Abstract
We standardized and assessed the performance of an in-house microtiter assay for determining the susceptibilities of Mycobacterium tuberculosis clinical isolates to isoniazid based on mycobacteriophage amplification technology. Seventy isolates (43 resistant and 27 sensitive according to the BACTEC 460 radiometric method and MIC determination) were studied. The isoniazid resistance molecular mechanism was previously determined by sequencing the entire katG gene and the mabA-inhA regulatory region. The sensitivity of the mycobacteriophage-based assay in detecting isoniazid resistance was 86.1%, the specificity achieved was 92.6%, and the overall accuracy was 88.6%. In order to assess the possible influence of resistance levels on the mycobacteriophage-based-assay sensitivity, the results were analyzed according to the isoniazid MICs. All the isolates exhibiting high-level resistance (MIC ≥ 2 μg/ml) were scored as resistant by the mycobacteriophage-based assay (100% concordance), and 95% showed mutations or deletions in the catalytic domain of the katG gene. In contrast, 26.1% of the low-level-resistance strains (MICs, 0.25 to 1 μg/ml) were misclassified, and 66.7% had alterations in the mabA-inhA regulatory region. The mycobacteriophage-based assay could be used as a rapid method to detect the isoniazid susceptibility pattern, although data from those areas with high rates of low-level-resistance strains should be interpreted with caution. The features of the assay make it suitable for widespread application due to its low technical demand and cost.
Despite intensive research done to improve tuberculosis (TB) diagnosis and drug susceptibility testing techniques, TB still remains one of the most threatening curable infectious diseases. Providing rapid diagnosis and antibiotic resistance detection systems is essential to prevent transmission, although these tools should be affordable in terms of cost and technical demand for those countries with limited resources and a high incidence of TB. In the vast majority of developing countries, conventional mycobacteriological techniques for culture isolation and antibiotic susceptibility testing are slow. More-rapid and newer methods such as the BACTEC 460 method (8) and the mycobacterium growth indicator tube method (22) are expensive and require sophisticated technology. There is an urgent need for accurate, simple, rapid, and inexpensive drug susceptibility testing methods adequate for widespread implementation.
Several studies have reported promising results using mycobacteriophages for the rapid, simple, and inexpensive determination of drug susceptibilities (5, 6, 7, 23), especially for isoniazid (INH) and rifampin (RIF) resistance detection. These mycobacteriophage-based assays (MBAs) depend upon the ability of resistant mycobacteria to support phage replication after being exposed to drugs, while sensitive bacilli are not able to support phage infection. Extracellular phages are inactivated with a virucidal agent, whereas intracellular phages are protected and replicate, causing their lysis and the release of a new phage progeny detected by the production of plaques on a fast-growing Mycobacterium smegmatis lawn.
We standardized and assessed an in-house microtiter assay for INH susceptibility determination using an MBA by testing Mycobacterium tuberculosis clinical isolates and comparing the results to those obtained with the BACTEC 460 method and MIC determination. The influences of INH resistance levels and their molecular mechanism on the MBA performance were also investigated.
MATERIALS AND METHODS
Isolates.
Seventy M. tuberculosis clinical isolates were collected from specimens from TB patients attending the six tertiary hospitals of the Barcelona, Spain, area from October 1995 to September 1997. The strains were identified by using DNA probes (AccuProbe; GenProbe, Inc., San Diego, Calif.) and conventional biochemical tests. Forty-three strains were INH resistant, and 27 were INH sensitive, according to the BACTEC 460 radiometric method (Becton Dickinson, Towson, Md.) and MIC determination. The INH-sensitive strain H37Ra (ATCC 25171) and the INH-resistant strain H37Rv (ATCC 35822), both M. tuberculosis reference strains, were used in the standardization and assessment of the MBA. Susceptibility testing of all the clinical isolates recruited for the study was performed by the BACTEC 460 method as described by Heifets (8). MICs from the INH-resistant isolates were also obtained by using Middlebrook 7H10 medium incubated for 21 days with concentrations of INH of 0.125, 0.25, 0.5, 1, 2, 4, 8, 16, and 32 μg/ml. The catalase and peroxidase activities were semiquantitatively determined following the usual procedures (9).
The strains were cultured on Löwenstein-Jensen medium, and the colonies were scraped, resuspended in 0.5 ml of TE buffer (10 mM Tris, 1 mM EDTA [pH 8]), and killed by freezing at −70°C followed by heating at 80°C. The DNA was extracted using a previously described methodology (21). The entire katG gene (six fragments; 2,200 bp) and the mabA-inhA regulatory region (248 bp), which are related to INH resistance, were PCR amplified by using specific primer pairs as previously described (4). DNA sequencing was performed with the fmol DNA cycle sequencing system (Promega Corporation, Madison, WI) with ALF Express II (Amersham Pharmacia Biotech). Mutations in codon 463 of the katG gene were not taken into account.
Preparation of isolates for the MBA.
As a source of mycobacterial bacilli, fresh cultures from Löwenstein-Jensen medium were used. The mycobacterial growths were transferred to sterile screw-cap glass tubes containing six to eight glass beads in 2 ml of Middlebrook 7H9 broth (MAIM S.L., Barcelona, Spain) with 10% (vol/vol) oleic acid-albumin-dextrose-catalase (OADC) enrichment (MAIM S.L.). Suspensions were homogenized with a vortex mixer for 15 to 20 s. Large clumps were allowed to settle by allowing the suspensions to stand for 10 min. The supernatants were transferred to sterile tubes and adjusted to 106 CFU/ml with Middlebrook 7H9 broth with OADC.
Preparation of mycobacteriophage D29.
Mycobacteriophage D29 was produced as described by McNerney et al. (13). Mycobacteriophages were inoculated onto a lawn of M. smegmatis growth in Middlebrook 7H9 agar (MAIM S.L.) supplemented with OADC and 1 mM CaCl2 (Merck KgaA, Darmstadt, Germany). Phages were harvested after overnight incubation at 37°C with Middlebrook 7H9 broth with 1 mM CaCl2, passed through a 0.45-μm-pore-size filter, and stored at 4°C for up to 6 months. The mycobacteriophage stock was quantified by pipetting 10-μl aliquots of serial dilutions onto a lawn of M. smegmatis growth. The phage suspension was diluted to achieve a working titer of 107 PFU/ml prior to the mycobacteriophage assay.
INH solution for the MBA.
INH (Sigma-Aldrich Chemicals GmbH, Steinheim, Germany) was made up as a 10-mg/ml stock solution in sterile distilled water and stored at −20°C until use. The working concentration was reached by dilution with Middlebrook 7H9 broth-OADC-CaCl2 to 4 μg/ml and further diluted to 2 μg/ml (optimal concentration, determined by a checkerboarding assay) when mixed with the mycobacterial suspension in the microtiter plate.
MBA.
Based on work by Wilson et al. (23) and our own previous studies (7), the MBA was performed as follows. A 75-μl volume of M. tuberculosis suspension (106 CFU/ml) was placed in wells of sterile microtiter plates (Asahi Techno Glass, Funabasi, Japan) containing 75 μl of INH (4 μg/ml) and incubated for 72 h at 37°C. Fifty microliters of mycobacteriophage D29 (107 PFU/ml) was added, and the microtiter plates were incubated for 90 min at 37°C. Phages unable to infect the bacilli were inactivated by the addition of 0.1 ml of ferrous ammonium sulfate (30 mM), while the intracellular phages were protected and replicated within the mycobacteria, causing their lysis and the release of a new mycobacteriophage progeny. Phages were detected in 10-μl drops by the formation of plaques on the surface of a lawn of the fast-growing M. smegmatis host (Middlebrook 7H9 agar-OADC-CaCl2 with 108 CFU/ml of M. smegmatis) after overnight incubation at 37°C. Visualization of plaques on the lawn of M. smegmatis growth was enhanced by adding a blue food coloring (Supercook, Leeds, United Kingdom) to the molten Middlebrook 7H9 agar. Results were available with a total turnaround time of 4 days. A strain was considered resistant if more than 30 plaques on the M. smegmatis lawn were formed by the release of mycobacteriophages from viable M. tuberculosis bacilli after exposure to INH and was considered sensitive if fewer than 30 plaques were recorded. Positive controls (strains to be tested that were incubated with assay broth instead of INH) and a negative control (broth only) were assayed in each run of the MBA. Reference strains were assayed with new batches of reagents or mycobacteriophages in order to assess the test performance. The researchers performing the assay were blinded to the INH resistance or sensitivity of the strains to avoid predictability of results.
RESULTS
Seventy clinical isolates (43 resistant and 27 sensitive according to the BACTEC 460 method and MIC determination) were studied to assess the performance of the MBA in INH susceptibility determination. Interpretation of strains to be scored as susceptible or resistant was clear and easy (Fig. 1). The positive and negative process controls and the mycobacterial reference strains performed correctly when they were assayed (data not shown). Twenty-five of the 27 susceptible strains and 37 of the 43 resistant strains were correctly identified by the MBA, yielding an overall accuracy of 88.6% relative to the BACTEC 460 method (Table 1). The sensitivity of the MBA in detecting INH resistance was 86.1%, and the specificity achieved was 92.6%. In terms of plaque numbers, fewer than 20 plaques were observed in the cases of the correctly classified sensitive strains, except for one case in which 26 plaques were reported. Confluent lysis was observed in the two isolates misclassified as resistant. Regarding resistant isolates, the correctly determined strains produced plaques ranging from 35 in number to total or confluent lysis, while 0 to 23 plaques were found in the six discordant isolates.
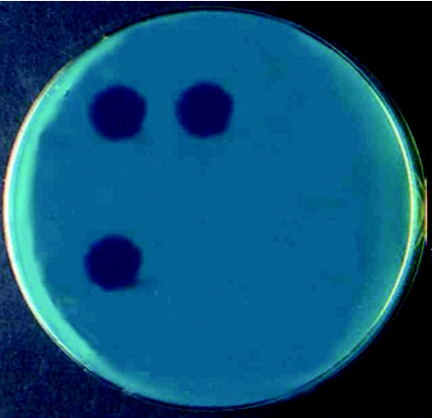
FIG. 1.
Interpretation of the MBA results with a lawn of fast-growing M. smegmatis. Upper left drop, INH-resistant strain incubated without INH (drug-free control); upper right drop, INH-resistant strain incubated with INH; bottom left drop, INH-sensitive strain incubated without INH (drug-free control); bottom right drop, INH-sensitive strain incubated with INH (drop not visible).
TABLE 1.
Phenotypic and genotypic analyses of the isoniazid-resistant isolates
| Clinical isolate | Phenotypic analysis
|
Genotypic analysis
|
||||||
|---|---|---|---|---|---|---|---|---|
| MBA resulta | MIC (μg/ml) | Activity scoresb
|
katG
|
mabA-inhA
|
||||
| Catalase | Peroxidase | Locationc | Alteration | Locationd | Alteration | |||
| 030/R | R | 0.25 | 3 | ++ | ||||
| 032/R | R | 0.25 | 3 | +++ | ||||
| 039/R | R | 0.25 | 1 | +++ | ||||
| 040/R | R | 0.25 | 1 | 0 | 94 | Asp→Asn | ||
| 046/R | R | 0.25 | 0 | +++ | −15 | C→T | ||
| 009/R | R | 0.25 | 2 | ++ | 728 | Trp→Tyr | ||
| 067/R | R | 0.25 | 0 | ++ | −15 | C→T | ||
| 061/R | R | 0.25 | 4 | +++ | −15 | C→T | ||
| 042/R | R | 0.25 | 2 | +++ | −8 | T→C | ||
| 034/R | R | 0.25 | 3 | +++ | −8 | T→C | ||
| 029/R | S | 0.5 | 1 | +++ | ||||
| 033/R | R | 0.5 | 3 | +++ | ||||
| 028/R | R | 0.5 | 0 | 0 | 172 | Ala→Thr | ||
| 047/R | R | 0.5 | 2 | +++ | −15 | C→T | ||
| 001/R | S | 0.5 | 3 | +++ | −15 | C→T | ||
| 014/R | R | 0.5 | 3 | + | −15 | C→T | ||
| 013/R | S | 0.5 | 4 | + | −15 | C→T | ||
| 011/R | S | 0.5 | 4 | + | −15 | C→T | ||
| 026/R | S | 0.5 | 4 | + | −17 | G→T | ||
| 059/R | S | 1 | 3 | ++ | ||||
| 036/R | R | 1 | 2 | + | 678 | Tyr→Cys | −15 | C→T |
| 003/R | R | 1 | 3 | ++ | −15 | C→T | ||
| 060/R | R | 1 | 2 | ++ | −15 | C→T | ||
| 017/R | R | 2 | 0 | 0 | 560 | Gly→Ala | ||
| 007/R | R | 2 | 1 | ++ | 204 | Trp→stop | ||
| 057/R | R | 2 | 0 | ++ | 189 | Asp→His | −15 | C→T |
| 004/R | R | 4 | 4 | 0 | 315 | Ser→Thr | ||
| 016/R | R | 8 | 2 | 0 | 315 | Ser→Arg | ||
| 008/R | R | 8 | 2 | 0 | 315 | Ser→Asn | ||
| 035/R | R | 8 | 0 | + | 315 | Ser→Thr | ||
| 063/R | R | 8 | 1 | + | 315 | Ser→Thr | ||
| 015/R | R | 8 | 2 | 0 | 315 | Ser→Thr | ||
| 044/R | R | 8 | 2 | ++ | 315 | Ser→Thr | ||
| 062/R | R | 8 | 2 | 0 | 315 | Ser→Thr | ||
| 023/R | R | 8 | 3 | 0 | 315 | Ser→Thr | ||
| 048/R | R | 8 | 3 | + | 315 | Ser→Thr | ||
| 002/R | R | 8 | 4 | +++ | 315 | Ser→Thr | ||
| 019/R | R | 8 | 0 | 0 | 315 | Ser→Thr | ||
| 012/R | R | 8 | 3 | + | 315 | Ser→Thr | ||
| 234 | Ala→Gly | |||||||
| 065/R | R | 16 | 0 | 0 | 315 | Ser→Thr | ||
| 005/R | R | >32 | 0 | 0 | 234 | Deletion | ||
| 155-158 | Deletion | |||||||
| 022/R | R | >32 | 0 | 0 | 152 | Deletion | ||
| 024/R | R | >32 | 0 | 0 | ||||
R, resistant to INH (presence of plaques); S, sensitive to INH (absence of plaques).
Catalase activity is reported in mm. Peroxidase activity is reported as follows: 0, no color; +, weak intense color; ++, medium intense color; +++, very intense color.
Codon affected.
Nucleotide position upstream of the gene initiation codon.
To assess the influence of resistance levels on the MBA performance, the results were analyzed according to the MICs of the INH-resistant strains (Table 1). All 20 isolates exhibiting high-level resistance (MIC ≥ 2 μg/ml) were scored as resistant by the MBA (100% concordance) (Table 2), while 6 of the 23 low-level-resistance strains (MICs, 0.25 to 1 μg/ml) were misclassified (74% accuracy) (Table 3). Nineteen of the 20 isolates (95%) presenting a high level of INH resistance had katG alterations consisting of small deletions (3 to 15 nucleotides) (n = 3) or nucleotide substitutions (n = 16) resulting in either amino acid replacement (n = 15) or stop mutation (n = 1) (Table 1). All but two strains had a single alteration in the katG gene. The genetic alterations affected the gene catalytic domain in 18 strains and the C-terminal region in only 1 strain. The most frequent mutation occurred at codon 315 (14 of 19; 73.6%). mabA-inhA regulatory region analysis showed a nucleotide substitution for 14 of the 23 strains with low-level INH resistance (60; 9%) with the following changes: C→T substitutions, involving a nucleotide at position 15 upstream of the mabA initiation codon −15 (11 of 23; 47.8%); T→C substitutions, involving nucleotide −8 (2 of 53; 8.7%); and G→T substitutions, involving nucleotide −17 (1 of 23; 4.3%). One strain with a C→T substitution involving nucleotide −15 had an additional mutation at C-terminal codon 678. Four of the six low-level-resistance strains (66.7%) misclassified as sensitive by the MBA had nucleotide substitutions at the mabA-inhA regulatory region. The two remaining strains were wild type at the analyzed targets (Table 3).
TABLE 2.
Mycobacteriophage-based assay results versus genotypes of the high-level-resistance isolates (MICs ≥ 2 μg/ml)
| INH resistance phenotypea | No. of isolates with indicated Genotype
|
Total no. of isolates | |||
|---|---|---|---|---|---|
| katG | katG + mabA-inhA | mabA-inhA | Wild type | ||
| INHr | 18 | 1 | 0 | 1 | 20 |
| INHs | 0 | 0 | 0 | 0 | 0 |
INHr, isoniazid resistant; INHs, isoniazid sensitive.
TABLE 3.
Mycobacteriophage-based assay results versus genotypes of the low-level-resistance isolates (MICs, 0.25 to 1 μg/ml)
| INH resistance phenotypea | No. of isolates with indicated genotype
|
Total no. of isolates | |||
|---|---|---|---|---|---|
| katG | katG + mabA-inhA | mabA-inhA | Wild type | ||
| INHr | 3 | 2 | 8 | 4 | 17 |
| INHs | 0 | 0 | 4 | 2 | 6 |
INHr, isoniazid resistant; INHs, isoniazid sensitive.
When the catalase and peroxidase activities of the resistant strains were correlated with the MBA results, catalase activity was detected in 73% of the strains, whereas peroxidase activity was found in 62% of these strains. The strains misclassified as sensitive by the MBA conserved catalase and peroxidase activities in all cases.
DISCUSSION
In the present paper, we compared the effectiveness of an in-house mycobacteriophage-based microtiter test in detecting INH susceptibility with that of the BACTEC 460 method and MIC determination by testing clinical isolates of M. tuberculosis with INH resistance molecular mechanisms previously determined by sequencing. We have reported here that the MBA shows high specificity and accuracy relative to the BACTEC 460 method, although the sensitivity of MBA in detecting INH resistance depends on the resistance level of the isolates (MBA demonstrated 100% concordance with the BACTEC 460 method in the case of strains demonstrating high MICs).
A possible explanation for the different results of the MBA depending on the level of INH resistance may be found by considering that the mechanisms of INH resistance are multifactorial and differ among strains with high and low resistance levels. Modifications of the katG gene catalytic domain, most frequently codon 315, are associated with high MICs. The strains with these mutations have high-level resistance in the presence of INH, being totally viable since INH is not activated, thus enabling the phage infection. The apparently conservative substitution of serine for threonine at codon 315 is of maximum benefit to M. tuberculosis since it reduces the activation of INH while maintaining a substantial catalase-peroxidase activity (14, 16, 17, 19). On the other hand, mutations affecting the C-terminal part of the katG gene are generally associated with low-level resistance (10, 11, 18) as well as mutations in the inhA operon regulatory region (which encodes the MabA and InhA enzymes involved in mycolic acid biosynthesis). Most of the low-level-resistance isolates in this work demonstrated mutations in the mabA-inhA regulatory region. These alterations result in the upregulation of gene expression and thus in increased amounts of these enzymes, which overwhelm the inhibitory action of INH (20). Since INH is in its active form, it may be acting on other possible targets (12) which render the bacilli not completely viable and incapable of supporting phage infection.
Consequently, although the overall performance of the MBA indicates that it could be used as a rapid method to assess INH susceptibility, care should be taken in interpreting MBA results in those areas with high rates of low-level-resistance strains, as they could be incorrectly identified as sensitive.
We reported the excellent performance of the MBA in determining RIF susceptibility in a previous study, achieving 100% sensitivity, specificity, and overall accuracy (7). Since INH and RIF are the most efficacious drugs included in standard TB therapy, the MBA could be considered a rapid and low-cost alternative to other usual drug susceptibility tests based on these promising results. In this work, as in the one mentioned above, the use of a microtiter plate-based methodology constituted a technical advantage over mycobacteriophage-based methodologies developed in other studies to assess drug susceptibilities (6, 13, 23). One the one hand, this format minimizes the risk of exposure to viable M. tuberculosis bacilli since small volumes of samples are assayed, and on the other hand, a large number of isolates can be processed by following a simple procedure.
The turnaround time needed to obtain drug susceptibility results is one of the major factors influencing the clinical outcome and control of transmission of multidrug-resistant TB. Liquid culture techniques (both radiometric and nonradiometric) have reduced the time needed to obtain susceptibility data in comparison to conventional techniques. Like the research that other authors (1, 3, 15) and members of our own group (2) have performed for the detection of M. tuberculosis in clinical specimens, research is required in order to standardize our in-house MBA for the direct detection of INH and RIF resistance in clinical specimens. As long as the mycobacteriophage technique is performed correctly with clinical samples, resistance results can be reported in 48 h for RIF and 4 days for INH by means of a simple-to-perform assay.
In summary, the MBA presented in this paper could be considered an alternative screening method to conventional drug susceptibility techniques and suitable for widespread implementation due to its technical characteristics: it does not require specialized equipment or reagents, the technical demand and cost are low, and the mycobacteriophage usage is simple (in terms of production, storage, and safety). Nevertheless, a limitation of the MBA in assessing INH susceptibility is its low sensitivity when testing strains with low-level INH resistance, with the possibility that erroneous data may result in those areas with a high number of isolates with such INH resistance patterns.
Laboratories located in countries with limited capital could benefit from this rapid and inexpensive assay, allowing a prompt characterization of INH and RIF resistance patterns which, in turn, would greatly improve the management of TB.
Acknowledgments
We thank R. McNerney, London School of Hygiene and Tropical Medicine, for supplying the mycobacteriophage D29 and for her kind help.
Members of the Mycobacteria Research Group of Barcelona include J. González (Hospital Clínic de Barcelona-IDIBAPS), N. Martí (Hospital Universitari Vall d'Hebrón), and M. Salvadó (Laboratori de Referència de Catalunya).
REFERENCES
- 1.Albert, H., A. Heydenrych, R. Brookes, R. J. Mole, B. Harley, E. Subotsky, R. Henry, and V. Azevedo. 2002. Performance of a rapid phage-based test, FASTPlaqueTB, to diagnose pulmonary tuberculosis from sputum specimens in South Africa. Int. J. Tuberc. Lung Dis. 6:529-537. [DOI] [PubMed] [Google Scholar]
- 2.Alcaide, F., N. Galí, J. Domínguez, P. Berlanga, S. Blanco, P. Orús, and R. Martín. 2003. Usefulness of a new mycobacteriophage-based technique for rapid diagnosis of pulmonary tuberculosis. J. Clin. Microbiol. 41:2867-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banaiee, N., M. Bobadilla-Del-Valle, S. Bardarov, Jr., P. F. Riska, P. M. Small, A. Ponce-de-Leon, W. R. Jacobs, Jr., G. F. Hatfull, and J. Sifuentes-Osornio. 2001. Luciferase reporter mycobacteriophages for detection, identification, and antibiotic susceptibility testing of Mycobacterium tuberculosis in Mexico. J. Clin. Microbiol. 39:3883-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cingolani, A., A. Antinori, M. Sanguinetti, L. Gillini, A. De Luca, B. Posteraro, F. Ardito, G. Fadda, and L. Ortona. 1999. Application of molecular methods for detection and transmission analysis of Mycobacterium tuberculosis drug resistance in patients attending a reference hospital in Italy. J. Infect. Dis. 179:1025-1029. [DOI] [PubMed] [Google Scholar]
- 5.Eltringham, I. J., F. A. Drobniewski, J. A. Mangan, P. D. Butcher, and S. M. Wilson. 1999. Evaluation of reverse transcription-PCR and a bacteriophage-based assay for rapid phenotypic detection of rifampin resistance in clinical isolates of Mycobacterium tuberculosis. J. Clin. Microbiol. 37:3524-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eltringham, L. J., S. M. Wilson, and F. A. Drobniewski. 1999. Evaluation of a bacteriophage-based assay (phage amplified biologically assay) as a rapid screen for resistance to isoniazid, ethambutol, streptomycin, pyrazinamide, and ciprofloxacin among clinical isolates of Mycobacterium tuberculosis. J. Clin. Microbiol. 37:3528-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galí, N., J. Domínguez, S. Blanco, C. Prat, M. D. Quesada, L. Matas, and V. Ausina. 2003. Utility of an in-house mycobacteriophage-based assay for rapid detection of rifampin resistance in Mycobacterium tuberculosis clinical isolates. J. Clin. Microbiol. 41:2647-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heifets, L. 1988. Qualitative and quantitative drug susceptibility tests in mycobacteriology. Am. Rev. Respir. Dis. 137:1217-1222. [DOI] [PubMed] [Google Scholar]
- 9.Heifets, L. B., and R. C. Good. 1994. Current laboratory methods for the diagnosis of tuberculosis, p. 85-110. In B. R. Bloom (ed.), Tuberculosis: pathogenesis, protection, and control. American Society for Microbiology, Washington, D.C.
- 10.Heym, B., P. M. Alzari, N. Honore, and S. T. Cole. 1995. Missense mutations in the catalase-peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Mol. Microbiol. 15:235-245. [DOI] [PubMed] [Google Scholar]
- 11.Heym, B., B. Saint-Joanis, and S. T. Cole. 1999. The molecular basis of isoniazid resistance in Mycobacterium tuberculosis. Tuber. Lung Dis. 79:267-271. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs, R. F. 1994. Multiple-drug-resistant tuberculosis. Clin. Infect. Dis. 19:1-8. [DOI] [PubMed] [Google Scholar]
- 13.McNerney, R., P. Kiepiela, K. S. Bishop, P. M. Nye, and N. G. Stoker. 2000. Rapid screening of Mycobacterium tuberculosis for susceptibility to rifampicin and streptomycin. Int. J. Tuberc. Lung Dis. 4:69-75. [PubMed] [Google Scholar]
- 14.Morlock, G. P., B. Metchock, D. Sikes, J. T. Crawford, and R. C. Cooksey. 2003. ethA, inhA, and katG loci of ethionamide-resistant clinical Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 47:3799-3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muzaffar, R., S. Batool, F. Aziz, A. Naqvi, and A. Rizvi. 2002. Evaluation of the FASTPlaqueTB assay for direct detection of Mycobacterium tuberculosis in sputum specimens. Int. J. Tuberc. Lung Dis. 6:635-640. [PubMed] [Google Scholar]
- 16.Pym, A. S., B. Saint-Joanis, and S. T. Cole. 2002. Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect. Immun. 70:4955-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramaswamy, S. V., R. Reich, S.-J. Dou, L. Jasperse, X. Pan, A. Wanger, T. Quitugua, and E. A. Graviss. 2003. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47:1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saint-Joanis, B., H. Souchon, M. Wilming, K. Johnsson, P. M. Alzari, and S. T. Cole. 1999. Use of site-directed mutagenesis to probe the structure, function and isoniazid activation of the catalase/peroxidase, KatG, from Mycobacterium tuberculosis. Biochem. J. 338:753-760. [PMC free article] [PubMed] [Google Scholar]
- 19.Scior, T., I. Meneses Morales, S. J. Garces Eisele, D. Domeyer, and S. Laufer. 2002. Antitubercular isoniazid and drug resistance of Mycobacterium tuberculosis—a review. Arch. Pharm. (Weinheim) 335:511-525. [DOI] [PubMed] [Google Scholar]
- 20.Telenti, A. 1998. Genetics and pulmonary medicine. 5. Genetics of drug resistant tuberculosis. Thorax 53:793-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Soolingen, D., P. E. de Haas, P. W. Hermans, and J. D. van Embden. 1994. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 235:196-205. [DOI] [PubMed] [Google Scholar]
- 22.Walters, S. B., and B. A. Hanna. 1996. Testing of susceptibility of Mycobacterium tuberculosis to isoniazid and rifampin by mycobacterium growth indicator tube method. J. Clin. Microbiol. 34:1565-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson, S. M., Z. Al-Suwaidi, R. McNerney, J. Porter, and F. A. Drobniewski. 1997. Evaluation of a new rapid bacteriophage-based method for the drug susceptibility testing of Mycobacterium tuberculosis. Nat. Med. 3:465-468. [DOI] [PubMed] [Google Scholar]