Abstract
Spiroplasma spp. have been proposed to be the etiological agents of the transmissible spongiform encephalopathies (TSEs). In a blind study, a panel of 20 DNA samples was prepared from the brains of uninfected hamsters or hamsters infected with the 263K strain of scrapie. The brains of the infected hamsters contained ≥1010 infectious doses/g. The coded panel was searched for bacterial 16S rRNA gene sequences, using primers selective for spiroplasma sequences, primers selective for mollicutes in general, and universal bacterial primers. After 35 PCR cycles, no samples were positive for spiroplasma or any other bacterial DNA, while control Spiroplasma mirum genomic DNA, spiked at 1% of the concentration required to account for the scrapie infectivity present, was readily detected. After 70 PCR cycles, nearly all samples yielded amplified products which were homologous to various bacterial 16S rRNA gene sequences, including those of frequent environmental contaminants. These sequences were seen in uninfected as well as infected samples. Because the concentration of scrapie infectivity was at a known high level, it is very unlikely that a bacterial infection at the same concentration could have escaped detection. We conclude that the infectious agent responsible for TSE disease cannot be a spiroplasma or any other eubacterial species.
The identity of the causal agents of the transmissible spongiform encephalopathies (TSEs)—neurodegenerative diseases which include scrapie in sheep, Creutzfeldt-Jakob disease (CJD) in humans, bovine spongiform encephalopathy in cattle, and chronic wasting disease in deer and elk—has been a major source of controversy since these diseases were shown to be transmissible (41, 43, 17, 38). Early experiments showing that TSE infectivity passed through bacterial filters (54) and was resistant to disinfection by common bacteriocides (27, 10, 41, 42, 9) seemingly eliminated bacteria as the agent. However, the discovery in the 1970s of spiroplasmas, very small, thermostable, wall-less, helical-fibrillar bacteria that pass through 0.2-μm bacterial filters and show remarkable resistance to many common biocides including heat (47), provided a possible bacterial candidate for the infectious agent of TSEs. There have also been reports of spiral structures resembling spiroplasmas in brain tissue from CJD patients (2, 23, 39, 33), although other investigators have suggested these structures might be artifactual (24, 28). Moreover, no spiroplasmas could be cultured from CJD brain tissue (32). Intracranial inoculation of Spiroplasma mirum into new-born hamsters (31), suckling rats (5), or suckling mice (16) produces central nervous system disease, though not a progressive spongiform encephalopathy (16).
More recently, Bastian et al. (3, 4) have reported the presence of spiroplasma-specific 16S rRNA genes in brain tissue taken at autopsy/necropsy from TSE-infected humans and animals. PCR amplification of 16S rRNA genes, or “ribotyping,” is a powerful method for detecting and identifying microbial agents in environmental and clinical specimens. Universal primers that are complementary to sequences that are highly conserved across all eubacteria can be used to span sequences that vary with class, genus, or species and uniquely identify specific bacteria (55, 50). Ribotyping has been successfully applied to the phylogenetic analysis and detection of species in the genus Spiroplasma (21) and in the closely related genus Mycoplasma (51, 48), whose members are even smaller than spiroplasmas.
The spiroplasma sequences reported by Bastian et al. were detected in humans, sheep and deer (3, 4), species for which the concentration of infectivity in brain and other tissues is uncertain but is suspected to be lower and more variable than that obtained in well-characterized experimental rodent models. The 263K strain of hamster-adapted scrapie used in this study reliably produces the highest titers in brain known for any TSE disease. We have searched for bacterial 16S ribosomal sequences in a coded panel of genomic DNA from scrapie-infected and uninfected hamster brains using both conventional and nested PCR, two different reagent formats, and three different oligonucleotide primer pairs. One set of primers was selective for the genus Spiroplasma. A second set was more broadly selective for bacteria in the Mollicutes class, to which both Spiroplasma and Mycoplasma belong. The third set were broad-range universal bacterial primers, known to detect virtually all known eubacteria.
MATERIALS AND METHODS
Nucleic acid extraction.
Using procedures that minimized microbial contamination, brains were collected in our biosafety level 3 facility from 10 normal golden Syrian hamsters and from 10 hamsters in the late stage of clinical infection with the hamster-adapted 263K strain of scrapie. Half of the brains (five normal and five scrapie-infected) were from animals obtained from Charles River, and the other half were from animals obtained from Harlan. Genomic DNA was extracted using DNAzol solution (guanidium thiocyanate; BRL, Life Technologies, MD) according to the manufacturer's directions and purified by digestion with proteinase K, RNase A, and extraction with phenol-chloroform-isoamyl alcohol. Aliquots of the DNA were digested with EcoRI and analyzed on a 1.0% agarose gel to assess the quality of the DNA. The restriction patterns were consistent with those of high-molecular-weight DNA. Each genomic DNA sample was diluted to 100 ng/μl in 50 mM Tris, 1 mM EDTA, pH 8.0.
The hamster DNA samples, labeled as to source, were sent to an independent laboratory at the National Heart, Lung, and Blood Institute (NHLBI), where the samples were randomized, relabeled, and sent back to our laboratory as a blind panel. All experiments were performed “blind”, with the disease status and source of the hamster DNA samples unknown to the investigators. After return from NHLBI, an aliquot from each sample was amplified using PrP or p53 primers to check the integrity of the sample DNA. All DNAs amplified well. The entire panel was then split into multiple replicates by dividing each coded sample into aliquots. The replicates were stored at −80°C until use. Once all PCR and sequencing results, including all replications, had been completed, recorded in writing, and presented to NHLBI, the code was broken and samples were identified as infected or normal.
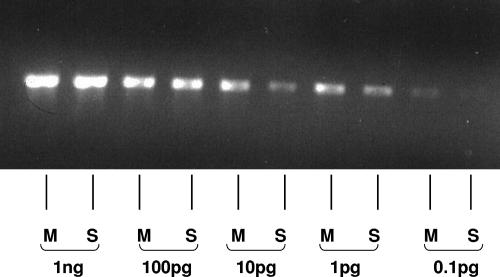
Positive control DNA was prepared from Spiroplasma mirum and Mycoplasma arginini. Each bacterium was grown to logarithmic stage in 100 ml of M1D medium (52) at 30°C. Genomic DNA was extracted using proteinase K digestion and phenol-chloroform as described previously (22). Tenfold serial dilutions from 1 ng to 10 fg of total DNA were used to optimize the sensitivity of detection under each set of PCR conditions, as illustrated in Fig. 1.
FIG. 1.
PCR amplification (30 cycles) of genomic DNA from M. arginini (M) and S. mirum (S) using universal primers, showing that the limit of detection of these agents by our methods is 1 to 0.1 pg.
PCR primers.
The sequences of spiroplasma and mollicute primer sets (Table 1) were provided by F. Bastian (personal communication). SpF corresponds to “F28”, and MoF and MoR correspond to “F1” and “R3,” respectively, of the work of Bastian and Foster (4). These primers correspond to regions of the 16S rRNA gene that are conserved or somewhat conserved (25, 37). Although the spiroplasma primer set was more specific for Spiroplasma species than was the mollicute primer set, other experiments have shown that the spiroplasma primers were not exclusively specific for spiroplasmas, in that they detect bacterial species in at least three mollicute genera other than Spiroplasma (G. Gasparich, unpublished).
TABLE 1.
Specificity and sequence of 16S rRNA gene primersa
| Name | Description | Sequence (5′ to 3′) | Location |
|---|---|---|---|
| UnF | Broad-range, universal bacterial primer, forward | GAGTTTGATCCTGGCTCAG | 9-27 |
| UnR | Broad-range, universal bacterial primer, reverse | GGACTACCAGGGTATCTAAT | 805-786 |
| MoF | Mollicute class-selective primer, forward | ACATAGGTGGCAAGCGTTATC | 534-554 |
| MoR | Mollicute class-selective primer, reverse | CTATTTGCTCCCTACGCTTTC | 784-764 |
| SpF | Spiroplasma genus-selective primer, forward | GCGCAGACGGTTTAACAAG | 578-596 |
| SpR | Spiroplasma genus-selective primer, reverse | TTCGCCACTGGTGTTCCTC | 729-747 |
The sequences of the broad-range universal primers (Table 1) are complementary to highly conserved regions of the 16S rRNA gene (25, 37) and are commonly used to detect bacteria nonspecifically (55, 46).
All oligonucleotides were synthesized on an Applied Biosystem, Inc., DNA synthesizer by the Biopolymer Laboratory of the University of Maryland, Baltimore.
PCRs.
Each PCR experiment included a negative control containing PCR-grade water as a DNA-free template and positive controls consisting of 0.1 pg and/or 1.0 pg of S. mirum genomic DNA and 1.0 pg of M. arginini genomic DNA. In addition to these controls, which were run with every amplification and on every gel, each coded sample in an experiment was amplified: alone, spiked with 1 pg S. mirum DNA, and spiked with 1 pg M. arginini DNA. A fourth aliquot of each sample was amplified with primers for either the PrP gene or the p53 gene to assess the quality of the genomic DNA and the presence of possible inhibitors of the PCR.
Amplifications were performed with a Perkin-Elmer GeneAmp PCR System 9700 instrument. To prevent cross-contamination, separate areas of the laboratory were used for handling the DNA and for assembling the PCRs. The hood in which PCRs were assembled and run was UV irradiated, and all PCR reagent mixtures were filtered through Centricon 100 columns (Millipore, Bedford, MA) before use.
Each amplification was conducted with 40 ng of hamster brain DNA. Amplification with the mollicute and spiroplasma primers followed the protocol described by Bastian et al. (4) using Pharmacia Ready-To-Go beads, which contain all reaction components except for template DNA and primers. Each bead was reconstituted with 25 μl of water, which was 0.6 μM in each primer; 40 ng of sample DNA (4 μl of a 10-ng/μl stock) was added to this. Cycling began with a 4-min melt at 95°C, followed by three cycles of 94°C (30 s), 59°C (30 s), 72°C (30 s), and then 32 cycles of 94°C (30 s), 56°C (30 s), 72°C (30 s), and a final extension at 72°C for 7 min. For nested PCR, a 1:10 dilution of the product of amplification with mollicute primers was used for a further amplification of 35 cycles using the spiroplasma primers under the same conditions.
Amplification with the universal primers used reagents from Perkin-Elmer Cetus, including AmpliTaq DNA Polymerase LD, optimized for detection of low-copy-number (<1,000) bacterial DNA sequences. For each sample, 40 ng of DNA was amplified in 50 μl of a reaction mixture whose final concentration was 50 mM KCl, 10 mM Tris HCl (pH 8.3), 1.5 mM MgCl2, 3.3 mg/ml bovine serum albumin, 0.6 μM (each) primer, 0.2 mM (each) deoxynucleoside triphosphate, and 1.25 U/50 μl AmpliTaq DNA Polymerase LD. The thermal profile employed a 4-min melt at 95°C, followed by 35 cycles of 94°C (30 s), 56°C (30 s), 72°C (30 s), and a final extension at 72°C for 7 min.
In some cases, additional cycles with the universal primers were performed using a 1:10 dilution of the first-round product and the same conditions as for the first round. In other cases, nested PCR was performed using the spiroplasma primers and a 1:10 dilution of the first-round product, following the Pharmacia Read-To-Go bead protocol.
Cloning and sequencing of amplified products.
PCR products were analyzed and purified by agarose gel electrophoresis on ethidium bromide-stained gels and cloned into the plasmid pCR 2.1 using an Original TA cloning kit (Invitrogen). Recombinant clones were picked and grown in 5 ml Luria-Bertani medium overnight, and recombinant plasmid DNAs were extracted using miniplasmid preparation kits (QIAGEN). Cloned products were sequenced in both directions using an ABI PRISM sequencer, and resulting sequences were analyzed for homologies to known genes, using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/blast).
Calculations of “infectious dose” equivalents of mollicute DNA.
The hamster brain contains approximately 2.5 mg of total DNA/g, and brains taken from scrapie-infected hamsters at late-stage disease contain ≥1010 infectious doses which cause disease in 50% of animals inoculated (ID50)/g(6, 36, 26). Thus, a 40-ng sample of genomic DNA from hamster brain, which is the sample size used in these PCR experiments, can be calculated to contain 1.6 × 105 ID50, or 1.1 × 105 infectious doses (ID), since 1 ID50 corresponds to 1.44 ID. The genome size of S. mirum is 1,300 kbp (15), and that of M. arginini is 610 kbp (51). Assuming an average molecular mass of 618 Da per bp, there are 750 genomes in 1 pg of S. mirum DNA and 1,560 genomes in 1 pg of M. arginini DNA. Thus, our methods readily detect 750 genomes of S. mirum and 1,560 genomes of M. arginini. If one TSE infectious dose is taken as one genome, then our methods can detect at least 1.1 × 105/750 = 150 times the amount of S. mirum DNA and at least 1.1 × 105/1600 = 70 times the amount of M. arginini DNA that is necessary to account for the infectivity in a 40-ng sample, if the extraction of mollicute DNA is assumed to be comparable in efficiency to that of hamster DNA. These calculations are summarized and extended to other mycoplasmas in Table 2.
TABLE 2.
Calculated detectability of representative mollicutes, if they were responsible for infectivity in TSE-infected braina
| Organism (description) | No. of kbp/genome (reference) | No. of genomes/pg | Fold excess |
|---|---|---|---|
| Spiroplasma citri (largest known spiroplasma) | 1,820 (56) | 535 | 207 |
| Spiroplasma mirum | 1,300 (15) | 750 | 148 |
| Spiroplasma platyhelix (smallest known spiroplasma) | 780 (53) | 1249 | 89 |
| Mycoplasma arginini | 610 (51) | 1598 | 69 |
| Mycoplasma genitalium (smallest known bacterium) | 580 (45) | 1680 | 66 |
Since 1 pg of S. mirum or M. arginini was readily detected in these studies, the number of genomes present in 1 pg was calculated for each mollicute shown, assuming an average molecular mass of 618 Daltons per bp. This was compared with 111,000, the number of agent genomes that must be present in 40 ng of DNA from scrapie-infected hamster brain, given that 1 g of brain contains 1010 ID50 of scrapie infectivity and yields 2.5 mg of total DNA and assuming that one genome is responsible for one infectious dose (which corresponds to 1.44 ID50). The ratio between the number of genomes in 1 pg of each molicute DNA (which, when spiked, was readily detectable by 35 cycles of PCR) and the number of genomes present in the 40-ng hamster brain DNA sample is the “fold excess”over the detection limit. This is a worst-case calculation. If the specific infectivity is less than 1 infection per organism or if the 16S rRNA gene copy number is greater than 1 (see reference 20), then the “fold excess” ratio will increase proportionately.
RESULTS
PCR with mollicute and spiroplasma primers.
PCR conditions using Pharmacia Ready-To-Go beads and mollicute-selective and spiroplasma-selective primers were optimized for the detection of the positive controls of S. mirum and M. arginini genomic DNA. The conditions chosen provided for detection and amplification of as little as 0.1 pg of S. mirum DNA alone, and detection of 1 pg of S. mirum and M. arginini DNA in 40 ng genomic DNA from hamster brain tissue was successful. The first round of amplification with the mollicute primer set yielded a PCR product of 250 bp with the S. mirum or M. arginini controls as a template. In tests of the integrity of the hamster genomic DNA, primers for the prion gene amplified an appropriately sized fragment from every sample (Fig. 2A).
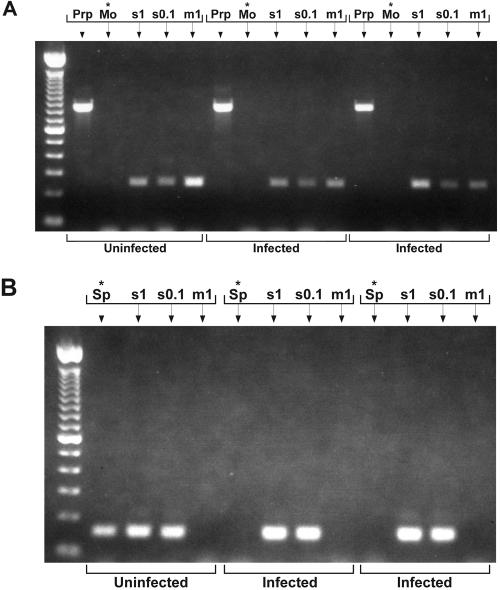
FIG. 2.
Gel bands of PCR products of three different samples from the coded panel. (A) First round of nested PCR (30 cycles). First lane shows 100-bp DNA ladder, included as molecular size marker. Lanes with an asterisk show sample plus mollicute primers (Mo) alone. All other lanes are controls, with PrP primers (PrP) to test for integrity of the sample DNA or mollicute primers plus spikes of 1 pg S. mirum DNA (s1), 0.1 pg S. mirum DNA (s0.1), or 1 pg M. arginini DNA (m1). No products were detected in any of the unspiked samples after first-round PCR with mollicute primers. (B) Second round of nested PCR (30 cycles). Lanes with an asterisk show sample plus spiroplasma primers (Sp) alone. All other lanes contain sample with the same spikes as in panel A. After second-round PCR with spiroplasma primers, product was found in one sample, which proved to be from uninfected normal hamster brain.
After 35 cycles of amplification with either the mollicute primers (Fig. 2A) or the spiroplasma primers (data not shown), there were no PCR products in any unspiked sample, indicating the absence of mollicute bacterial DNA at a level over 1 pg in any of the 20 coded samples.
To increase the sensitivity of detection, the product of the amplification with the mollicute primers was amplified with the spiroplasma primers (Fig. 2B). Nested PCR readily detected S. mirum at the 0.1-pg level, producing 150-bp fragments in both the buffer controls (data not shown) and when spiked into the brain DNA samples. The M. arginini control was not detected by the spiroplasma primers (Fig. 2B), even after amplification in the first round of PCR. When the sequence of the spiroplasma primers was checked against the 16S rRNA gene sequence of M. arginini, it was found that there were no homologies to the primers present in the M. arginini gene.
For about a fourth of the unspiked samples, the nested PCR generated product during the second round. This nested PCR, performed with mollicute primers followed by spiroplasma primers, was repeated four or more times for each sample in the panel, with similar results. In each case there were no PCR products in any unspiked sample after the first round, but about a fourth of the samples generated product during the second round. However, the particular samples which yielded product differed from experiment to experiment and included DNA from both infected and uninfected hamsters.
The PCR products from the second round of amplification were cloned and sequenced for two of the experiments. In one instance, one of the sequences isolated was 100% homologous to S. mirum DNA. Once the code was broken, the sample that produced this product proved to be an uninfected sample, and the S. mirum sequence was judged to result from an inadvertent contamination with the positive control S. mirum spike. None of the other cloned sequences matched sequences from any mollicutes, but they were homologous to 16S rRNA gene sequences from a variety of known and unknown bacteria, including Acidovorax, Variovorax, and Aquaspirillum, bacterial species which are naturally found in environmental water sources and which can be isolated from purified water systems (29, 13, 34, 30). These sequences were found as frequently in uninfected samples as in infected samples and only after 70 cycles of amplification. This is consistent with a very low level of nonspecific contamination of the laboratory or the PCR reagents by bacteria or their DNA.
PCR with broad-range universal bacterial primers.
To screen generically, we also amplified the panel using universal bacterial primers that detect virtually any eubacterium. The universal primers generated an 834-bp amplification product with the S. mirum and M. arginini controls. After 35 cycles of PCR using the universal primers, no amplified product was observed in any unspiked sample (Fig. 3A). This experiment was repeated four times or more for each sample, with no amplified product in any of the coded samples. We increased the sensitivity of detection with universal primers in two different ways: by an additional round of 35 PCR cycles using the same universal primers and by an additional round of 35 cycles using the spiroplasma primers and Pharmacia Ready-to-Go beads. In both cases, we used a 1:10 dilution of the PCR product from the first round of amplification as a template for the second. A second PCR round following universal primer amplification resulted in amplified products in almost every coded sample (Fig. 3B). When these products were cloned and sequenced, they showed sequence similarity to 16S rRNA genes from a variety of known and unknown bacterial sequences, including Acidovorax, Variovorax, Aquabacterium, and Aquaspirillum, as well as Comamonas, Bacillus, Clostridium, Pseudomonas, Staphylococcus, and Streptococcus species. As was the case for the nested PCR experiments with the mollicute and spiroplasma primers, there was no correlation between the bacterial sequence and infection status of the sample. Moreover, repeat PCR of the same sample did not amplify the same bacterial sequence. Only one sample produced a sequence related to any known mollicute species. Again, the sequence was 100% homologous to the S. mirum DNA used as a spike. Although the sample producing this sequence proved to be an infected sample, the sequence was found only once out of several amplifications of the same sample. We therefore judged this to be another instance of contamination with minute amounts of the spike, which was not apparent after 35 PCR cycles and became apparent only after 70 cycles.
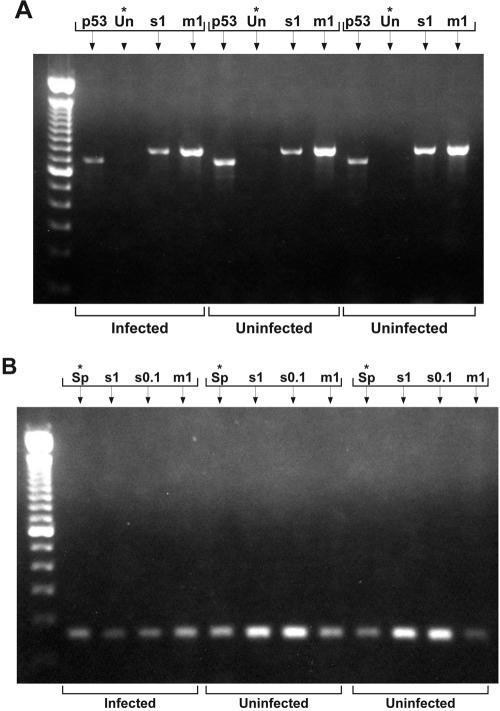
FIG. 3.
Gel bands of PCR products of three samples. (A) After 30 cycles of PCR. Lanes with an asterisk show sample plus universal primers (Un) alone. All other lanes are controls, with p53 primers (p53) to test sample DNA or universal primers and spikes labeled as in Fig. 2. (Universal primers did not detect 0.1 pg S. mirum DNA against the template background.) No products were detected in unspiked samples after the first round with universal primers. (B) After the second round of PCR (30 cycles). Lanes with an asterisk show sample plus spiroplasma primers alone. Other lanes show sample with spikes as in Fig. 2A. Product was detected in every sample, including two uninfected samples as well as infected samples.
The much greater variety of bacterial species and the increased incidence of signal in these experiments, compared with the nested PCR experiments using mollicute and spiroplasma primers, were judged to be a result of more nonspecific primer annealing of the universal primers in the first round. That is, a wider variety of environmental contaminants was identified by the universal primers. The spiroplasma primers of the second round recognized a considerable number of these, due to homology between the primers and the known 16S rRNA gene sequences of the bacteria. The results of this second series of PCR experiments are consistent with those of the first, however, and indicate that there is no bacterial species that is consistently or reliably associated with 263K scrapie infection in hamsters.
DISCUSSION
To establish whether bacteria could be the etiological agents of TSE diseases, we used PCR to search for 16S rRNA gene bacterial sequences in DNA from the well-characterized hamster 263K scrapie model, which produces predictably high concentrations of TSE infectivity in the brains of clinically affected animals. We optimized our PCR conditions to reliably detect bacterial DNA at about 1% of the concentration that would be present, if bacteria were responsible for the TSE infectivity that we know to be present (see calculations below, in discussion on “Bacteria as infectious agent in TSE disease”). That is, our assays had 150 to 300 times the sensitivity needed to reliably detect the presence of the bacterial 16S rRNA genes in 35 cycles of PCR amplification. We validated our detection limits by spiking every sample in every determination with S. mirum and M. arginini DNA at concentrations equivalent to ∼1% of the concentration necessary to account for the TSE infectivity represented in the samples. We amplified the DNA using primers developed specifically to detect spiroplasma rRNA genes and mycoplasma rRNA genes, as well as universal primers that detect all eubacteria including the mollicutes. We tested two distinct amplification protocols. We conducted all of our experiments with a standard panel of 20 blinded samples. We found no evidence that any eubacterium, including spiroplasmas or mycoplasmas, was consistently associated with scrapie-infected tissue.
This finding is at odds with that of Bastian and colleagues (3, 4), who have reported finding spiroplasma 16S rRNA gene sequences in most samples from CJD-infected humans, scrapie-infected sheep, and chronic wasting disease-infected cervids but not in control samples from the same species. Our DNA extraction method, the PCR protocol using Pharmacia Ready-to-Go beads, and the spiroplasma and mollicute primers were chosen to be essentially the same as those used by Bastian et al. It is highly unlikely that the TSE strains of these different hosts would differ from hamster scrapie in the nature of the infectious agent or in the involvement of a bacterial factor. Indeed, the 263K strain of scrapie is derived from a naturally occurring strain of scrapie in goats. Furthermore, the arguments against bacterial involvement in TSE disease that are based on quantitative considerations (see “Bacteria as the infectious agent in TSE disease” below) apply to all strains. On the contrary, the experimental hamster 263K scrapie model used in our study offers several critical advantages for demonstrating a bacterial etiology over the natural infections studied by Bastian and colleagues. (i) First and foremost, the concentration of 263K scrapie infectivity in the brains of hamsters in the advanced stages of the symptomatic infection is known and reproducible (11). As a consequence, the sensitivity required for detection of an equivalent concentration of bacterial rRNA genes, if bacteria are responsible for the infection, is also known. This allowed us to calibrate the PCR assay with concentration standards from representative mollicutes and adjust the sample mass to give a large excess of sensitivity over that needed to detect the available infectivity. Positive controls spiked at ∼1% of the infectivity concentration validated the requisite sensitivity for every sample. The failure to amplify any bacterial signal with any of the bacterial primer sets under conditions that readily amplified the controls argues strongly against bacteria as the source of TSE infectivity. (ii) The high concentration of infectivity in hamster 263K scrapie, 1010 ± 0.3 ID50 per gram of brain, results in a high signal-to-noise ratio over environmental contaminants. Nested PCR is unnecessary for detection of bacterial rRNA genes at this concentration or even at a concentration 100-fold lower than this. This eliminates the necessity of differentiating the ubiquitous environmental contaminants detected by nested amplifications from a signal of interest. It also makes the results far less vulnerable to minute inadvertent contaminations by positive control sequences used to calibrate and validate the assay. (iii) Both the growth environment and tissue collection for an experimental TSE model can be carefully controlled to minimize contamination. The hamsters in this experiment were raised under microisolators in a pathogen-controlled biosafety level 3 facility and the tissues harvested in the same environment. Even under these conditions and even with meticulous effort to minimize bacterial contamination, we still detected very low levels of contaminating bacterial sequences when the PCR was pushed to high cycle numbers. This is consistent with the experience of many other investigators who have reported trace contaminants after extensive PCR amplifications (49, 46, 18) and is evidence of the ubiquity of bacterial DNA. (iv) Laboratory controls can be perfectly matched by birth cohort and growth conditions to the infected animals. In contrast, the control tissues used by Bastian et al. (3, 4) were from different populations and were collected under different conditions from those for the infected samples. This may have introduced systematic errors that account for the lack of amplifications in the controls and near-perfect correlations with the infected samples. Since all amplifications used mycoplasma and spiroplasma selective primers, where amplifications occurred, mollicute sequences were favored. Universal primers might have revealed a much broader range of bacterial sequences in the same samples. (v) The use of laboratory animals facilitates the construction and interpretation of blind panels of coded samples. Conducting the analyses on coded specimens assures an objective and unbiased result.
Bacteria as the infectious agent in TSE disease.
Other investigators have hypothesized that bacteria cause TSE diseases either by triggering autoimmunity by molecular mimicry with bacterial antigens (19), as the agents of a toxicosis (44), or as the source of other long-term sequelae of peripheral bacterial infections (12). However, none of these proposals can be easily reconciled with the failure to demonstrate bacteria in the brains of TSE-infected rodents, given the high concentrations of TSE infectivity found there. Unlike an antigen or toxin, the pathogenic agent is self replicating and regenerating after a 100-billion-fold dilution.
The hamster brain contains approximately 2.5 mg of total DNA/g, and brains taken from scrapie-infected hamsters at late-stage disease contain ≥1010 ID50/g (6, 36, 26). The concentration of infectivity relative to hamster DNA is therefore 4 × 103 ID50/ng DNA. If pathogen and host DNA are extracted with the same efficiency, a 40-ng sample of DNA from an infected brain would contain a minimum of 230,000 pathogen genomes (where 1 genome corresponds to 1 ID [= 1.44 × 1 ID50]). If the efficiency of infection is less than 1.0 (which is likely) or the number of ribosomal gene copies per genome is greater than one (typically only one or two copies for mollicutes [1]), then the number of ribosomal gene copies per 40 ng of DNA would increase proportionately. Thus, 230,000 gene copies is a minimum estimate of the pathogen's representation in the sample, if it were responsible for the TSE infectivity. In contrast, Table 2 shows that only 750 S. mirum genomes or 1,560 M. arginini genomes were required to obtain strong PCR amplifications from either mollicute.
The amplification with universal primers extends our results to eubacteria in general. For a bacterium such as E. coli, which has 7 copies of the 16S rRNA gene per genome (20), as few as 100 genomes would be readily detected by these methods. This is 1,000 times fewer than the minimum number of genomes needed to account for the infectivity. Thus, no eubacterium amplifiable by the universal primers was present at sufficient concentrations to account for the TSE infectivity.
This conclusion is consistent with many other observations that make a bacterial etiology unlikely. One of the most compelling is our own demonstration that highly dispersed preparations of hamster scrapie infectivity readily pass through 15-nm Planova nanofilters (Asahi Corporation) that block Phi X-174, a 24-nm icosahedral bacteriophage (L. Gregori, R. G. Rohwer, et al., in preparation).
These experiments, which rule out bacteria as the etiologic agent of TSE diseases, do not rule out viruses. A number of investigators have argued persuasively that there is no compelling evidence that TSE infections are not caused by viruses or virus-size exogenous nucleus acid and that since a viral etiology is the prevailing paradigm for filterable pathogens, it should not be abandoned without incontrovertible proof to the contrary (14, 40, 43, 35, 17). The smallest viruses, at a concentration of 1.1 × 105 genomes, would be difficult to detect de novo without specific knowledge of their genome sequence or unique proteins. However, given this knowledge, the nucleic acid in 1.1 × 105 genomes of such a virus would be easily detectible by a PCR-based assay.
The investigation of natural infections, their epidemiology, pathology, and pathogenesis, is essential to a comprehensive and contextual understanding of infectious diseases. However, basic questions like the identity of the etiologic agent are better answered in the controlled environment of well-characterized experimental models. This is because the concentration of TSE infectivity in natural infections is either unknown or only poorly established from cross-species titrations into rodents or monkeys and will vary with specific agent strains, host genetics, the stage in disease progression at which the tissues are taken, and the specific piece of tissue analyzed. For example, the concentration of TSE-associated amyloid varies markedly from region to region in the brain of human TSE patients (8). Without knowledge of the infectivity concentration, a failure to amplify rRNA genes from natural cases is intrinsically uninterpretable, since the infectivity, if present, may be at a concentration below the threshold for detection. Conversely, there is no way to validate the credibility of a positive amplification without knowing whether or not the concentration of infectivity is within the detection range of the assay. Moreover, both autopsies and field collections of animal specimens are especially vulnerable to contamination. The extreme sensitivity of nested or high-cycle PCR predisposes to the detection both of environmental contaminants and of low-level contamination by positive controls. An agent that is discovered in natural infections that cannot be confirmed in well-defined, high-titer experimental tissues is unlikely to be valid.
Acknowledgments
This work was supported by the Research Service of the VA Medical Center, Baltimore, and funded by an interagency transfer from the NHLBI to the VA Medical Center, Baltimore. The work was also supported by grant R01-HL63930 from NHLBI to R.G.R. and by contract N01-NS-0-2327 from NHLBI/NINDS to the Baltimore Research and Education Foundation, Inc., at the VA Medical Center, Baltimore.
REFERENCES
- 1.Amikam, D., G. Glaser, and S. Razin. 1984. Mycoplasmas (Mollicutes) have a low number of rRNA genes. J. Bacteriol. 158:376-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bastian, F. O. 1979. Spiroplasma-like inclusions in Creutzfeldt-Jakob disease. Arch. Pathol. Lab. Med. 103:665-669. [PubMed] [Google Scholar]
- 3.Bastian, F. O., S. Dash, and R. F. Garry. 2004. Linking chronic wasting disease to scrapie by comparison of Spiroplasma mirum ribosomal DNA sequences. Exp. Mol. Pathol. 77:49-56. [DOI] [PubMed] [Google Scholar]
- 4.Bastian, F. O., and J. W. Foster. 2001. Spiroplasma sp. 16S rDNA in Creutzfeldt-Jakob disease and scrapie as shown by PCR and DNA sequence analysis. J. Neuropathol. Exp. Neurol. 60:613-620. [DOI] [PubMed] [Google Scholar]
- 5.Bastian, F. O., D. M. Purnell, and J. G. Tully. 1984. Neuropathology of spiroplasma infection in the rat brain. Am. J. Pathol. 114:496-514. [PMC free article] [PubMed] [Google Scholar]
- 6.Blum, M., M. O. Budnick, E. M. Chait, W. E. Vaz, C. MacAuley, and R. G. Rohwer. 1998. A bovine spongiform encephalopathy validation study for aprotinin and bovine serum albumin. Bio. Pharm. 11:28-34. [Google Scholar]
- 7.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, P., K. Kenney, B. Little, J. Ironside, R. Will, L. Cervenakova, R. J. Bjork, R. A. San Martin, J. Safar, R. Roos, et al. 1995. Intracerebral distribution of infectious amyloid protein in spongiform encephalopathy. Ann. Neurol 38:245-253. [DOI] [PubMed] [Google Scholar]
- 9.Brown, P., R. G. Rohwer, and D. C. Gajdusek. 1986. Newer data on the inactivation of scrapie virus or Creutzfeldt-Jakob Disease virus in brain tissue. J. Infect. Dis. 153:1145-1148. [DOI] [PubMed] [Google Scholar]
- 10.Brown, P., R. G. Rohwer, E. M. Green, and D. C. Gajdusek. 1982. Effect of chemicals, heat and histopathologic processing on high-infectivity hamster-adapted scrapie virus. J. Infect. Dis. 145:683-687. [DOI] [PubMed] [Google Scholar]
- 11.Brown, P., R. G. Rohwer, M. C. Moreau-Dubois, E. M. Green, and D. C. Gajdusek. 1981. Use of the golden Syrian hamster in the study of scrapie virus. Adv. Exp. Med. Biol. 134:365-373. [DOI] [PubMed] [Google Scholar]
- 12.Broxmeyer, L. 2004. Is mad cow disease caused by a bacteria? Med. Hypotheses 63:731-739. [DOI] [PubMed] [Google Scholar]
- 13.Brummer, I. H., A. Felske, and I. Wagner-Dobler. 2003. Diversity and seasonal variability of β-proteobacteria in biofilms of polluted rivers: analysis by temperature gradient gel electrophoresis and cloning. Appl. Environ Microbiol. 69:4463-4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunori, M., M. C. Silvestrini, and M. Pocchiari. 1988. The scrapie agent and the prion hypothesis. Trends Biochem. Sci. 13:309-313. [DOI] [PubMed] [Google Scholar]
- 15.Carle, P., F. Laigret, J. G. Tully, and J. M. Bove. 1995. Heterogeneity of genome sizes within the genus Spiroplasma. Int. J. Syst. Bacteriol. 45:178-181. [DOI] [PubMed] [Google Scholar]
- 16.Chastel, C., F. Le Goff, and I. Humphery-Smith. 1991. Multiplication and persistence of Spiroplasma melliferum strain A56 in experimentally infected suckling mice. Res. Microbiol. 142:411-417. [DOI] [PubMed] [Google Scholar]
- 17.Chesebro, B. 2003. Introduction to the transmissible spongiform encephalopathies or prion diseases. Br. Med. Bull. 66:1-20. [DOI] [PubMed] [Google Scholar]
- 18.Corless, C. E., M. Guiver, R. Borrow, V. Edwards-Jones, E. B. Kaczmarski, and A. J. Fox. 2000. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J. Clin. Microbiol. 38:1747-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebringer, A., C. Thorpe, J. Pirt, C. Wilson, P. Cunningham, and C. Ettelaie. 1997. Bovine spongiform encephalopathy: is it an autoimmune disease due to bacteria showing molecular mimicry with brain antigens? Environ. Health Perspect. 105:1172-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fogel, G. B., C. R. Collins, J. Li, and C. F. Brunk. 1999. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb. Ecol. 38:93-113. [DOI] [PubMed] [Google Scholar]
- 21.Gasparich, G. E. 2002. Spiroplasmas: evolution, adaptation and diversity. Front. Biosci. 7:d619-d640. [DOI] [PubMed] [Google Scholar]
- 22.Gasparich, G. E., K. J. Hackett, E. A. Clark, J. Renaudin, and R. F. Whitcomb. 1993. Occurrence of extrachromosomal deoxyribonucleic acids in spiroplasmas associated with plants, insects, and ticks. Plasmid 29:81-93. [DOI] [PubMed] [Google Scholar]
- 23.Gray, A., R. J. Francis, and C. L. Scholtz. 1980. Spiroplasma and Creutzfeldt-Jakob disease. Lancet ii:152. [DOI] [PubMed] [Google Scholar]
- 24.Gray, E. G. 1986. Spongiform encephalopathy: a neurocytologist's viewpoint with a note on Alzheimer's disease. Neuropathol. Appl. Neurobiol. 12:149-172. [DOI] [PubMed] [Google Scholar]
- 25.Gray, M. W., D. Sankoff, and R. J. Cedergren. 1984. On the evolutionary descent of organisms and organelles: a global phylogeny based on a highly conserved structural core in small subunit ribosomal RNA. Nucleic Acids Res. 12:5837-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gregori, L., J. A. Maring, C. MacAuley, B. Dunston, M. Rentsch, C. Kempf, and R. G. Rohwer. 2004. Partitioning of TSE infectivity during ethanol fractionation of human plasma. Biologicals 32:1-10. [DOI] [PubMed] [Google Scholar]
- 27.Hartley, E. G. 1967. Action of disinfectants on experimental mouse scrapie. Nature 213:1135. [DOI] [PubMed] [Google Scholar]
- 28.Humphery-Smith, I., C. Chastel, and F. Le Goff. 1992. Spiroplasmas and spongiform encephalopathies. Med. J. Aust. 156:142. [PubMed] [Google Scholar]
- 29.Kawai, M., E. Matsutera, H. Kanda, N. Yamaguchi, K. Tani, and M. Nasu. 2002. 16S ribosomal DNA-based analysis of bacterial diversity in purified water used in pharmaceutical manufacturing processes by PCR and denaturing gradient gel electrophoresis. Appl. Environ Microbiol. 68:699-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawai, M., J. Yamagishi, N. Yamaguchi, K. Tani, and M. Nasu. 2004. Bacterial population dynamics and community structure in a pharmaceutical manufacturing water supply system determined by real-time PCR and PCR-denaturing gradient gel electrophoresis. J. Appl. Microbiol. 97:1123-1131. [DOI] [PubMed] [Google Scholar]
- 31.Kirchhoff, H., T. Kuwabara, and M. F. Barile. 1981. Pathogenicity of Spiroplasma sp. strain SMCA in Syrian hamsters: clinical, microbiological, and histological aspects. Infect. Immun. 31:445-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leach, R. H., W. B. Matthews, and R. Will. 1983. Creutzfeldt-Jakob disease. Failure to detect spiroplasmas by cultivation and serological tests. J. Neurol. Sci. 59:349-353. [DOI] [PubMed] [Google Scholar]
- 33.Liberski, P. P., and S. Mori. 1997. The Echigo-1: a panencephalopathic strain of Creutzfeldt-Jakob disease: a passage to hamsters and ultrastructural studies. Folia Neuropathol. 35:250-254. [PubMed] [Google Scholar]
- 34.Mailloux, B. J., and M. E. Fuller. 2003. Determination of in situ bacterial growth rates in aquifers and aquifer sediments. Appl. Environ Microbiol. 69:3798-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manuelidis, L. 2003. Transmissible encephalopathies: speculations and realities. Viral Immunol. 16:123-139. [DOI] [PubMed] [Google Scholar]
- 36.Miekka, S. I., R. Y. Forng, R. G. Rohwer, C. MacAuley, R. E. Stafford, S. L. Flack, M. MacPhee, R. S. Kent, and W. N. Drohan. 2003. Inactivation of viral and prion pathogens by gamma-irradiation under conditions that maintain the integrity of human albumin. Vox Sang. 84:36-44. [DOI] [PubMed] [Google Scholar]
- 37.Neefs, J. M., Y. Van de Peer, P. De Rijk, S. Chapelle, and R. De Wachter. 1993. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 21:3025-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prusiner, S. B. 1991. Molecular biology of prion diseases. Science 252:1515-1522. [DOI] [PubMed] [Google Scholar]
- 39.Reyes, J. M., and E. M. Hoenig. 1981. Intracellular spiral inclusions in cerebral cell processes in Creutzfeldt-Jakob disease. J. Neuropathol. Exp. Neurol. 40:1-8. [PubMed] [Google Scholar]
- 40.Rohwer, R. G. 1991. The scrapie agent—a virus by any other name, p. 195-232. In B. Chesebro (ed.), Current topics in microbiology and immunology, vol. 172. Springer-Verlag, Heidelberg, Germany. [PubMed] [Google Scholar]
- 41.Rohwer, R. G. 1984. Scrapie infectious agent is virus-like in size and susceptibility to inactivation. Nature 308:658-662. [DOI] [PubMed] [Google Scholar]
- 42.Rohwer, R. G. 1984. Virus-like sensitivity of the scapie agent to heat inactivation. Science 223:600-602. [DOI] [PubMed] [Google Scholar]
- 43.Somerville, R. A. 2002. TSE agent strains and PrP: reconciling structure and function. Trends Biochem. Sci. 27:606-612. [DOI] [PubMed] [Google Scholar]
- 44.Stockdale, T. 1997. Are bacterial toxins involved in the aetiology of transmissible spongiform encephalopathies? Nutr. Health 11:301-307. [DOI] [PubMed] [Google Scholar]
- 45.Su, C. J., and J. B. Baseman. 1990. Genome size of Mycoplasma genitalium. J. Bacteriol. 172:4705-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanner, M. A., B. M. Goebel, M. A. Dojka, and N. R. Pace. 1998. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl. Environ Microbiol. 64:3110-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tully, J. G., and R. F. Whitcomb. 1990. The genus Spiroplasma, 2nd ed. Springer Verlag, New York, N.Y.
- 48.van Kuppeveld, F. J., J. T. van der Logt, A. F. Angulo, M. J. van Zoest, W. G. Quint, H. G. Niesters, J. M. Galama, and W. J. Melchers. 1992. Genus- and species-specific identification of mycoplasmas by 16S rRNA amplification. Appl. Environ Microbiol. 58:2606-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 50.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weisburg, W. G., J. G. Tully, D. L. Rose, J. P. Petzel, H. Oyaizu, D. Yang, L. Mandelco, J. Sechrest, T. G. Lawrence, J. Van Etten, et al. 1989. A phylogenetic analysis of the mycoplasmas: basis for their classification. J. Bacteriol. 171:6455-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitcomb, R. F. 1983. Culture and media for spiroplasmas. Methods Mycoplasmol. 1:147-158. [Google Scholar]
- 53.Williamson, D. L., J. R. Adams, R. F. Whitcomb, J. G. Tully, P. Carle, M. Konai, J. M. Bove, and R. B. Henegar. 1997. Spiroplasma platyhelix sp. nov., a new mollicute with unusual morphology and genome size from the dragonfly Pachydiplax longipennis. Int. J. Syst. Bacteriol. 47:763-766. [DOI] [PubMed] [Google Scholar]
- 54.Wilson, D. R., R. D. Anderson, and W. Smith. 1950. Studies in scrapie. J. Comp. Pathol. 60:267-282. [DOI] [PubMed] [Google Scholar]
- 55.Wilson, K., R. Blitchington, and R. Greene. 1990. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J. Clin. Microbiol. 28:1942-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye, F., F. Laigret, J. C. Whitley, C. Citti, L. R. Finch, P. Carle, J. Renaudin, and J. M. Bove. 1992. A physical and genetic map of the Spiroplasma citri genome. Nucleic Acids Res. 20:1559-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]