Abstract
Porphyromonas gingivalis is an important periodontal pathogen that can be isolated from both active and inactive periodontal lesions. Apparently, differences in virulence between P. gingivalis strains exist, but the mechanisms underlying these differences are not yet fully understood. To obtain more information about pathogenicity and virulence of P. gingivalis, it is relevant to assess the genetic population structure of the species and to examine the occurrence of putative virulence factors against the genetic background. Presently, multilocus sequence typing (MLST) is the best method for analyzing bacterial population structures. Forty P. gingivalis strains from worldwide sources were analyzed by MLST. Internal 310- to 420-bp DNA fragments of the eight ubiquitous chromosomal genes, ftsQ, hagB, gdpxJ, pepO, mcmA, recA, pga, and nah, were amplified by PCR and then sequenced. The number of alleles at individual loci ranged from 2 to 19, and a total of 33 allelic profiles, or sequence types (STs), were identified. Nucleotide variation between alleles was located at one or a few sites. Identical or similar STs were found in isolates from different geographic regions. Our results showed signs of a clonal population structure with a level of recombination not as high as that previously suggested for the species. We also found that P. gingivalis isolates from individual patients were genetically heterogeneous.
Destructive periodontal disease represents several entities with different clinical presentations and probably different pathogenic mechanisms. However, all periodontal diseases are infectious (43). Accumulated evidence shows that Porphyromonas gingivalis is frequently detected in patients with various forms of periodontitis, and it is considered a major periodontal pathogen (15, 34, 43, 44). In addition, P. gingivalis has been reported to cause extraoral infections, such as lung and brain abscess and pulmonary infection (20, 39). It has also been suggested by several researchers that P. gingivalis may contribute to the development of atheromas in cardiovascular disease (4, 36).
The fact that P. gingivalis can be present both in periodontal pockets undergoing destruction and in healthy gingival sulci suggests a heterogeneous species with subpopulations of high and low pathogenicities (7, 12, 25, 32). It has also been claimed that differences in progression rates of infections may be due to differences in virulence of the infecting strains (13, 14, 18, 24).
A number of virulence factors are known for P. gingivalis, including fimbriae, lipopolysaccharide, collagenase, and cysteine proteinases with trypsin-like activity (15, 35, 38, 47). These factors may contribute to neutrophil accumulation at infected sites as well as to pocket deepening and gingival bleeding on probing, which are important clinical signs of periodontitis (32). To evaluate whether differences in virulence in P. gingivalis strains reflect the existence of particular pathogenic, evolutionary lineages, it is necessary to obtain more information about the population structure of this species.
Since the genetic diversity among isolates of P. gingivalis is a reflection of sequence differences in their chromosomal genes, several molecular typing methods have been used as tools for characterization of individual strains (12, 26, 27, 30, 35). However, the discriminatory powers and reproducibilities of these methods are variable (5, 10, 12).
Pulsed-field gel electrophoresis and multilocus sequence typing (MLST) are the two methods most commonly used to discriminate among isolates below the species level. Through pulsed-field gel electrophoresis, one is able to identify highly variable and rapidly evolving individual loci within a bacterial species for the study of short-term epidemiology (28). In contrast, MLST detects allelic variation at multiple housekeeping loci that are accumulating very slowly in the bacterial population. The discriminatory power of MLST is achieved by the DNA sequence analysis of 7 to 10 fragments of housekeeping genes (approximately 450 bp), which can be used to detect the degree of genetic diversity within the bacterial population of interest (5, 28). The technique is well suited to study large strain collections of broad origin (long-term epidemiology). MLST schemes have been created for a variety of bacteria, such as Neisseria meningitidis (28), Streptococcus pneumoniae (8), Streptococcus pyogenes (9), Haemophilus influenzae (29), Campylobacter jejuni (6), Staphylococcus aureus (10), Salmonella spp. (23), and Bacillus cereus (17). Recently, Koehler et al. (22) developed an MLST scheme for P. gingivalis. Their results, based on the analysis of 19 strains limited to Germany, indicated a panmictic population structure with frequent recombinations. This is in agreement with earlier phylogenetic studies of P. gingivalis based on multilocus enzyme electrophoresis (MLEE), ribotyping, and random amplified polymorphic DNA (1, 2, 12, 26, 30).
The purpose of the present investigation was to extend the MLST study of P. gingivalis by applying a modified MLST scheme to a larger collection of isolates from worldwide geographic regions. Our results indicate that some clones of P. gingivalis can be geographically widespread and that the level of recombination is not as high as previously suggested (22). We also found that P. gingivalis isolates from individual patients were genetically heterogeneous.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A total of 40 P. gingivalis strains were analyzed. Thirty-nine strains were obtained from the collection of the Department of Oral Microbiology, Academic Center for Dentistry Amsterdam (ACTA), Amsterdam, The Netherlands, and included strains from worldwide sources (Table 1). Thirty-seven of these strains were recovered from patients with chronic, marginal periodontitis, and two were strains used in an experimental study of periodontitis in monkeys. Twenty-six strains were recognized by their designation number from the study of Loos et al. (26), and eight of these were also included in the study by Ménard and Mouton (30). The 13 remaining strains were isolated from four different periodontitis patients from Indonesia (Table 1). The strain A7A1-28 was referred to by Loos et al. (26) as ATCC 53977, also analyzed by Koehler et al. (22). This isolate and isolates CLN 17-6-1 and CLN 16-6-4 were recovered from the same individual, a patient with non-insulin-dependent diabetes mellitus (26). In addition, we included one P. gingivalis isolate from the Department of Oral Biology, Dental Faculty, University of Oslo, Oslo, Norway, recovered from a patient with chronic, marginal periodontitis.
TABLE 1.
Origins and STs of 40 P. gingivalis isolates examined by MLST
| Strain designation | ST no. | Origin | Type of infection | Isolation site | Geographic origin |
|---|---|---|---|---|---|
| A7A1-28 | 1 | Human | Periodontitis | Subgingival | Sacaton, AZ |
| CLN17-6-1 | 1 | Human | Periodontitis | Subgingival | Sacaton, AZ |
| CLN16-6-4 | 1 | Human | Periodontitis | Subgingival | Sacaton, AZ |
| W12 | 25 | Human | Periodontitis | Subgingival | Birmingham, AL |
| 16V2K-10 | 33 | Monkey | Experimental periodontitis | Subgingival | Buffalo, NY |
| 18Y2K-8 | 32 | Monkey | Experimental periodontitis | Subgingival | Buffalo, NY |
| AJW4 | 45 | Human | Periodontitis | Subgingival | Buffalo, NY |
| 34-4-4 | 31 | Human | Periodontitis | Subgingival | Minneapolis, MN |
| BH18/10 | 44 | Human | Periodontitis | Subgingival | Winnipeg, Manitoba, Canada |
| BH6/26 | 50 | Human | Periodontitis | Subgingival | Winnipeg, Manitoba, Canada |
| 195PG1 | 39 | Human | Periodontitis | Subgingival | Indonesia |
| 195PG2 | 28 | Human | Periodontitis | Subgingival | Indonesia |
| 195PG3 | 38 | Human | Periodontitis | Subgingival | Indonesia |
| 196PG1 | 28 | Human | Periodontitis | Subgingival | Indonesia |
| 196PG2 | 28 | Human | Periodontitis | Subgingival | Indonesia |
| 212PG1 | 36 | Human | Periodontitis | Subgingival | Indonesia |
| 212PG2 | 35 | Human | Periodontitis | Subgingival | Indonesia |
| 212PG3 | 34 | Human | Periodontitis | Subgingival | Indonesia |
| 212PG4 | 49 | Human | Periodontitis | Subgingival | Indonesia |
| 213PG1 | 22 | Human | Periodontitis | Subgingival | Indonesia |
| 213PG2 | 37 | Human | Periodontitis | Subgingival | Indonesia |
| 213PG3 | 22 | Human | Periodontitis | Subgingival | Indonesia |
| 213PG4 | 22 | Human | Periodontitis | Subgingival | Indonesia |
| B42 | 29 | Human | Odontogenic abscess | Incision | Hofu, Japan |
| B129 | 48 | Human | Odontogenic abscess | Incision | Hofu, Japan |
| ESO127 | 8 | Human | Periodontitis | Subgingival | Okayama, Japan |
| ESO75 | 26 | Human | Periodontitis | Subgingival | Okayama, Japan |
| OMG406 | 27 | Human | Periodontitis | Subgingival | Kenya |
| SP1-6 | 21 | Human | Periodontitis | Subgingival | Oslo, Norway |
| OMG268 | 46 | Human | Periodontitis | Subgingival | Gothenburg, Sweden |
| 816B | 30 | Human | Periodontitis | Subgingival | Malmö, Sweden |
| 332-2 | 23 | Human | Juvenile periodontitis | Subgingival | Umeå, Sweden |
| 295-1 | 47 | Human | Periodontitis | Subgingival | Umeå, Sweden |
| 102 | 42 | Human | Root canal infection | Root canal | Umeå, Sweden |
| Be-c | 25 | Human | Root canal infection | Root canal | Umeå, Sweden |
| OMZ481 | 20 | Human | Periodontitis | Subgingival | Zurich, Switzerland |
| OMZ482 | 41 | Human | Periodontitis | Subgingival | Zurich, Switzerland |
| OMZ479 | 40 | Human | Periodontitis | Subgingival | Zurich, Switzerland |
| 13JC | 24 | Human | Periodontitis | Submarginal | Rennes, France |
| 102822 | 43 | Human | Periodontitis | Subgingival | Unknown |
Together with these 40 isolates, we received 3 isolates, from sheep, dog, and cat, respectively, that were included and characterized in the study by Loos et al. (26). Identification of these isolates by sequencing of the 16S rRNA gene (37), however, revealed another Porphyromonas species, Porphyromonas gulae (11).
The strains were received as pure cultures in anaerobic transport tubes containing blood agar and were immediately on arrival plated onto Columbia agar plates and incubated anaerobically (90% N2, 5% H2, 5% CO2) at 37°C (Anoxomat WS9000; Mart, Lichtenvoorde, The Netherlands). After incubation for 7 to 14 days, the cultures were harvested for DNA extraction.
Preparation of DNA, PCR amplification, and selection of genes.
One loop of bacterial growth was suspended in 450 μl of distilled water, and DNA was extracted by using the hexadecyltrimethyl-ammonium bromide procedure (40). The DNA obtained from each sample was diluted 1:5 with distilled, sterile water. PCR amplification of eight ubiquitous genes (Table 2) was performed with MicroAmp reaction tubes (Applied Biosystems) and a GeneAMP system 9700 (ABI, Foster City, CA). The reaction volume of 50 μl consisted of 1 μl diluted (1:5) DNA, 5 μl 10× PCR buffer, 1 μl (10 μM) of forward and reverse primers (MWG-Biotech Ag), 4 μl of 10 μM deoxynucleoside triphosphate mix solution (Applied Biosystems), 0.25 μl of DNA Ampli Taq DNA polymerase (Applied Biosystems), and 37.75 μl distilled, sterile water. PCR primers and amplifying conditions were as described by Koehler et al. (22). PCR amplification products were examined by agarose gel electrophoresis in Tris-borate-EDTA buffer. Gels were stained for 10 min in a solution of ethidium bromide, washed for 10 min in deionized, distilled water, and photographed under UV light (312 nm). Fragments, 310 to 420 bp, from eight (ftsQ, hagB, gdpxJ, pepO, mcmA, recA, pga, and nah) of the 10 chromosomal P. gingivalis genes chosen by Koehler et al. (22), were used.
TABLE 2.
Allele distribution at eight loci in 33 STs of P. gingivalisa
| Allele no.a | Locus
|
|||||||
|---|---|---|---|---|---|---|---|---|
| pga | hagB | recA | mcmA | ftsQ | gdpxJ | nah | pepO | |
| 1 | 1 | 13 | 15 | 4 | 1 | 31 | 5 | |
| 2 | 1 | 8 | 2 | |||||
| 3 | 15 | 2 | 1 | 9 | 2 | 3 | 1 | |
| 4 | 1 | 5 | 3 | |||||
| 5 | 2 | 6 | 3 | 2 | 4 | 2 | ||
| 6 | 1 | 1 | 2 | 6 | 4 | |||
| 7 | 5 | 2 | 1 | 2 | ||||
| 8 | 2 | 1 | 3 | |||||
| 9 | 1 | 3 | ||||||
| 10 | 2 | 1 | 2 | 2 | ||||
| 11 | 1 | |||||||
| 12 | 1 | 2 | 2 | |||||
| 13 | 1 | 1 | 1 | |||||
| 14 | 1 | 2 | 1 | 1 | 1 | |||
| 15 | 1 | 1 | 1 | 2 | 1 | |||
| 16 | 1 | 1 | 1 | 6 | 1 | 5 | ||
| 17 | 1 | 1 | 2 | 4 | 2 | |||
| 18 | 1 | 3 | 2 | 1 | 1 | |||
| 19 | 1 | 1 | 1 | 1 | ||||
| 20 | 3 | 1 | 1 | 1 | ||||
| 21 | 1 | 1 | 1 | 1 | ||||
| 22 | 1 | 1 | 1 | 1 | ||||
| 23 | 1 | 1 | ||||||
| 24 | 1 | |||||||
| 25 | 1 | |||||||
| 26 | 1 | |||||||
Alleles are according to Koehler et al. (22). New alleles are numbered consecutively in column 1 and marked in bold in other columns.
Nucleotide sequencing.
The PCR products were purified by using an exonuclease-alkaline phosphate kit (Exo SAP-IT; USB Corporation, Cleveland, OH), as described by the manufacturer. The primers for PCR amplification were diluted at 1.6 μM and used to sequence the PCR products on both strands. DNA sequencing was performed with an ABI Prism Big Dye Terminator (v.1.1) cycle sequencing ready reaction kit (v.2.0) according to the manufacturer's recommendation, and sequence reactions were analyzed with an ABI 377 DNA sequencer (Applied Biosystems) using 5% Long Ranger electrophoresis gel (Cambrex Bio Science Rockland, Inc., Rockland, MA).
Data analyses.
After electrophoresis, the complementary strands were assembled, and the sequences were edited using the Auto Assembler v.2.1 program (Applied Biosystems, Foster City, CA). For each gene, the sequence fragments of the 40 isolates were compared via the Internet, using the nucleotide-nucleotide BLAST program (http://www.ncbi.nlm.nih.gov/BLAST).
Alleles identical to those found in GenBank were put in an MLST scheme and assigned the same numbers used by Koehler et al. (22). New alleles were given consecutive and higher numbers by computation with a UNIX computer script developed by Nicolas J. Tourasse (School of Pharmacy, University of Oslo), as utilized by Helgason et al. (17).
On the basis of the combination of its alleles at the eight loci, each strain was assigned an allelic profile, and distinct profiles were designated multilocus sequence types (STs) (17). The standard MLST statistics, numbers of polymorphic sites, average G+C contents, allele frequencies, and phylogenetic tree were computed by utilization of the START software v.1.05 (21). To assess their genetic relationships, a dendrogram based on the allelic profile of each isolate was constructed by the unweighted-pair group method with arithmetic averages (45, 46). In addition, the START software was utilized for the calculation of the index of association (IA) (42) and for the calculation of pairwise ratios of nonsynonymous to synonymous substitutions (dN/dS) (31). Split decomposition analysis was performed with SplitsTree v.4.0 for all genes and gene fragments (19).
The genetic diversity for each locus (h) among the isolates was calculated as h = 1 − Σ xi2 [n/(n−1)], where xi is the frequency of the ith allele at the locus and n is the number of isolates (27).
RESULTS
Allelic profiles and STs of 40 P. gingivalis isolates.
The fragments analyzed in this study ranged from 310 bp (nah) to 420 bp (ftsQ and mcmA) (Table 3). The average G+C content for each gene was estimated, from 48.5% for the pepO gene to 57.9% for the hagB gene. For comparison, the average G+C content for the whole genome of strain W83 (33) is 48.3%.
TABLE 3.
Genetic diversity at eight loci in 33 STs of 40 isolates of P. gingivalis
| Locus | Fragment length (bp) | No. of alleles | No. of polymorphic sites | % Variable nucleotide sites | Avg G + C content (%) | Avg dN/dS ratio | Genetic diversity |
|---|---|---|---|---|---|---|---|
| pga | 400 | 13 | 10 | 2.5 | 53.7 | 0.0889 | 0.747 |
| hagB | 380 | 15 | 7 | 1.8 | 57.9 | 0.3277 | 0.808 |
| recA | 400 | 7 | 2 | 0.5 | 53.1 | 0.0000 | 0.695 |
| mcmA | 420 | 12 | 6 | 1.5 | 51.5 | 0.0244 | 0.855 |
| ftsQ | 420 | 19 | 19 | 4.5 | 49.1 | 0.2897 | 0.918 |
| gdpxJ | 380 | 16 | 19 | 5.0 | 55.5 | 0.2507 | 0.906 |
| nah | 380 | 2 | 1 | 0.3 | 51.6 | 0.085 | |
| pepO | 400 | 15 | 9 | 2.3 | 48.5 | 0.1691 | 0.899 |
The average number of alleles per locus for the eight genes was 12.4 (range, 2 to 19). No insertions or deletions were observed for any of the sequences, and for all eight loci the 40 sequences were aligned without gaps and numbered as shown in Table 4. The sequences revealed from 0.3% (nah) to 5.0% (gdpxJ) variable sites, and the number of polymorphic sites ranged between 1 and 19 for the different genes. Variation between alleles was usually located at one or a few nonadjacent nucleotide sites. The average dN/dS values are shown in Table 3. All values are <1, indicating that most of the sequence variability identified is selectively neutral. For the eight loci, the amount of synonymous substitutions were at least three times more frequent than the amino acid changes at any locus (1/0.3277, hagB gene) (Table 3).
TABLE 4.
Allele numbers at eight loci in 33 STs of 40 P. gingivalis isolates
| ST no.a | No. of alleles at locus
|
|||||||
|---|---|---|---|---|---|---|---|---|
| pga | hagB | recA | mcmA | ftsQ | gdpxJ | nah | pepO | |
| 1 | 1 | 1 | 1 | 1 | 1 | 12 | 1 | 1 |
| 8 | 10 | 7 | 5 | 8 | 7 | 4 | 1 | 6 |
| 20 | 11 | 1 | 6 | 6 | 3 | 10 | 1 | 2 |
| 21 | 7 | 21 | 1 | 1 | 25 | 10 | 1 | 2 |
| 22 | 5 | 1 | 2 | 15 | 10 | 4 | 1 | 19 |
| 23 | 5 | 1 | 2 | 4 | 14 | 16 | 1 | 1 |
| 24 | 3 | 1 | 5 | 4 | 16 | 6 | 1 | 1 |
| 25 | 3 | 2 | 5 | 4 | 16 | 6 | 1 | 1 |
| 26 | 9 | 15 | 5 | 3 | 5 | 5 | 1 | 1 |
| 27 | 16 | 1 | 4 | 4 | 5 | 8 | 1 | 16 |
| 28 | 12 | 16 | 2 | 4 | 20 | 12 | 1 | 18 |
| 29 | 7 | 1 | 2 | 17 | 24 | 22 | 1 | 8 |
| 30 | 3 | 1 | 2 | 16 | 3 | 6 | 1 | 8 |
| 31 | 3 | 18 | 2 | 3 | 22 | 21 | 1 | 6 |
| 32 | 7 | 14 | 2 | 3 | 18 | 17 | 1 | 17 |
| 33 | 7 | 1 | 2 | 3 | 18 | 17 | 1 | 17 |
| 34 | 3 | 20 | 1 | 18 | 16 | 4 | 1 | 16 |
| 35 | 3 | 20 | 1 | 18 | 16 | 3 | 1 | 16 |
| 36 | 3 | 20 | 1 | 18 | 6 | 3 | 1 | 16 |
| 37 | 3 | 3 | 1 | 3 | 16 | 20 | 1 | 3 |
| 38 | 3 | 1 | 5 | 3 | 17 | 6 | 1 | 10 |
| 39 | 3 | 1 | 1 | 3 | 17 | 6 | 1 | 10 |
| 40 | 3 | 1 | 1 | 3 | 6 | 19 | 1 | 17 |
| 41 | 3 | 1 | 1 | 14 | 19 | 18 | 1 | 17 |
| 42 | 3 | 12 | 1 | 1 | 17 | 3 | 1 | 6 |
| 43 | 3 | 17 | 7 | 3 | 21 | 5 | 1 | 20 |
| 44 | 17 | 3 | 1 | 5 | 23 | 5 | 1 | 8 |
| 45 | 15 | 22 | 1 | 17 | 16 | 9 | 1 | 22 |
| 46 | 3 | 13 | 1 | 13 | 15 | 9 | 5 | 16 |
| 47 | 7 | 12 | 1 | 5 | 15 | 9 | 1 | 15 |
| 48 | 14 | 19 | 3 | 5 | 26 | 6 | 1 | 14 |
| 49 | 10 | 7 | 5 | 8 | 7 | 23 | 1 | 6 |
| 50 | 13 | 14 | 1 | 1 | 17 | 5 | 5 | 21 |
STs are numbered according to Koehler et al. (22). New STs are numbered consecutively.
All together, 33 unique STs were identified in this study (Table 4). Of these, 29 (87.8%) were only identified once. Three STs were each represented by three (9.0%) of the isolates examined (ST 1, ST 22, and ST 28), and ST 25 was identified in two isolates (6.0%) (Table 1). The three ST 1 isolates were from the same patient and the three ST 22 isolates were from the same patient, while the three ST 28 isolates were recovered from two different Indonesian patients. The two ST 25 isolates were collected in the United States and Sweden, respectively.
Analysis of multiple isolates from the same patient revealed that some individuals (strains 195PG, 196PG, 212PG, and 213PG) harbored isolates of different STs, with four identified in one patient (strain 212PG), three in another patient (strain 195PG), and two in a third patient (strain 213PG). ST 8, represented by strain ESO127 isolated in Japan, was also present in the collection from Germany analyzed by Koehler et al. (22).
The allele distribution among STs in the population showed for each locus one or two dominant alleles with a frequency from 10% to 95% (Table 2). The rest were more evenly distributed in the population. For one locus (nah), only two alleles were present; the most frequent was detected in 95% of the isolates (Table 3). The genetic diversity at individual loci ranged from 0.085 (nah) to 0.918 (ftsQ), with a mean of 0.739.
Cluster analysis.
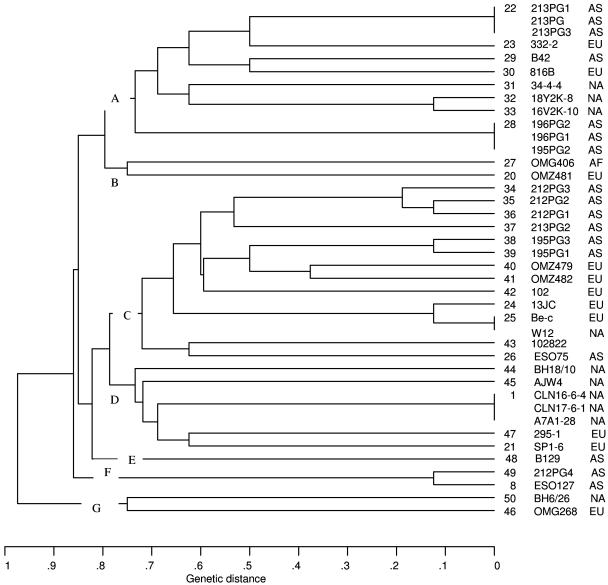
The 33 STs clustered together at a genetic distance of 0.9. At a distance of 0.75, corresponding to an average difference between STs at six of the eight loci studied, seven clusters were seen (clusters A to G) (Fig. 1). With the exceptions of lineage E, which included the ST of the strain from Japan, and lineage F, which included two STs of strains from Indonesia and Japan, the remaining clusters comprised STs of strains isolated in different continents.
FIG. 1.
Genetic relationships among 40 P. gingivalis isolates. A distance matrix was calculated on the basis of the differences in allelic profiles of the isolates. The dendrogram was generated from the distance matrix by the unweighted-pair group method with arithmetic averages. Clusters at a genetic distance of 0.75 were designated by letters A to G. Columns at right of tree give (from left to right, respectively) allele numbers, strain designations, and continents of origin of the strains (EU, Europe; NA, North America; AF, Africa; and AS, Asia).
Congruence and recombination analysis.
To analyze whether recombination is frequent in P. gingivalis, a split decomposition analysis was performed. The algorithm utilized is able to create a tree-like structure when the descent is clonal and a bush-like structure when recombination plays a major role in the evolutionary history of the genes investigated. The test was performed for all isolates with each of the gene fragments individually (data not shown). Some of the genes displayed a typical clonal descent (hagB, pepO, and ftsQ), while others showed evidence of a more bush-like structure (recA, gdpxJ, mcmA, and pga). A split decomposition using concatenated allele sequences of the eight gene fragments from the 40 isolates is shown in Fig. 2.
FIG. 2.
Split decomposition analysis of fragments of eight P. gingivalis genes from 40 isolates. See Table 1 for strain designations.
Based on allelic profile data, the IA gives an estimate of the degree of association between alleles at different loci. Using one representative of each ST, the IA was calculated to 0.384, indicating a significant linkage disequilibrium in the population and a weak clonal population structure.
DISCUSSION
In this study, we applied the MLST technique to the periodontal pathogen P. gingivalis, with a modified setup compared to the design utilized by Koehler et al. (22). Only one MLST investigation for this species had been published when we started our study. We selected 8 of the 10 genes from the original scheme (22), mainly because our first PCR amplification products for the EF-Tu gene and the dnaK gene were not satisfactory. MLST schemes for pathogenic microorganisms should include at least seven genes spread around the chromosome (28). The eight genes used by us were well spread on the chromosome of strain W83, for which the full genome sequence is available (33); thus, our modifications in the setup were justified.
To obtain a better understanding of the population structure of P. gingivalis, we increased the number of isolates and widened the geographic origin of patients. Koehler et al. (22) analyzed isolates from 19 different and epidemiologically unrelated periodontitis patients from two regions in Germany, while the number in our study was 40 isolates from nine different countries in four continents. For comparison, Loos et al. (26) investigated 88 human and 12 animal isolates by MLEE from 11 countries in five continents, while Ménard and Mouton (30) applied random amplified polymorphic DNA to 97 isolates of P. gingivalis from 79 humans and 32 strains from 30 animals from five continents.
While the number of isolates from different patients that we analyzed was smaller than in the studies by Loos et al. (26) and Ménard and Mouton (30), our strain collection can be compared to theirs with respect to geographic origin. It is also valuable to utilize a new molecular technique with isolates that have been extensively studied earlier with other techniques, in order to assess the discriminatory power of the new method.
Our study supported the findings by these investigators concerning the Porphyromonas isolates from animal origins. For the three isolates from dog, cat, and sheep, originally assigned to P. gingivalis, PCR was unsuccessful for some of the genes, and when PCR products were obtained, sequence data were difficult to align with the alleles of the human strains because of diverging sequence in large fragments of the genes. Therefore, we performed PCR amplification and sequencing of the 16S rRNA genes of these three isolates and identified them as P. gulae by use of the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/). This result confirmed the findings by Loos et al. (26), who, on the basis of MLEE, suggested that these strains belonged to another Porphyromonas species. Interestingly, we had no problems with the interpretation of the sequencing data of the isolates from monkeys, Macaca fascicularis. In the dendrogram (Fig. 1), these two isolates (16V2K-10 and 18Y2K-8) were closely related and clustered in group A among some of the human isolates. In the study by Loos et al. (26), these isolates were clustered and assigned to group E, in division I, also closely related to the human strains. The study of Ménard and Mouton using random amplified DNA came to the same conclusion (30). Their finding that Old World monkeys seem to harbor human genotype strains was corroborated by us.
Thirty-three new allelic profiles, or STs, were identified in our investigation. Surprisingly, our results showed that three Indonesian patients harbored two, three, and four STs in the mouth, respectively. Loos et al. (27) found one patient who carried two distinct clonal types as assessed by DNA fingerprinting, but most studies of P. gingivalis suggest the presence of a single or very limited number of genotypes in one individual at a time (27).
Koehler et al. (22) concluded, from the identification of 19 unique STs, that P. gingivalis is a heterogeneous species undergoing frequent recombination. Different STs in individual patients and our finding that 29 STs were identified only once supported their result of a high genetic heterogeneity in the species. In this study, we detected between 2 and 19 alleles per locus, which demonstrated variable levels of polymorphism for the various genes. The average number of alleles per locus relative to the strains analyzed (12.4/40) was somewhat higher (31%) than that determined by studies of other species. For example, 25.1 alleles per locus were identified for 295 S. pneumoniae strains (143 STs) (8.5%), and 33.3 alleles per locus were found with 194 isolates of C. jejuni (17.2%). On the other hand, Helgason et al. (17) found a higher number of alleles relative to the strains analyzed (33.9%) upon investigating the B. cereus group. Our findings reflect an extensive diversity of P. gingivalis.
For all of the eight loci, the amount of synonymous substitutions was at least three times (1/0.3277, hagB gene) larger than the amount of nonsynonymous substitutions (Table 3). This finding indicates that most of the observed variability in the genes investigated was selectively neutral at the protein level and confirms the selection of the loci suitable for MLST analysis (28). The hagB gene, however, codes for a protein related to hemagglutination, and one might question whether it can be characterized as a core housekeeping gene, defined as ubiquitous within the population, encoding proteins essential for central metabolism, and evolving at a moderate rate (5).
Split decomposition analysis of separate genes, comparative analysis of congruence between the different graphs obtained, and examination of association among loci in allelic profiles (IA) suggested that genetic exchanges through recombination may not be as extensive for P. gingivalis as previously suggested by some authors (12, 22). A study by Califano et al. (3), investigating insertion element distribution in a P. gingivalis population, supports our conclusion.
Evidence of some degree of clonality was supported by a coefficient of linkage disequilibrium (42), 0.384, which is significantly different from zero and by the recovery of strains with identical or similar genotypes in different geographic areas. In addition, no evidence of intragenic recombination (no mosaic genes) (41) was seen. In our study, we included isolates from different parts of the world (Europe, Africa, North America, and Asia), while the other investigation included strains only from Germany. These differences could explain some of the discrepancies of the results of the two studies. Our IA value was also higher than that obtained by Frandsen et al. (12), who analyzed four housekeeping genes from 57 P. gingivalis strains of various origins.
The standardized index of association (IAS), which takes into consideration the number of loci studied (16), has been claimed to give a better estimate of linkage equilibrium than the simple IA value. This parameter was estimated by Koehler et al. (22) to be 0.0898 for 19 isolates analyzed at 10 loci. Our IAS for the 33 unique STs was calculated to be 0.0548, a value comparable to that obtained for the P. gingivalis population in the Koehler study. This value is closer to that estimated for N. meningitidis (0.14), which has a weak clonal population structure, than to the value estimated for Neisseria gonorrhoeae (0.005), a panmictic species undergoing extensive recombination (22).
Our study provides further evidence of a high genetic diversity of the species, but in spite of extensive recombination, some degree of clonality exists in the P. gingivalis population. Thus, a typing scheme such as MLST might be very valuable for the study of the eventual association of virulence with specific clones. We would like to propose a seven-locus scheme, as developed for many other bacterial species, including the most polymorphic loci in our study. While nah was represented by four alleles in the strains from Germany (22), only two alleles were found in our global strain collection. Therefore, the MLST scheme should use the following loci: ftsQ, hagB, gdpxJ, pepO, mcmA, recA, and pga.
Acknowledgments
We thank the Dental Faculty, University of Oslo, Oslo, Norway, for financial support, Anne-Marie Klem and Torill Alvestad for skillful technical assistance, and Erlendur Helgason and Nicolas J. Tourasse (School of Pharmacy, University of Oslo) for help and advice with computation of the data.
REFERENCES
- 1.Ali, R. W., A. C. Johannessen, G. Dahlen, S. S. Socransky, and N. Skaug. 1997. Comparison of the subgingival microbiota of periodontally healthy and diseased adults in northern Cameroon. J. Clin. Periodontol. 24:830-835. [DOI] [PubMed] [Google Scholar]
- 2.Ali, R. W., L. Martin, A. D. Haffajee, and S. S. Socransky. 1997. Detection of identical ribotypes of Porphyromonas gingivalis in patients residing in the United States, Sudan, Romania and Norway. Oral Microbiol. Immunol. 12:106-111. [DOI] [PubMed] [Google Scholar]
- 3.Califano, J. V., T. Arimoto, and T. Kitten. 2003. The genetic relatedness of Porphyromonas gingivalis clinical and laboratory strains assessed by analysis of insertion sequence (IS) element distribution. J. Periodontal Res. 38:411-416. [DOI] [PubMed] [Google Scholar]
- 4.Chun, Y. H., K. R. Chun, D. Olguin, and H. L. Wang. 2005. Biological foundation for periodontitis as a potential risk factor for atherosclerosis. J. Periodontal Res. 40:87-95. [DOI] [PubMed] [Google Scholar]
- 5.Cooper, J. E., and E. J. Feil. 2004. Multilocus sequence typing—what is resolved? Trends Microbiol. 12:373-377. [DOI] [PubMed] [Google Scholar]
- 6.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dzink, J. L., S. S. Socransky, and A. D. Haffajee. 1988. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J. Clin. Periodontol. 15:316-323. [DOI] [PubMed] [Google Scholar]
- 8.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 9.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fournier, D., C. Mouton, P. Lapierre, T. Kato, K. Okuda, and C. Ménard. 2001. Porphyromonas gulae sp. nov., an anaerobic, gram-negative coccobacillus from the gingival sulcus of various animal hosts. Int. J. Syst. Evol. Microbiol. 51:1179-1189. [DOI] [PubMed] [Google Scholar]
- 12.Frandsen, E. V., K. Poulsen, M. A. Curtis, and M. Kilian. 2001. Evidence of recombination in Porphyromonas gingivalis and random distribution of putative virulence markers. Infect. Immun. 69:4479-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genco, C. A., C. W. Cutler, D. Kapczynski, K. Maloney, and R. R. Arnold. 1991. A novel mouse model to study the virulence of and host response to Porphyromonas (Bacteroides) gingivalis. Infect. Immun. 59:1255-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grenier, D., and D. Mayrand. 1987. Selected characteristics of pathogenic and nonpathogenic strains of Bacteroides gingivalis. J. Clin. Microbiol. 25:738-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haffajee, A. D., and S. S. Socransky. 1994. Microbial etiological agents of destructive periodontal diseases. Periodontol. 2000 5:78-111. [DOI] [PubMed] [Google Scholar]
- 16.Haubold, B., and R. R. Hudson. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16:847-848. [DOI] [PubMed] [Google Scholar]
- 17.Helgason, E., N. J. Tourasse, R. Meisal, D. A. Caugant, and A.-B. Kolstø. 2004. Multilocus sequence typing scheme for bacteria of the Bacillus cereus group. Appl. Environ. Microbiol. 70:191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holt, S. C., J. Ebersole, J. Felton, M. Brunsvold, and K. S. Kornman. 1988. Implantation of Bacteroides gingivalis in nonhuman primates initiates progression of periodontitis. Science 239:55-57. [DOI] [PubMed] [Google Scholar]
- 19.Huson, D. H. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 20.Iida, Y., K. Honda, T. Suzuki, S. Matsukawa, T. Kawai, T. Shimahara, and H. Chiba. 2004. Brain abscess in which Porphyromonas gingivalis was detected in cerebrospinal fluid. Br. J. Oral Maxillofac. Surg. 42:180. [DOI] [PubMed] [Google Scholar]
- 21.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 22.Koehler, A., H. Karch, T. Beikler, T. F. Flemmig, S. Suerbaum, and H. Schmidt. 2003. Multilocus sequence analysis of Porphyromonas gingivalis indicates frequent recombination. Microbiology 149:2407-2415. [DOI] [PubMed] [Google Scholar]
- 23.Kotetishvili, M., O. C. Stine, A. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2002. Multilocus sequence typing for characterization of clinical and environmental Salmonella strains. J. Clin. Microbiol. 40:1626-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laine, M. L., and A. J. van Winkelhoff. 1998. Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol. Immunol. 13:322-325. [DOI] [PubMed] [Google Scholar]
- 25.Lamell, C. W., A. L. Griffen, D. L. McClellan, and E. J. Leys. 2000. Acquisition and colonization stability of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in children. J. Clin. Microbiol. 38:1196-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loos, B. G., D. W. Dyer, T. S. Whittam, and R. K. Selander. 1993. Genetic structure of populations of Porphyromonas gingivalis associated with periodontitis and other oral infections. Infect. Immun. 61:204-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loos, B. G., A. J. Van Winkelhoff, R. G. Dunford, R. J. Genco, J. De Graaff, D. P. Dickinson, and D. W. Dyer. 1992. A statistical approach to the ecology of Porphyromonas gingivalis. J. Dent. Res. 71:353-358. [DOI] [PubMed] [Google Scholar]
- 28.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meats, E., E. J. Feil, S. Stringer, A. J. Cody, R. Goldstein, J. S. Kroll, T. Popovic, and B. G. Spratt. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 41:1623-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ménard, C., and C. Mouton. 1995. Clonal diversity of the taxon Porphyromonas gingivalis assessed by random amplified polymorphic DNA fingerprinting. Infect. Immun. 63:2522-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 32.Neiders, M. E., P. B. Chen, H. Suido, H. S. Reynolds, J. J. Zambon, M. Shlossman, and R. J. Genco. 1989. Heterogeneity of virulence among strains of Bacteroides gingivalis. J. Periodontal Res. 24:192-198. [DOI] [PubMed] [Google Scholar]
- 33.Nelson, K. E., R. D. Fleischmann, R. T. DeBoy, I. T. Paulsen, D. E. Fouts, J. A. Eisen, S. C. Daugherty, R. J. Dodson, A. S. Durkin, M. Gwinn, D. H. Haft, J. F. Kolonay, W. C. Nelson, T. Mason, L. Tallon, J. Gray, D. Granger, H. Tettelin, H. Dong, J. L. Galvin, M. J. Duncan, F. E. Dewhirst, and C. M. Fraser. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185:5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishihara, T., and T. Koseki. 2004. Microbial etiology of periodontitis. Periodontol. 2000 36:14-26. [DOI] [PubMed] [Google Scholar]
- 35.Ozmeric, N., H. R. Preus, and I. Olsen. 2000. Genetic diversity of Porphyromonas gingivalis and its possible importance to pathogenicity. Acta Odontol. Scand. 58:183-187. [DOI] [PubMed] [Google Scholar]
- 36.Paquette, D. W. 2004. The periodontal-cardiovascular link. Compend. Contin. Educ. Dent. 25:681-682, 685-692. [PubMed] [Google Scholar]
- 37.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pavloff, N., P. A. Pemberton, J. Potempa, W. C. Chen, R. N. Pike, V. Prochazka, M. C. Kiefer, J. Travis, and P. J. Barr. 1997. Molecular cloning and characterization of Porphyromonas gingivalis lysine-specific gingipain. A new member of an emerging family of pathogenic bacterial cysteine proteinases. J. Biol. Chem. 272:1595-1600. [DOI] [PubMed] [Google Scholar]
- 39.Scannapieco, F. A., R. B. Bush, and S. Paju. 2003. Associations between periodontal disease and risk for nosocomial bacterial pneumonia and chronic obstructive pulmonary disease. A systematic review. Ann. Periodontol. 8:54-69. [DOI] [PubMed] [Google Scholar]
- 40.Smith, G. L., S. S. Socransky, and C. M. Smith. 1989. Rapid method for the purification of DNA from subgingival microorganisms. Oral Microbiol. Immunol. 4:47-51. [DOI] [PubMed] [Google Scholar]
- 41.Smith, J. M. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126-129. [DOI] [PubMed] [Google Scholar]
- 42.Smith, J. M., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Socransky, S. S., and A. D. Haffajee. 1994. Evidence of bacterial etiology: a historical perspective. Periodontol. 2000 5:7-25. [DOI] [PubMed] [Google Scholar]
- 44.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 45.Takezaki, N., and M. Nei. 1996. Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144:389-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takezaki, N., A. Rzhetsky, and M. Nei. 1995. Phylogenetic test of the molecular clock and linearized trees. Mol. Biol. Evol. 12:823-833. [DOI] [PubMed] [Google Scholar]
- 47.Travis, J., R. Pike, T. Imamura, and J. Potempa. 1997. Porphyromonas gingivalis proteinases as virulence factors in the development of periodontitis. J. Periodontal Res. 32:120-125. [DOI] [PubMed] [Google Scholar]