively (instead of 5.8/100,000 and 25.3/100,000), values comparable to other French metropolitan departments (3, 15, 27).
The present study gives a first outline of the diversity of the population structure of M. tuberculosis in patients from the three French departments of the Americas between 1994 and 2003. The use in combination of spoligotyping and VNTR methods gives a greater accuracy of isolate clustering and more evidence of possible links between epidemiologically related patients. The global percentage of clustered isolates, using the combination of spoligotyping and VNTR, was 59% (345 out of 585) compared to 77.3% (452 out of 585) for spoligotyping alone (Table 4). Using the N-1 formula (24), the recent transmission rate in our study was calculated to be 47.4 for the whole region (Table 4). If the genotyping results were considered for the three departments individually, the global clustering rate for Martinique and Guadeloupe was 27.1% and 37.7%, respectively (or 34.8% if taken together for FWI), compared to a high of 64.5% in French Guiana. Thus, the clustering rate in FWI was similar to those reported from Paris (36% [15]), New-York City (37.5% [2]), and San Francisco (40% [24]). This remarkable difference was also reflected by the rate of recent transmission that was as high as 49.3% in French Guiana, compared to 16.9% and 27.2% in Martinique and Guadeloupe, respectively. Thus, the rate of recent transmission in FWI is relatively low, with values for Martinique being close to those reported in low-incidence areas like Sweden (14.1% [4]), and London, England (14.4% [17]), and those for Guadeloupe being similar to Denmark (24% [34]) and Paris, France (25% 15). On the contrary, the rate of recent transmission of TB in French Guiana with a value of around 50% is close to data reported from high-incidence areas like Equatorial Guinea (28), Zimbabwe (10), Cape Town, South Africa (31), and Antananarivo, Madagascar (19). However, these clustering rates should be interpreted carefully, because clustering may be observed between individuals without any recent transmission links or established contacts (8). In our study, the contact tracing was attempted in Guadeloupe since 1999 but was insufficient to establish transmission pathways other than those evident by demographic data such as members of a same family, ethnic community, or living in the same area. Contact tracing was also attempted by local health authorities in French Guiana, but the data were not exhaustive enough to establish definite links in most of the cases. Nonetheless, a careful analysis of the available epidemiological and demographic data of the clustered isolates defined possible links between patients (see the supplemental material).
It is well known that a considerable risk of developing TB exists among immigrants arriving from high-TB-burden countries and that this risk persists even for some decades following entry in low-incidence countries (6, 32). This would explain why, despite a high proportion of clustered isolates in French Guiana, the only evident link among clustered isolates was the foreign origin (Haiti). The rest of the characteristics among clustered cases were essentially similar to those recently reported in a molecular epidemiology study from Haiti (11). This supports the hypothesis that the disease encountered in foreign-born patients from high-TB-incidence countries in our settings is more likely to have arisen from reactivation of latent infection acquired in their country of origin (18).
According to a previous study (20), the rate of TB-HIV coinfection in the three regions studied was important (around 25%) and predominated among males within the age group 25 to 44 years (70% of HIV-positive cases). In the present study, this information was not available for all the patients; nonetheless, a total of 126 patients with known HIV-positive serology corresponded to 17% of all culture-positive cases (results not shown). Among these, 68% were males, 50% belonged to the age group 25 to 44, and 48% were foreign born.
The total proportion of 2.5% and 1.2% MDR-TB isolates in French Guiana and Guadeloupe, respectively, was significantly higher than the rate of 0.6% in France between 1992 and 1999 (21). In our case, the MDR-TB cases were essentially attributable to secondary drug resistance among foreign-born patients from countries with a high incidence of MDR-TB, such as Haiti, Brazil, Surinam, or Guyana (11, 13). Calculated on the basis of a total of 126 out of 744 patients with known HIV-positive serology in the present study, 16.7% of the patients harbored drug-resistant isolates and 4.8% were infected with MDR-TB isolates (the similar values for the remaining 618 out of 744 patients without known HIV serology or HIV-negative status were 13.9% and 1.1%, respectively). This suggests that high HIV seropositivity, TB incidence, and precariousness among immigrant populations in our case constitute the major hurdle to TB elimination and should be the focus of future TB control strategy. Indeed, HIV has been associated with epidemic MDR-TB transmission in a number of high-TB-burden countries, like Haiti and India (5, 11). Even in regions with low TB incidence, HIV is one of the main factors having a high impact on the epidemiology, history, and clinical evolution of tuberculosis (5).
As summarized in Table 3, the spoligotype profiles observed in our study allowed the classification of 77.9% of the isolates into the four major phylogenetic families (Haarlem, LAM, T, and X) corresponding to 456 isolates, a feature that had been previously established by comparing the TB population structure in Cuba, Guadeloupe, and Haiti (9). These spoligotypes were already reported from neighboring Caribbean islands, and recent studies have underlined the potential interregional transmission between the Caribbean islands (12, 20). The two major clades encountered are the T family, with 30.1% of all isolates, and Haarlem, with 21.7% of isolates. Isolates from the Haarlem family represent isolates of European descent (23, 26) due to the past colonization history of these islands and continuing business, administrative, political, and touristic links with Europe. The T family corresponds to isolates whose ancestry remains ill defined (23, 26). The LAM family is prevalent in Latin America and the Mediterranean region and represents 19.7% of isolates, and its presence may be explained by European colonization (3, 9), particularly of the Spanish empire. The percentage of isolates belonging to the X family, presumably of Anglo-Saxon descent (22), was higher in Guadeloupe than in Martinique and French Guiana (Table 3). Regarding its affinity for the Caribbean, ST 14 must be highlighted because of its prevalence in Guadeloupe (78.8% of all the ST 14 isolates in SpolDB4 are from Guadeloupe). Nonetheless, the number of isolates with ST 14 decreased during the last years (from four to seven isolates per year to only one isolate per year since 2001), which may be a sign of extinction of the type in Guadeloupe.
In addition to the European colonization, the role of the indigenous populations escaping colonizers in the spread of TB may not be excluded. For example, Amerindian populations like the Arawaks and Caribs from Venezuela, without immune protection to diseases that were brought by colonizers, fled to other areas, thus migrating and dispersing throughout the Caribbean (http://www.centerlink.org/Cultures.html). During this period, they may have involuntarily introduced LAM and Haarlem isolates to neighboring islands. A recent study has underlined the presence of Haarlem, LAM, and X as major clades in Haiti (11), most of which are also found in the present study, a fact explained by the high level of immigration from Haiti. However, concerning the immigrants from Haiti, the immigrant either could migrate with its isolate or could be infected in the destination country. According to the international database, the former may probably be true for ST 193 (one patient from French Guiana and eight from Haiti), ST 295 (one from Guadeloupe and five from Haiti), ST 633 and 729 (one from Guadeloupe and three from Haiti), and ST 1087 (one from Guadeloupe and one from Haiti). On the other hand, ST 51, which is not described in the Haitian study (11) but is present in the Haitian population of Guadeloupe and French Guiana (cluster S; see the supplemental material), may represent ongoing infections in these French departments. Nonetheless, the role of homoplasia in such a case may not be fully excluded (33).
The ancestral EAI clade contained 28 isolates, with only 4 isolates (1.9%) in FWI versus 24 (6.4%) in French Guiana, and a single isolate of the CAS family was found in Guadeloupe. This result was quite amazing in the light of massive past immigration in this region from India about 150 years ago, which could have resulted in a higher frequency of EAI and CAS clades that predominate in the Indian subcontinent today (12, 23). Somehow, this highly regulated immigration did not result in imported cases of TB, hence the relative absence of isolates specific to India like ST 26 (CAS family). Interestingly, the number of isolates of the EAI family has increased in recent years in French Guiana. This could be explained by the fact that this family is prevalent in Southeast Asia, East Africa, and some parts of Europe (12), and these isolates could have been introduced because of recent migration from Asia, particularly the Hmong community (http://www.hmongcenter.org/hmoninfrengu.html). On the other hand, the Beijing family (ST 1), which is also highly prevalent in Asia, is not well represented in FWI or French Guiana. Nonetheless, the number of Beijing isolates, which have been linked to a greater ability to be transmitted throughout the world (30), has recently increased in the last 2 years in French Guiana (one isolate in 1997, three isolates in 2002, and two in 2003), but not a single isolate was found in FWI in the present study. This recent introduction of Beijing isolates in French Guiana could be explained by the presence of Asian-born populations originating in Vietnam, Singapore, Hong Kong, Taiwan, and continental China, which settled in French Guiana at the beginning of the 20th century (http://www.tlfq.ulaval.ca/axl/amsudant/guyanefr2.htm) and the recent arrival of Hmongs in the last 3 decades. Although these populations lived for a long time within themselves without much contact with the outside world, they recently opened to the rest of the Asian community in the world thanks to globalization and new business opportunities with the Chinese mainland.
In conclusion, all three French departments studied show characteristics of an ancient tuberculosis introduction that occurred about 400 years ago by migration flows at the time of colonization (essentially isolates of European or Anglo-Saxon descent such as Haarlem, LAM, and X [25]). Nonetheless, major differences observed between FWI and French Guiana concerned the incidence of new cases, the rate of recent transmission, and drug resistance, which were all significantly higher in French Guiana than in FWI. These could be easily explained on the basis of the population structure, particularly the high proportion of foreign-born populations in French Guiana. One may hypothesize that with the continuing human migratory flow to French Guiana (and to a lesser degree to Guadeloupe) and the increased intermingling of the populations, the genotypic diversity of M. tuberculosis isolates encountered may reach a plateau in the future. One has to be extremely vigilant that a newly arrived clone does not become predominant in coming years because of the increased rates of MDR-TB. This can be avoided by better access to health care for populations at risk of contracting or developing the disease, particularly the immigrants from high-TB-burden countries and those living in Amazonian forests and other distant parts of French Guiana (e.g., clandestine workers such as gold diggers living near forest rivers). The existence of such crowded communities living in unhealthy conditions and their migration to other parts of the country may favor a rapid spread of TB and MDR-TB, with a negative impact on health control in French Guiana.
Supplementary Material
Acknowledgments
We are grateful to the following colleagues for their precious collaboration: M. Théodore, J.-M. Perez, G. Larifla, M. Lévy, A. Accipe, M. Fassih, C. Saint-Martin, J.-J. Wanou, O. Faure, F. Cnudde, D. Caparros, G. Cadelis, I. Lamaury, and M. Strobel in Guadeloupe; D. Vigée, J. Lafaye, L. Ghisalberti, J. Jouannelle, C. Olive, R. Théodose, J. de Thoré, J. Zecler, I. Kaffa, P. Ray, A. Blateau, and P. Chaud in Martinique; and B. Cottrelle, D. Louvel, V. Aït Ouada, B. Carme, J. Thonnon, R. Pradineau, G. Guillot, A.-M. Bourbigot, and N. Quintard in French Guiana. The technical expertise of K. S. Goh, M. Berchel, and F. Prudenté (Institut Pasteur de Guadeloupe) for identification and drug susceptibility testing of the isolates studied is gratefully acknowledged.
This work was supported by the International network of the Pasteur Institutes, Institut Pasteur, Paris, France. K.B. benefited from a Ph.D fellowship partially financed by the European Social Funds, provided through the Regional Council of Guadeloupe.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Aït Ouada, V. 2002. La tuberculose en Guyane. Doctorat en Médecine. Université de Montpellier-I, Montpellier, France.
- 2.Alland, D., G. E. Kalkut, A. R. Moss, R. A. McAdam, J. A. Hahn, W. Bosworth, E. Drucker, and B. R. Bloom. 1994. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N. Engl. J. Med. 330:1710-1716. [DOI] [PubMed] [Google Scholar]
- 3.Brudey, K., I. Filliol, C. Sola, C. Bebear, S. Elia-Pasquet, J. Texier-Maugein, and N. Rastogi. 2003. Molecular characterization and biodiversity of Mycobacterium tuberculosis in the Antilles-Guiana region and comparative analysis in a metropolitan region, Aquitaine. Pathol. Biol. (Paris) 51:282-289. [DOI] [PubMed] [Google Scholar]
- 4.Brudey, K., M. Gordon, P. Mostrom, L. Svensson, B. Jonsson, C. Sola, M. Ridell, and N. Rastogi. 2004. Molecular epidemiology of Mycobacterium tuberculosis in western Sweden. J. Clin. Microbiol. 42:3046-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corbett, E. L., C. J. Watt, N. Walker, D. Maher, B. G. Williams, M. C. Raviglione, and C. Dye. 2003. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch. Intern. Med. 169:1009-1021. [DOI] [PubMed] [Google Scholar]
- 6.Cowie, R. L., and J. W. Sharpe. 1998. Tuberculosis among immigrants: interval from arrival in Canada to diagnosis. A 5-year study in southern Alberta. Can. Méd. Assoc. J. 158:599-602. [PMC free article] [PubMed] [Google Scholar]
- 7.Deschamps, M. M., D. W. Fitzgerald, J. W. Pape, and W. D. Johnson, Jr. 2000. HIV infection in Haiti: natural history and disease progression. AIDS 14:2515-2521. [DOI] [PubMed] [Google Scholar]
- 8.Diel, R., S. Schneider, K. Meywald-Walter, C. M. Ruf, S. Rusch-Gerdes, and S. Niemann. 2002. Epidemiology of tuberculosis in Hamburg, Germany: long-term population-based analysis applying classical and molecular epidemiological techniques. J. Clin. Microbiol. 40:532-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duchêne, V., S. Ferdinand, I. Filliol, J. F. Guegan, N. Rastogi, and C. Sola. 2004. Phylogenetic reconstruction of Mycobacterium tuberculosis within four settings of the Caribbean region: tree comparative analyse and first appraisal on their phylogeography. Infect. Genet. Evol. 4:5-14. [DOI] [PubMed] [Google Scholar]
- 10.Easterbrook, P. J., A. Gibson, S. Murad, D. Lamprecht, N. Ives, A. Ferguson, O. Lowe, P. Mason, A. Ndudzo, A. Taziwa, R. Makombe, L. Mbengeranwa, C. Sola, N. Rastogi, and F. Drobniewski. 2004. High rates of clustering of isolates causing tuberculosis in Harare, Zimbabwe: a molecular epidemiological study. J. Clin. Microbiol. 42:4536-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferdinand, S., C. Sola, B. Verdol, E. Legrand, K. S. Goh, M. Berchel, A. Aubéry, M. Timothée, P. Joseph, J. W. Pape, and N. Rastogi. 2003. Molecular characterization and drug-resistance of Mycobacterium tuberculosis isolated from patients in an AIDS counselling center in Port-au-Prince, Haiti: a one-year study. J. Clin. Microbiol. 41:694-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filliol, I., J. R. Driscoll, D. van Soolingen, B. N. Kreiswirth, K. Kremer, G. Valétudie, D. D. Anh, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, M. C. Gutierrez, W. H. Haas, H. Heersma, G. Källenius, E. Kassa-Kelembho, T. Koivula, H. M. Ly, A. Makristathis, C. Mammina, G. Martin, P. Moström, I. Mokrousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. N. Niobe-Eyangoh, J. W. Pape, V. Rasolofo-Razanamparany, M. Ridell, M. L. Rossetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. H. d. Waard, C. Sola, and N. Rastogi. 2002. Global distribution of Mycobacterium tuberculosis spoligotypes. Emerg. Infect. Dis. 8:1347-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filliol, I., S. Ferdinand, C. Sola, J. Thonnon, and N. Rastogi. 2002. Spoligotyping and IS6110-RFLP typing of Mycobacterium tuberculosis from French Guiana: a comparison of results with international databases underlines interregional transmission from neighbouring countries. Res. Microbiol. 153:81-88. [DOI] [PubMed] [Google Scholar]
- 14.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 15.Gutierrez, M. C., V. Vincent, D. Aubert, J. Bizet, O. Gaillot, L. Lebrun, C. LePendeven, M. P. LePennec, D. Mathieu, C. Offredo, B. Pangon, and C. Pierre-Audigier. 1998. Molecular fingerprinting of Mycobacterium tuberculosis and risk factors for tuberculosis transmission in Paris, France, and surrounding area. J. Clin. Microbiol. 36:486-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamerbeek, J., L. Schouls, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. D. A. van Embden. 1997. Simultaneous detection and isolate differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maguire, H., J. W. Dale, T. D. McHugh, P. D. Butcher, S. H. Gillespie, A. Costetsos, H. Al-Ghusein, R. Holland, A. Dickens, L. Marston, P. Wilson, R. Pitman, D. Strachan, F. A. Drobniewski, and D. K. Banerjee. 2002. Molecular epidemiology of tuberculosis in London 1995-7 showing low rate of active transmission. Thorax 57:617-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray, M. 2002. Molecular epidemiology and the dynamics of tuberculosis transmission among foreign-born people. Can. Méd. Assoc. J. 167:355-356. [PMC free article] [PubMed] [Google Scholar]
- 19.Rasolofo Razanamparany, V., D. Menard, G. Auregan, B. Gicquel, and S. Chanteau. 2002. Extrapulmonary and pulmonary tuberculosis in Antananarivo (Madagascar): high clustering rate in female patients. J. Clin. Microbiol. 40:3964-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rastogi, N., L. Schlegel, F. Pfaff, I. Jeanne, C. Magnien, G. Lajoinie, C. Saez, S. Firmin, V. Mazille, and M. Theodore. 1998. La tuberculose dans la région Antilles-Guyane: situation épidémiologique de 1994 à 1996. Bull. Epidemiol. Hebdo. 11:45-47. [Google Scholar]
- 21.Robert, J., D. Trystram, C. Truffot-Pernot, and V. Jarlier. 2003. Multidrug-resistant tuberculosis: eight years of surveillance in France. Eur. Respir. J. 22:833-837. [DOI] [PubMed] [Google Scholar]
- 22.Sebban, M., I. Mokrousov, N. Rastogi, and C. Sola. 2002. A data-mining approach to spacer oligonucleotide typing of Mycobacterium tuberculosis. Bioinformatics 18:235-243. [DOI] [PubMed] [Google Scholar]
- 23.Singh, U. B., N. Suresh, N. V. Bhanu, J. Arora, H. Pant, S. Sinha, R. C. Aggarwal, S. Singh, J. N. Pande, C. Sola, and P. Seth. 2004. Predominant tuberculosis spoligotypes, Delhi, India. Emerg. Infect. Dis. 10:1138-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Small, P. M., P. C. Hopewell, S. P. Singh, A. Paz, J. Parsonnet, D. C. Ruston, G. F. Schecter, C. L. Daley, and G. K. Schoolnik. 1994. The epidemiology of tuberculosis in San Francisco. N. Engl. J. Med. 330:1703-1709. [DOI] [PubMed] [Google Scholar]
- 25.Sola, C., A. Devallois, L. Horgen, J. Maïsetti, I. Filliol, E. Legrand, and N. Rastogi. 1999. Tuberculosis in the Caribbean: using spacer oligonucleotide typing to understand isolate origin and transmission. Emerg. Infect. Dis. 5:404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sola, C., I. Filliol, C. Guttierez, I. Mokrousov, V. Vincent, and N. Rastogi. 2001. Spoligotype database of Mycobacterium tuberculosis: biogeographical distribution of shared types and epidemiological and phylogenetic perspectives. Emerg. Infect. Dis. 7:390-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sola, C., I. Filliol, J. Maisetti, B. Carbonnelle, and N. Rastogi. 2003. Epidemiological study of tuberculosis in the area of Angers, France, as studied by 3 PCR-based fingerprinting methods. Pathol. Biol. (Paris) 51:13-20. [DOI] [PubMed] [Google Scholar]
- 28.Tudo, G., J. Gonzalez-Martin, R. Obama, J. M. Rodriguez, J. R. Franco, M. Espasa, P. P. Simarro, G. Escaramis, C. Ascaso, A. Garcia, and M. T. Jimenez De Anta. 2004. Molecular epidemiology of tuberculosis in the Bata and Malabo districts of Equatorial Guinea. Int. J. Tuberc. Lung Dis. 8:1458-1463. [PubMed] [Google Scholar]
- 29.van Soolingen, D., P. E. de Haas, P. W. M. Hermans, and J. D. A. van Embden. 1994. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 235:196-205. [DOI] [PubMed] [Google Scholar]
- 30.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verver, S., R. M. Warren, Z. Munch, E. Vynnycky, P. D. van Helden, M. Richardson, G. D. van der Spuy, D. A. Enarson, M. W. Borgdorff, M. A. Behr, and N. Beyers. 2004. Transmission of tuberculosis in a high incidence urban community in South Africa. Int. J. Epidemiol. 33:351-357. [DOI] [PubMed] [Google Scholar]
- 32.Vos, A. M., A. Meima, S. Verver, C. W. Looman, V. Bos, M. W. Borgdorff, J. D. Habbema. 2004. High incidence of pulmonary tuberculosis persists a decade after immigration, The Netherlands. Emerg. Infect. Dis. 10:736-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warren, R. M., E. M. Streicher, S. L. Sampson, G. D. van der Spuy, M. Richardson, D. Nguyen, M. A. Behr, T. C. Victor, and P. D. van Helden. 2002. Microevolution of the direct repeat region of Mycobacterium tuberculosis: implications for interpretation of spoligotyping data. J. Clin. Microbiol. 40:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, Z. H., P. E. de Haas, C. H. Wachmann, D. van Soolingen, J. D. van Embden, and A. B. Andersen. 1995. Molecular epidemiology of tuberculosis in Denmark in 1992. J. Clin. Microbiol. 33:2077-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.