Abstract
The present study represents the first application of multilocus sequence typing to retrospectively investigate a suspected outbreak of Candida albicans bloodstream infection cases that occurred in the same hospital ward between July 1987 and October 1991. Results demonstrated that eight bloodstream infections were caused by the same strain, endemic in the ward, over a 4-year period.
Typing methods used in surveillance studies have shown that outbreaks of Candida bloodstream infections (BSI) may be caused by strains endemic in hospitals. Several molecular methods, such as restriction fragment length polymorphism analysis (9), Southern blot hybridization with discriminating probes (6), electrophoretic karyotyping (11), and random amplified polymorphic DNA analysis (4), have been applied to characterize related and unrelated Candida albicans isolates in epidemiological studies. Multilocus sequence typing (MLST), based on DNA sequence analysis of nucleotide polymorphisms within housekeeping gene fragments, has emerged as an alternative typing tool (1). MLST has been proven to be a highly discriminating and stable method for the characterization of various microbial pathogens, including C. albicans (1, 3, 5, 8). A collaborative consensus has recently been published, and a set of seven housekeeping gene fragments were recommended for the typing of C. albicans isolates (2). We applied this optimized typing method, as well as PCR fingerprinting (7), to retrospectively investigate a suspected outbreak of C. albicans BSI that occurred more than 15 years ago in a surgical ward.
The suspected outbreak involved six patients and occurred in a surgical ward of IRCCS Ospedale Maggiore di Milano from November 1988 to January 1989. Analysis of the laboratory database revealed 10 other cases of C. albicans BSI in the same ward between July 1987 and October 1991. All of the patients had received parenteral nutrition. Isolates from 11 out of these 16 patients, cultured from blood and/or vascular catheter tips or residual parenteral nutrition fluid, were available in our culture collection. A total of 16 strains isolated from these 11 patients were investigated. In addition, 18 C. albicans strains isolated from the blood (11 from patients hospitalized in other wards of the same hospital and 7 from patients in other hospitals) were selected as control strains.
Genomic DNA was extracted as previously described (10), and a PCR-fingerprinting mixture was made to include the following: 100 pmol of the repetitive oligonucleotide (GTG)5 (7); 400 μM (each) dATP, dCTP, dGTP, and dTTP (Boehringer Mannheim GmbH, Mannheim, Germany); 3 mM MgCl2 (Applied Biosystems, Monza, Italy); 10× reaction buffer (500 mM KCl, 100 mM Tris-HCl [pH 8.3]; Applied Biosystems); 2.5 U of AmpliTaq DNA polymerase (Applied Biosystems); and 400 ng of the DNA sample. PCR was performed using a GeneAmp PCR system 2400 thermal cycler (Applied Biosystems), with an initial cycle of 5 min at 94°C; 38 cycles of 30 s at 94°C, 30 s at 50°C, and 60 s at 72°C; and a final cycle of 5 min at 72°C. The amplification products were visualized by electrophoresis on 1.4% agarose gels in 1× Tris-borate-EDTA (0.089 M Trizma base, 0.089 M boric acid, 0.002 M EDTA [pH 8.4]; Sigma-Aldrich, Milano, Italy) at 60 V for 2 h 30 min and stained with ethidium bromide (Sigma-Aldrich). The fingerprints were analyzed and compared using Diversity One software (PDI, Huntington Station, NY).
Fourteen isolates from 11 patients of the surgical ward and seven isolates from 7 patients of other wards or hospitals were selected for MLST analysis. MLST was performed as described elsewhere (2). For each strain, seven C. albicans gene fragments were sequenced (C. albicans AAT1a [CaAAT1a], CaACC1, CaADP1, CaMP1, CaSYA1, CaVPS13, and CaZWF1b) and a total of 76 polymorphic sites were screened. Sequence comparison and phylogenetic tree construction were performed using MEGA software (www.megasoftware.net). Isolates with a percentage of similarity lower than 97% were considered to be unrelated. All of the isolates from the surgical ward were further investigated by sequencing two other gene fragments (CaGLN4 and CaRPN2) in order to compare their allelic profiles with 140 C. albicans profiles reported in the MLST database (http://calbicans.mlst.net).
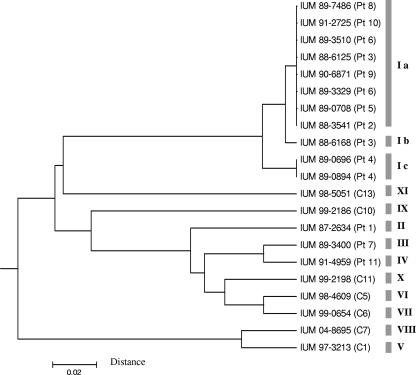
The results of molecular typing are shown in Table 1. PCR-fingerprinting analysis revealed that 12 out of 14 isolates from 11 patients hospitalized in the surgical ward had the B genotype. This genotype was not identified in patient 1 or 7 or in the 14 control isolates from patients hospitalized in other wards and hospitals. MLST analysis of the 21 selected strains showed that 8 of the 12 isolates with the B genotype also had the same Ia allelic profile (Table 2). Of the remaining four isolates with the B genotype, one from patient 11 (IUM 91-4959) had an unrelated allelic profile (IV), while the other three had sequence types closely related to the Ia profile: the Ic profile (97% similarity) in the case of isolates from the blood and the vascular catheter tip of patient 4 (IUM 89-0696 and IUM 89-0894) and the Ib profile (99% similarity) observed in isolates from the blood of patient 3 (IUM 88-6168) (Fig. 1). The other strain, isolated from the vascular catheter tip of patient 3 (IUM 88-6125), had the Ia profile. Strain IUM 87-2634 from patient 1 and strain IUM 89-3400 from patient 7, as well as the seven control strains tested (IUM 97-3213, IUM 98-4609, IUM 98-5051, IUM 99-0654, IUM 99-2186, IUM 99-2198, and IUM 04-8695), not only did not present the B genotype, but also had different multilocus profiles. Strains with an Ia, Ib, or Ic MLST profile also had the same sequences as did the gene fragments CaGLN4 and CaRPN2. The sequences of the I profile strains proved to be different from those of the 140 C. albicans profiles reported in the MLST database.
TABLE 1.
Origins and genotypes of the 34 Candida albicans strains tested
| Hospital/warda | Patient or controlb | Strain | Date of isolation (day.mo.yr) | Source of sample | Fingerprinting genotype | MLST genotype |
|---|---|---|---|---|---|---|
| 1/1 | P1 | IUM 87-2634 | 20.07.87 | Blood | A | II |
| P2 | IUM 88-3541 | 21.06.88 | Vascular catheter tip | B | Ia | |
| P3 | IUM 88-6125 | 15.11.88 | Vascular catheter tip | B | Ia | |
| IUM 88-6168 | 14.11.88 | Blood | B | Ib | ||
| P4 | IUM 89-0696 | 27.01.89 | Blood | B | Ic | |
| IUM 89-0894 | 30.01.89 | Vascular catheter tip | B | Ic | ||
| P5 | IUM 89-0708 | 24.01.89 | Parenteral nutrition fluid | B | Ia | |
| P6 | IUM 89-3329 | 17.05.89 | Blood | B | Ia | |
| IUM 89-3386 | 18.05.89 | Vascular catheter tip | B | |||
| IUM 89-3510 | 25.05.89 | Vascular catheter tip | B | Ia | ||
| IUM 89-3615 | 30.05.89 | Vascular catheter tip | B | |||
| P7 | IUM 89-3400 | 19.05.89 | Vascular catheter tip | D | III | |
| P8 | IUM 89-7486 | 18.12.89 | Vascular catheter tip | B | Ia | |
| P9 | IUM 90-6871 | 27.11.90 | Blood | B | Ia | |
| P10 | IUM 91-2725 | 23.05.91 | Vascular catheter tip | B | Ia | |
| P11 | IUM 91-4959 | 08.10.91 | Vascular catheter tip | B | IV | |
| 1/2 | C1 | IUM 97-3213 | 01.09.97 | Blood | H | V |
| 1/3 | C2 | IUM 97-3348 | 15.09.97 | Blood | L | |
| 1/4 | C3 | IUM 98-1643 | 28.04.98 | Blood | P | |
| C4 | IUM 99-2974 | 02.07.99 | Blood | C | ||
| 1/5 | C5 | IUM 98-4609 | 05.12.98 | Blood | I | VI |
| 1/6 | C6 | IUM 99-0654 | 09.02.99 | Blood | L | VII |
| C7 | IUM 04-8695 | 23.06.04 | Blood | N | VIII | |
| C8 | IUM 04-8645 | 21.06.04 | Blood | O | ||
| 1/7 | C9 | IUM 99-1444 | 01.04.99 | Blood | I | |
| 1/8 | C10 | IUM 99-2186 | 20.05.99 | Blood | V | IX |
| 1/9 | C11 | IUM 99-2198 | 18.05.99 | Blood | M | X |
| 2 | C12 | IUM 98-4982 | 03.08.92 | Blood | T | |
| 3 | C13 | IUM 98-5051 | 19.02.98 | Blood | U | XI |
| 4 | C14 | IUM 98-5059 | 18.05.98 | Blood | F | |
| 5 | C15 | IUM 99-3228 | 07.09.99 | Blood | S | |
| 6 | C16 | IUM 99-4839 | 02.11.99 | Blood | Q | |
| 7 | C17 | IUM 99-5779 | 06.07.99 | Blood | H | |
| 8 | C18 | IUM 99-5798 | 23.05.99 | Blood | R |
Ward 1 was the surgical ward; wards 2 to 9 were other wards of the same hospital.
P, patient; C, control.
TABLE 2.
MLST genotypes of the 21 Candida albicans isolates studied
| Strain (source)a | Genotype
|
MLST genotype | ||||||
|---|---|---|---|---|---|---|---|---|
| CaADP1 | CaSYA1 | CaVPS13 | CaACC1 | CaAAT1a | CaZWF1 | CaMP1 | ||
| IUM 87-2634 (P1) | 15 | 9 | 45 | 4 | 13 | 10 | 2 | II |
| IUM 88-3541 (P2) | 22 | 11 | 46 | 11 | 13 | 13 | 14 | Ia |
| IUM 88-6125 (P3) | 22 | 11 | 46 | 11 | 13 | 13 | 14 | Ia |
| IUM 88-6168 (P3) | 22 | 25 | 46 | 11 | 13 | 13 | 14 | Ib |
| IUM 89-0696 (P4) | 22 | 11 | 46 | 11 | 15 | 13 | 14 | Ic |
| IUM 89-0894 (P4) | 22 | 11 | 46 | 11 | 15 | 13 | 14 | Ic |
| IUM 89-0708 (P5) | 22 | 11 | 46 | 11 | 13 | 13 | 14 | Ia |
| IUM 89-3329 (P6) | 22 | 11 | 46 | 11 | 13 | 13 | 14 | Ia |
| IUM 89-3510 (P6) | 22 | 11 | 46 | 11 | 13 | 13 | 14 | Ia |
| IUM 89-3400 (P7) | 15 | 28 | 45 | 12 | 3 | 23 | 2 | III |
| IUM 89-7486 (P8) | 22 | 11 | 46 | 11 | 13 | 13 | 14 | Ia |
| IUM 90-6871 (P9) | 22 | 11 | 46 | 11 | 13 | 13 | 14 | Ia |
| IUM 91-2725 (P10) | 22 | 11 | 46 | 11 | 13 | 13 | 14 | Ia |
| IUM 91-4959 (P11) | 15 | 28 | 45 | 13 | 16 | 10 | 2 | IV |
| IUM 97-3213 (C1) | 13 | 29 | 45 | 14 | 17 | 2 | 1 | V |
| IUM 98-4609 (C5) | 15 | 9 | 14 | 5 | 3 | 27 | 2 | VI |
| IUM 99-0654 (C6) | 15 | 9 | 11 | 2 | 3 | 26 | 2 | VII |
| IUM 04-8695 (C7) | 13 | 4 | 47 | 11 | 1 | 24 | 1 | VIII |
| IUM 99-2186 (C10) | 12 | 31 | 5 | 2 | 4 | 28 | 4 | IX |
| IUM 99-2198 (C11) | 15 | 9 | 21 | 5 | 3 | 29 | 2 | X |
| IUM 98-5051 (C13) | 24 | 30 | 20 | 13 | 4 | 25 | 15 | XI |
P, patient; C, control strains.
FIG. 1.
Dendrogram of the genetic relations between 21 isolates of Candida albicans, based on the seven housekeeping loci investigated. The dendrogram was constructed using the unweighted pair group method with arithmetic averages and the matrix of distances. MLST genotypes are indicated on the right.
In the last decade, various molecular biology methods have been applied in epidemiological studies to investigate the origins of suspected nosocomial outbreaks and to study the genetic relationships among isolates from patients, health care workers, and/or the hospital environment. Recently, MLST was proposed as the reference method for typing C. albicans strains. To our knowledge, the present study is the first application of MLST analysis to identify a nosocomial outbreak of C. albicans BSI. The use of molecular analysis of the available strains allowed us to demonstrate that the suspected candidemia outbreak involving patients in the period November 1988 to January 1989, as well as five additional cases identified in an extended analysis, were caused by a single C. albicans strain that was endemic in the ward. The strain isolated from the vascular catheter of one of the patients (patient 7) who developed candidemia in the epidemic period, as well as strains from patients 1 and 11, presented completely different genotypes and were not correlated to the epidemic. Therefore, a single genotype was proven to persist in the ward from June 1988 to May 1991, causing candidemia in eight patients. As all of the patients had received parenteral nutrition, and the isolate was also cultured from the parenteral nutrition fluid that was administered to one of them (patient 5), contamination of the fluids by a health care worker involved in the preparation of the parenteral nutrition bags was suspected. A microevolutionary process might have occurred in the isolates from patient 4, who was infected by a closely related strain with the Ic profile that differed by four nucleotides in the CaAAT1a gene fragment, and also might have occurred in one of the two isolates from patient 3. In this patient, the isolate from the vascular catheter tip (IUM 88-6125) presented the MLST Ia genotype, while the strain from the blood (IUM 88-6168) presented a profile that differed by two nucleotides in the CaSYA1 gene fragment. The MLST results agreed with the fingerprinting data, except in the case of one isolate (IUM 91-4959 from patient 11) whose allelic profile had only a 92% similarity with the Ia genotype.
In the present study, the application of MLST typing, with its high degree of intraspecies discriminatory power, proved to be extremely useful in identifying a strain endemic in a surgical ward. The absence of the Ia genotype from the isolates from other wards and hospitals, as well as the divergence of its allelic profile from those reported in the MLST database, confirm the uniqueness of this endemic strain. In conclusion, this retrospective analysis showed a long period of circulation for a single isolate in one ward, although it was impossible to identify the external source of infection due to the lack of isolates from health care workers. As standardized typing methods such as MLST are available, it would be appropriate to conserve strains that are now usually discarded soon after identification so that they can be typed if an outbreak is suspected. In addition, samples from administered fluids, hospital devices, and health care workers' skin and mucous membranes should be cultured in order to identify the source of infection and route of transmission so that tailored measures may be implemented to control the epidemic.
REFERENCES
- 1.Bougnoux, M.-E., S. Morand, and C. d'Enfert. 2002. Usefulness of multilocus sequence typing for characterization of clinical isolates of Candida albicans. J. Clin. Microbiol. 40:1290-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bougnoux, M.-E., A. Tavanti, C. Bouchier, N. A. R. Gow, A. Magnier, A. D. Davidson, M. C. J. Maiden, C. d'Enfert, and F. C. Odds. 2003. Collaborative consensus for optimized multilocus sequence typing of Candida albicans. J. Clin. Microbiol. 41:5265-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eggimann, P., J. Garbino, and D. Pittet. 2003. Epidemiology of Candida species infections in critically ill non-immunosuppressed patients. Lancet Infect. Dis. 3:685-702. [DOI] [PubMed] [Google Scholar]
- 4.Robert, F., F. Lebreton, M. E. Bougnoux, A. Paugam, D. Wassermann, M. Schlotterer, C. Tourte-Schaefer, and J. Dupouy-Camet. 1995. Use of random amplified polymorphic DNA as a typing method for Candida albicans in epidemiological surveillance of a burn unit. J. Clin. Microbiol. 33:2366-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robles, J. C., L. Koreen, S. Park, and D. S. Perlin. 2004. Multilocus sequence typing is a reliable alternative method to DNA fingerprinting for discriminating among strains of Candida albicans. J. Clin. Microbiol. 42:2480-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmid, J., Y. P. Tay, L. Wan, M. Carr, D. Parr, and W. McKinney. 1995. Evidence for nosocomial transmission of Candida albicans obtained by Ca3 fingerprinting. J. Clin. Microbiol. 33:1223-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schonian, G., O. Meusel, H. J. Tietz, W. Meyer, Y. Graser, I. Tausch, W. Presber, and T. G. Mitchell. 1993. Identification of clinical strains of Candida albicans by DNA fingerprinting with the polymerase chain reaction. Mycoses 36:171-179. [DOI] [PubMed] [Google Scholar]
- 8.Tavanti, A., N. A. R. Gow, S. Senesi, M. C. J. Maiden, and F. C. Odds. 2003. Optimization and validation of multilocus sequence typing for Candida albicans. J. Clin. Microbiol. 41:3765-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vazquez, J. A., V. Sanchez, C. Dmuchowski, L. M. Dembry, J. D. Sobel, and M. J. Zervos. 1993. Nosocomial acquisition of Candida albicans: an epidemiologic study. J. Infect. Dis. 168:195-201. [DOI] [PubMed] [Google Scholar]
- 10.Viviani, M. A., H. Wen, A. Roverselli, R. Caldarelli-Stefano, M. Cogliati, P. Ferrante, and A. M. Tortorano. 1997. Identification by polymerase chain reaction fingerprinting of Cryptococcus neoformans serotype AD. J. Med. Vet. Mycol. 35:355-360. [PubMed] [Google Scholar]
- 11.Voss, A., R. J. Hollis, M. A. Pfaller, R. P. Wenzel, and B. N. Doebbeling. 1994. Investigation of the sequence of colonization and candidemia in nonneutropenic patients. J. Clin. Microbiol. 32:975-980. [DOI] [PMC free article] [PubMed] [Google Scholar]