Abstract
We developed a novel PCR-restriction fragment length polymorphism test for the ccrB gene by using HinfI and BsmI for rapid typing of staphylococcal cassette chromosome mec (SCCmec). When tested with reference strains and methicillin-resistant Staphylococcus aureus isolates, the method proved to be valid and useful for rapid identification of four SCCmec types, especially type IV.
Methicillin-resistant Staphylococcus aureus (MRSA) has been a common worldwide hospital pathogen. In recent years, MRSA has also emerged as a significant community pathogen, especially in previously healthy children with no recognizable risk factors, and is predominantly associated with skin and soft-tissue infections (2). Some strains of community-acquired MRSA (CA-MRSA) are particularly virulent and capable of causing life-threatening diseases (14). Clinicians should consider MRSA as a potential pathogen in patients with suspected S. aureus infections in a community setting, should obtain appropriate material for bacterial culture, and should follow up on the results of susceptibility testing of all S. aureus isolates (2, 4). In addition, development of efficient molecular typing methods is needed for local monitoring of the prevalence of CA-MRSA infection or propagation of CA-MRSA strains and infection control.
The genetic basis of the methicillin resistance of MRSA is the presence of mecA, a gene coding for the low-affinity penicillin-binding protein PBP2′ (15, 18), carried by a mobile genetic element designated staphylococcal cassette chromosome mec (SCCmec) (7, 9, 11). The SCCmec element is characterized by the presence of terminal inverted and direct repeats, two site-specific recombinases (ccrA and ccrB), and the mecA gene complex (7, 11). Elucidation of the structure of SCCmec has disclosed four structural types of MRSA (9). Three types are typically found among hospital-acquired MRSA (HA-MRSA) isolates. Type I (34 kb) was identified in a 1960s isolate (strain NCTC10442); type II (53 kb) was identified in a 1982 isolate (strain N315) which is ubiquitous in Japan, Korea, and the United States; and type III (67 kb) was identified in a 1985 isolate (strain 85/2082) which is prevalent in Germany, Austria, India, and other South Asian and Pacific areas (6, 12). Type IV (20 to 24 kb) is generally carried by CA-MRSA isolates (9, 12), and at least four subtypes have been reported (9). In 2004, type V was reported in an isolate of CA-MRSA, in which the only difference was the presence of a restriction-modification system composed of the ccrC gene and the surrounding open reading frames (10).
Conventional multiplex PCR (13, 17) or quantitative PCR (qPCR) (3) assays for rapid SCCmec typing have been developed, based on sequence variations in the mecA complex and/or the ccr gene complex. These methods have proved useful as tools for rapid tentative identification (17) or molecular epidemiological screening of the SCCmec types (3). However, conventional PCR assays using five to nine primer pairs in a single-tube assay can give various sensitivities and a chance of contamination, requiring further probe hybridization for the target genes. The qPCR assay requires expensive reagents and instruments, limiting its use in many microbiology laboratories. Thus, the development of an alternative rapid and cost-effective confirmatory test would be of value.
In this study, we developed a novel rapid SCCmec typing method by PCR amplification of the ccrB gene in combination with restriction fragment length polymorphism (RFLP) with restriction endonucleases HinfI and BsmI. The ccr gene complex, ccrA and ccrB, encodes products responsible for integration and excision of the SCCmec element (8). The ccrB gene was chosen as the target gene to be amplified because it has highly conserved sequences compared to the ccrA gene. To design primers, the ccrB genes of SCCmec types I (GenBank accession number AB033763), II (GenBank accession number D86934), III (GenBank accession number AB037671), and IV (GenBank accession numbers AB097677, AB096217, AB063172, and AB063173) were aligned with ClustalW to localize homologous regions. The forward primer was 5′-GGC TAT TAT CAA GGC AAT TTA CC-3′, and the reverse primer was 5′-ACT TTA TCA CTT TTG ACT ATT TCG-3′ (PrimerExpress 2.0 software; PE Biosystems, Foster City, Calif.) (Fig. 1). Chromosomal DNA from MRSA was extracted with the Wizard genomic DNA preparation kit (Promega, Madison, Wis.) by using lysostaphin (0.5 mg/ml) and RNase (0.3 mg/ml); 1 μg of each sample was used as template DNA. Thermal cycling was set at 30 cycles of 30 s for denaturation at 94°C, 1 min for annealing at 50°C, and 2 min for elongation at 72°C. Purified PCR products were digested with HinfI and BsmI (New England Biolabs Inc.) and analyzed by 2% agarose gel electrophoresis.
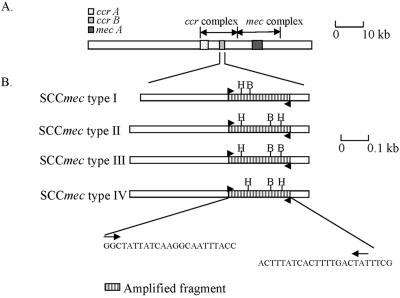
FIG. 1.
Physical map of SCCmec structure showing the ccrB gene and restriction sites for PCR-RFLP analysis. (A) Genomic structure of SCCmec. Locations of the mec complex and ccr complex are illustrated. (B) Sequence of the ccrB gene amplified by PCR in this study. Thick arrows indicate the primers. H, HinfI; B, BsmI.
The PCR-RFLP method was validated with two sets of representative strains of SCCmec types I to IV (HPV107, BK2464, ANS46, MW2, NCTC10442, N315, 85/2082, and JCSC4044). As shown in Fig. 2, four distinctive RFLP patterns were observed. We then applied the PCR-RFLP SCCmec typing method to 42 MRSA isolates obtained from clinical cultures during a 6-month period; they were selected on the basis of antibiogram results. Results showed 9 isolates of SCCmec type IV (21%), 22 of type II (53%), and 11 of type III (26%). These results matched exactly those obtained by the multiplex PCR assay (18; data not shown). Interestingly, two out of nine MRSA-SCCmec type IV isolates were from patients with cellulitis or necrotizing fasciitis in a community setting. The other MRSA-SCCmec type IV isolates were from clinical samples in a hospital setting, suggesting that CA- and HA-MRSA are likely to coexist in our hospital (Table 1). A similar situation has been previously reported in hospitals in the United States (1). The genetic background of nine MRSA-SCCmec type IV isolates was further analyzed by multilocus sequence typing and pulsed-field gel electrophoresis (PFGE). All SCCmec type IV MRSA isolates from the hospital setting showed PFGE type A (sequence type 72 [ST72], 1-4-1-8-4-4-3), except for one isolate (PFGE type C; ST239, 2-3-1-1-4-4-3), while isolates from the community setting revealed PFGE type B (ST1, 1-1-1-1-1-1-1) or PFGE type D (ST375, 19-23-15-2-19-20-42). This suggests that the PFGE type A strain may propagate in our hospital environment.
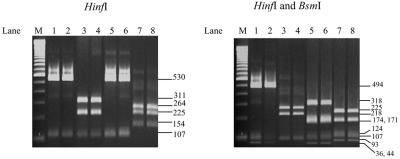
FIG. 2.
Agarose gel electrophoresis showing PCR-RFLP patterns of the 643-bp amplified ccrB gene fragment from reference MRSA strains of four SCCmec types digested by HinfI (left) and by HinfI and BsmI (right). Lanes: 1, HPV107; 2, NCTC10442; 3, BK2464; 4, N315; 5, ANS46; 6, 85/2082; 7, MW2; 8, JCSC4044; M, 100-bp ladder marker.
TABLE 1.
Results of antibiogram analysis and genotyping of nine MRSA isolates of SCCmec type IV identified by PCR-RFLP analysis of the ccrB gene
| Strain | Origin | Sample | Antibiogramdresult
|
MLSTa(STe, allelic profilef) | PFGE patterng | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CEF | CIP | CLI | ERY | GEN | OXA | PEN | TZP | TET | VAN | |||||
| K-1 | HA | Sputum | R | S | S | R | S | R | R | R | S | S | 72, 1-4-1-8-4-4-3 | A |
| K-2 | HA | Wound | I | S | S | R | S | R | R | R | S | S | 72, 1-4-1-8-4-4-3 | A |
| K-3 | HA | Sputum | S | S | S | R | S | R | R | I | S | S | 72, 1-4-1-8-4-4-3 | A |
| K-4 | HA | Urine | S | S | S | R | S | R | R | I | S | S | 72, 1-4-1-8-4-4-3 | A |
| K-5 | CA | Woundb | S | S | R | S | S | R | R | R | S | S | 375, 19-23-15-2-19-20-42 | D |
| K-6 | HA | Ear discharge | S | S | S | R | S | R | R | R | S | S | 72, 1-4-1-8-4-4-3 | A |
| K-7 | HA | Nasal swab | S | S | S | R | R | R | R | R | S | S | 72, 1-4-1-8-4-4-3 | A1 |
| K-8 | CA | Woundc | S | S | S | R | S | R | R | I | S | S | 1, 1-1-1-1-1-1-1 | B |
| K-9 | HA | Nasal swab | S | S | S | R | I | R | R | I | S | S | 239, 2-3-1-1-4-4-3 | C |
MLST, multilocus sequence typing.
Cellulities.
Necrotizing fasciitis.
Antibiotic abbreviations: CEF, cephalothin; CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; GEN, gentamicin; OXA, oxacillin; PEN, penicillin; TZP, piperacillin-tazobactam; TET, tetracycline; VAN, vancomycin. R, resistant; S, susceptible; I, intermediate.
ST, sequence type.
Allelic profile: arcC aroE glpF gmk pta tpi yqiL.
According to criteria of Tenover et al. (19).
Although the PCR-RFLP method used in this study includes two steps of restriction digestion, with HinfI followed by BsmI, one-step restriction digestion with HinfI is enough for the identification of SCCmec type II and IV strains. The second step with BsmI digestion is only necessary for discriminating type I and III strains. Since no SCCmec type I strains were found among 42 clinical MRSA isolates tested, identifying SCCmec types II to IV by one-step digestion with HinfI seems possible. With one-step digestion, we have successfully discriminated nine isolates of SCCmec type IV.
This study demonstrates the value of a novel PCR-RFLP analysis of the ccrB gene as a rapid typing method for the SCCmec elements (types I to IV) in HA- or CA-MRSA strains. Since the ccrC gene of type V shows approximately 37.4% homology to the ccrB gene of types I to IV at the nucleic acid level, the PCR-RFLP method is not likely to identify type V. We confirmed that the PCR-RFLP method is a highly specific, simple, time-effective alternative to multiplex PCR methods (3, 13, 17). The PCR-RFLP test can be achieved with just one pair of primers and one or two restriction enzymes ($3 to $4 per sample), and more than 100 samples can be processed in a day. The cost and efficiency of the PCR-RFLP method are comparable to those of the qPCR method published previously (3). Especially, it seems to be a very useful tool for rapid identification of SCCmec type IV MRSA in clinical laboratories, since serious MRSA infection has been increasingly reported in persons without identified predisposing risk factors, including recent healthcare exposure (5).
Acknowledgments
We thank Keiichi Hiramatsu (Juntendo University, Tokyo, Japan) and Herminia de Lencastre (Instituto de Technologia Quimica e Biologica, Oeiras, Portugal) for reference strains. We thank Hee Sun Sim for excellent technical assistance.
REFERENCES
- 1.Charlebois, E. D., D. R. Bangsberg, N. J. Moss, M. R. Moore, A. R. Moss, H. F. Chambers, and F. Perdreau-Remington. 2002. Population-based community prevalence of methicillin-resistant Staphylococcus aureus in the urban poor of San Francisco. Clin. Infect. Dis. 34:425-433. [DOI] [PubMed] [Google Scholar]
- 2.Eady, E. A., and J. H. Cove. 2003. Staphylococcal resistance revisited: community-acquired methicillin resistant Staphylococcus aureus—an emerging problem for the management of skin and soft tissue infections. Curr. Opin. Infect. Dis. 16:103-124. [DOI] [PubMed] [Google Scholar]
- 3.Francois, P., G. Renzi, D. Pittet, M. Bento, D. Lew, S. Harbarth, P. Vaudaux, and J. Chrenzel. 2004. A novel multiplex real-time PCR assay for rapid typing of major staphylococcal cassette chromosome mec elements. J. Clin. Microbiol. 42:3309-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hageman, J. C., M. Morrison, L. T. Sanza, K. Como-Sabetti, J. A. Jernigan, K. Harriman, L. H. Harrison, R. Lynfield, and M. M. Farley for the Active Bacterial Core Surveillance Program of the Emerging Infections Program Network. 2005. Methicillin-resistant Staphylococcus aureus disease in three communities. N. Engl. J. Med. 352:1436-1444. [DOI] [PubMed] [Google Scholar]
- 5.Herold, B. C., L. C. Immergluck, M. C. Maranan, D. S. Lauderdale, R. E. Gaskin, S. Boyle-Vavra, C. D. Leitch, and R. S. Daum. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593-598. [DOI] [PubMed] [Google Scholar]
- 6.Hiramatsu, K., L. Cui, M. Kuroda, and T. Ito. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9:486-493. [DOI] [PubMed] [Google Scholar]
- 7.Ito, T., Y. Katayama, and K. Hiramatsu. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 43:1449-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito, T., K. Okuma, X. X. Ma, H. Yuzawa, and K. Hiramatsu. 2003. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist. Updates 6:41-52. [DOI] [PubMed] [Google Scholar]
- 10.Ito, T., X. X. Ma, F. Yakeuchi, K. Okuma, H. Yyzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcal cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katayama, Y., F. Takeuchi, T. Ito, X. X. Ma, Y. Ui-Mizutani, I. Kobayashi, and K. Hiramatsu. 2003. Identification in methicillin-susceptible Staphylococcus hominis of an active primordial mobile genetic element for the staphylococcal cassette chromosome mec of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 185:2711-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maguire, G. P., A. D. Arthur, P. J. Boustead, B. Dwyer, and B. J. Currie. 1998. Clinical experience and outcomes of community-acquired and nosocomial methicillin-resistant Staphylococcus aureus in a northern Australian hospital. J. Hosp. Infect. 38:273-281. [DOI] [PubMed] [Google Scholar]
- 15.Matsuhashi, M., M. D. Song, F. Ishimoto, M. Wachi, M. Doi, M. Inoue, K. Ubukata, N. Yamashita, and M. Konno. 1986. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to β-lactam antibiotics in Staphylococcus aureus. J. Bacteriol. 167:975-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reference deleted.
- 17.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song, M. D., M. Wachi, M. Doi, F. Ishino, and M. Matsuhashi. 1987. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 221:167-171. [DOI] [PubMed] [Google Scholar]
- 19.Tenover, F. C., R. D. Arbeit, and R. V. Goering. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]