Abstract
Agrobacterium tumefaciens strains C58, A136, and BG53 are chloramphenicol resistant, and each contains the catB gene originally identified by Tennigkeit and Matzuran (Gene 99:113-116, 1991). The chloramphenicol acetyltransferase activity in all of the strains is chloramphenicol inducible. Examination of the catB gene in strain BG53 indicates that it is regulated by an attenuation mechanism similar to translation attenuation that regulates inducible catA genes resident in gram-positive bacteria and the inducible cmlA gene that confers chloramphenicol resistance in Pseudomonas spp.
Bacteria that are resistant to chloramphenicol are commonly encountered in nature and result from either of two genetically dominant resistance mechanisms: alteration in permeability to the antibiotic or antibiotic inactivation (4, 6, 12, 17, 21). Antibiotic inactivation is the more common resistance mechanism and is due to the presence of a plasmid- or chromosome-linked cat gene that encodes chloramphenicol acetyltransferase (CAT), an enzyme that acetylates chloramphenicol, rendering the antibiotic unable to interfere with translation (19). Virtually all of the cat genes identified in gram-positive bacteria are inducibly expressed, and chloramphenicol is the inducer (19). Extensive studies of two of these genes, catA86 and catA112, have demonstrated that regulation occurs by a mechanism known as translation attenuation (15). In brief, cat genes are continuously transcribed but the transcripts are not translated because the ribosome binding site (RBS) for the cat coding sequence is sequestered in RNA secondary structure (15). Immediately upstream from, and overlapping with, the region of secondary structure is a short open reading frame (ORF) known as the leader ORF. Ribosome stalling in the leader ORF alters the RNA secondary structure, exposing the RBS for the cat coding sequence, resulting in its translation. The sequence of the leader peptide and the antibiotic inducer chloramphenicol appear to dictate the site of ribosome stalling in the leader (14).
In 1991, Tennigkeit and Matzura (22) sequenced a cat gene cloned from Agrobacterium tumefaciens C58. The deduced translation product differed obviously from previously studied CAT proteins, and the A. tumefaciens cat gene was therefore designated catB to distinguish it from the other cat genes then known, which were given the designation catA (24). catB-like genes have since been identified in several other bacteria, including gram-positive bacteria (16, 24). catB genes specify CAT enzymes that acetylate chloramphenicol by using acetyl coenzyme A as a coenzyme. This is the same overall reaction catalyzed by catA-specified enzymes. catB in A. tumefaciens strains is induced by chloramphenicol, and we demonstrate here that the regulation is due to an attenuation mechanism.
CAT activity in A. tumefaciens strain BG53 is induced approximately 10-fold by a subinhibitory level of chloramphenicol (2 μg/ml; data not shown) or the nonacetylatable analog fluorothiamphenicol (0.2 μg/ml) (Fig. 1A). Comparable induction results were obtained with strains C58 and A136. Using primers at the extreme 5′ and 3′ ends of the catB-containing fragment originally sequenced by Tennigkeit and Matzura (22), we PCR amplified catB-containing fragments from the DNAs of strains A136, C58, and BG53 and cloned these into the pNoTA/T7 vector (5′→3′). The nucleotide sequences of each of the PCR-generated clones were identical to that reported by Tennigkeit and Matzura, with the exception that our sequences contained a single additional G residue 40 nucleotides downstream from the catB translation termination codon. The recent sequencing of the genome of A. tumefaciens C58 (25) demonstrated that the catB gene in that strain is on the 2.1-Mb linear chromosome (2, 13). We have confirmed the same location for catB in strain BG53 by hybridization (M. S. Rahman and R. T. Hill, data not shown).
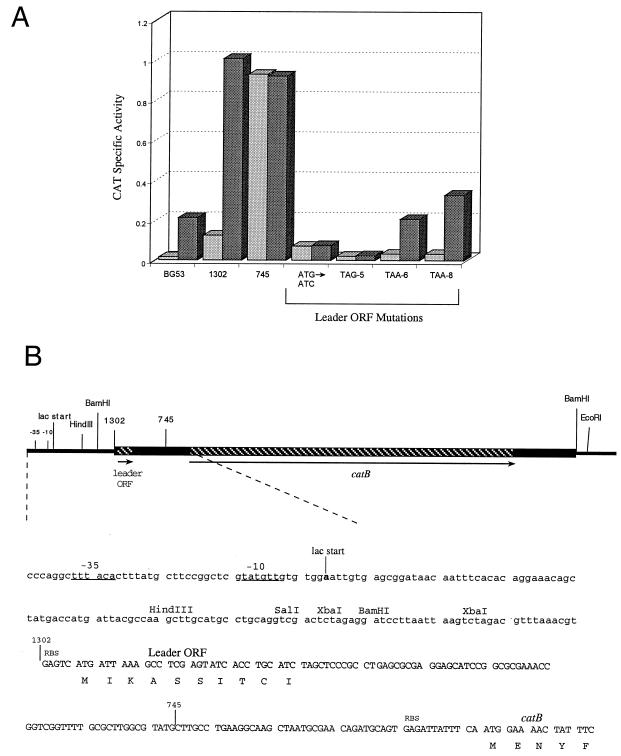
FIG. 1.
Induction of catB and organization of the regulatory region. (A) Induction assays of various catB-containing strains of A. tumefaciens. BG53 is a wild-type strain of A. tumefaciens and contains a resident catB gene in the 2.1-Mb linear chromosome. All other induction assays were performed with strain LBA4404 with or without plasmids p1302, p745, and p1302 with the mutations ATG→ATC (a mutation of the leader initiation codon), TAG-5, TAA-6, and TAA-8 (stop codon mutations at leader codons 5, 6, and 8, respectively). Cells were grown to mid-log phase and exposed to 0.2 μg of fluorothiamphenicol per ml for 2 h at 30°C. Cells were lysed, and CAT assays were performed at 25°C as previously described (1, 18). Protein was measured by the Bradford method (5), and specific activities are expressed as micromoles of chloramphenicol acetylated per minute per milligram of protein. Mutations were made in E. coli by using the Quick Change Site Directed Mutagenesis Kit (Stratagene). Light bars represent CAT levels of cells grown without inducer, and dark bars show CAT levels of cells grown with fluorothiamphenicol. In the experiment depicted, CAT assays were performed in duplicate and the duplicates differed by less than 4%. The entire experiment was repeated more than six times with results comparable to those shown. (B) Diagram of the cloned catB gene in pBin19 and the sequence of the region involved in regulation. The diagram depicts the 1302 clone at the BamHI site of pBin19 with the location of the vector lac promoter and the site of transcription initiation indicated. The sequence shows the catB cloned fragment that begins at the site designated 1302; the endpoint of the 745 deletion derivative is designated. The sequence between the BamHI site and the site designated 1302 is from the linker in pNoTA/T7, and the sequence upstream of the BamHI site corresponds to the pBin19 vector plasmid. The leader ORF coding region consists of 10 sense codons preceded by an RBS, GAG, that is likely very weak because the spacing between this site and the initiator ATG is only two nucleotides. The first five codons of the catB structural gene are designated and are preceded by an RBS, GAG, spaced nine nucleotides upstream from the initiator ATG. The site of transcription initiation was determined by primer extensions with a 30-mer primer complementary to the leader ORF region of the mRNA.
A catB-containing fragment amplified from BG53 DNA was transferred from the pNoTA/T7 vector into the unique BamHI site in the binary pBin19 plasmid (3, 11). The orientation chosen placed cat gene transcription under the control of the lac promoter present in the pBin19 vector. This construction, designated p1302, was transformed into competent A. tumefaciens LBA4404 (Life Technologies), a strain that is chloramphenicol sensitive. Selection was for the plasmid-linked neomycin resistance (at 50 μg/ml) marker. In LBA4404, CAT activity specified by p1302 was induced 10-fold by chloramphenicol (data not shown) or fluorothiamphenicol (Fig. 1A). This level of CAT is fivefold greater than that observed in extracts of induced BG53.
To ensure that the CAT enzyme was indeed specified by the catB gene identified by sequencing, we isolated CAT protein from extracts of LBA4404(p1302) by using a chloramphenicol affinity column (23). The sequence of the 12 N-terminal amino acid residues determined by automated Edman degradation (MENYFESPFRGI) was in agreement with the first 12 codons of the catB sequence (Fig. 1B; reference 22).
Inspection of the p1302 sequence upstream of the catB gene showed a 10-codon leader ORF that overlapped an adjacent, downstream region predicted to encode complex secondary structure in the corresponding mRNA (see Fig. 2). By PCR, we constructed a version of the catB gene lacking 99 nucleotides at the 5′ end of the 1302 clone. The deletion version of the 1302 clone was designated clone 745 (Fig. 1) and lacked the leader ORF and the region predicted to form a domain of secondary structure. p745, which is the 745 clone in pBin19, expressed catB constitutively in LBA4404 (Fig. 1A).
FIG. 2.
Folding of the mRNA corresponding to the regulatory region of catB. Computer folding utilized algorithms of Zuker et al. (26). The leader sequence is overlined, and the catB coding region and the 5′ end point of the 745 deletion are indicated. The calculated ΔG of the structure is −64 kcal/mol.
If the leader sequence were involved in the regulation process, a mutation that prevents leader translation would be predicted to block inducibility. The leader start codon was therefore changed from ATG to ATC, and this prevented induction (Fig. 1A). To determine the number of leader codons essential to induction, we individually replaced leader codon 5, 6, or 8 with a translation stop codon (TAG, TAA, or TAA, respectively). A stop codon at leader codon 5 blocked induction, but induction was observed when the stop codon was at codon 6 or 8 (Fig. 1A). It is evident that the stop codons at positions 6 and 8 resulted in only partial induction, with TAA-8 showing higher induction than TAA-6. These results are similar to those obtained in studies of the leaders of inducible catA genes from Bacillus pumilus and Staphylococcus aureus (1, 7, 9). For both catA86 and catA112, only the first five leader codons are essential for induction and mutating leader codon 6 to a stop codon results in full induction rather than the partial induction observed with catB.
Inducible catA genes are regulated at the level of translation and not at the level of transcription (10). Inspection of the region of secondary structure of catB mRNA shows that it includes the predicted RBS for the catB coding sequence (Fig. 2). Moreover, the effects of mutations in the leader ORF on catB induction argue strongly for an attenuation form of regulation. In theory, if translation attenuation regulated catB, the corresponding mRNA would be present both upstream and downstream of the domain of secondary structure in uninduced, as well as induced, cells. By contrast, if transcription attenuation regulated the gene, catB mRNA would only be detected upstream of the secondary structure in uninduced cells; induction would relieve the termination, and the mRNA would also be detected downstream of the secondary structure. Accordingly, we probed for catB mRNA corresponding to sequences immediately upstream and downstream of the region of secondary structure in induced and uninduced cells. Probe 1 is complementary to the leader region of the mRNA, and probe 2 is complementary to catB codons 6 to 13. As shown in the scan of dot blots (Fig. 3), mRNA in LBA4404(p1302) corresponding to the leader region of the mRNA and to the catB coding sequence (probes 1 and 2) was detected in induced and uninduced cells although the level of mRNA was significantly elevated in the induced cells. Densitometer scans of the blots demonstrated that the level of RNA detected by probe 1 was 4-fold greater in induced cells and probe 2 detected 3.9-fold more RNA in induced cells than in uninduced cells. Generally similar results were obtained by using RNA from induced and uninduced BG53 cells, although the overall mRNA levels were much reduced. LBA4404 carrying constitutively expressed deletion mutant plasmid p745 contained about equal levels of cat mRNA, detected with probe 2, whether or not the cells had been exposed to the inducer (Fig. 3); probe 1 detected no signal since the corresponding sequences are absent from the 745 clone.
FIG. 3.
Dot blot analysis of catB mRNA from BG53 or from LBA4404 carrying either p1302 or p745. RNA (10 μg) from uninduced cells (designated −) or cells grown with 0.2 μg of fluorothiamphenicol per ml for 2 h (designated +) was bound to GeneScreen Nylon Membrane (NEN Life Science Products). The filters were hybridized with end-labeled primers in 50% formamide-5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-1% sodium dodecyl sulfate-5× Denhardt's solution-100 μg of calf thymus DNA per ml at 42°C and washed at the same temperature. Probe 1 was a 30-mer complementary to the leader sequence, and probe 2 was a 24-mer probe complementary to codons 6 to 13 of the catB structural gene. Analysis was performed on a Storm PhosphorImager (Molecular Dynamics).
We suggest that catB is regulated by translation attenuation on the basis of three lines of evidence. First, catB is induced when the promoter for the gene is replaced with a heterologous promoter; primer extension studies using probe 1 demonstrated that transcription of catB in p1302 initiates at the lac promoter in the vector (data not shown). Second, a leader consisting of 10 codons can be identified. A mutation that converts the predicted leader initiator codon from ATG to ATC prevents induction, and stop codon mutations placed at different locations in the leader either abolish or reduce inducibility, depending on their location. Third, mRNA for the catB gene can be detected in both the uninduced and induced states. The level of the mRNA detected in induced cells is clearly greater than that detected in uninduced cells. This result parallels results of similar studies with catA112, and it has been shown that the act of translation of the catA112 coding sequence dramatically increases the stability of the mRNA (8).
Previous studies have shown that translation attenuation regulates inducible catA genes from gram-positive bacteria and has been reported to regulate the Pseudomonas cmlA gene (20), a gene that governs permeability to chloramphenicol. Since this form of regulation also controls the catB gene, it appears that translation attenuation is a preferred control device for responding to chloramphenicol.
Acknowledgments
We thank E. Nester for providing strains of A. tumefaciens and Arun Pal for participating in the early phase of this work.
This investigation was supported by Public Health Service grant GM56381 from the National Institutes of Health to P.S.L. and an award from the VIRTUE Program, Wallenberg Foundation, to R.T.H.
Footnotes
Contribution no. 570 from the Center of Marine Biotechnology.
REFERENCES
- 1.Alexieva, Z., E. J. Duvall, N. P. Ambulos, Jr., U. J. Kim, and P. S. Lovett. 1988. Chloramphenicol induction of cat-86 requires ribosome stalling at a specific site in the leader. Proc. Natl. Acad. Sci. USA 85:3057-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allardet-Servent, A., S. Michaux-Charachon, E. Jumas-Bilak, L. Karaya, and M. Ramuz. 1993. Presence of one linear and one circular chromosome in the Agrobacterium tumefaciens C58 genome. J. Bacteriol. 175:7869-7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bevan, M. 1984. Binary Agrobacterium vectors from plant transformation. Nucleic Acids Res. 12:8711-8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bissonette, L., S. Champetier, P. P. Buisson, and P. H. Roy. 1991. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane proteins. J. Bacteriol. 173:4493-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-252. [DOI] [PubMed] [Google Scholar]
- 6.Burns, J. L., C. E. Rebens, P. M. Mandelman, and A. L. Smith. 1986. Cloning and expression in Escherichia coli of a gene encoding nonenzymatic chloramphenicol resistance. Antimicrob. Agents Chemother. 29:445-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dick, T., and H. Matzura. 1988. Positioning ribosomes on leader mRNA for translational activation of the message of an inducible Staphylococcus aureus cat gene. Mol. Gen. Genet. 214:108-111. [DOI] [PubMed] [Google Scholar]
- 8.Dreher, J., and H. Matzura. 1991. Chloramphenicol-induced stabilization of cat messenger RNA in Bacillus subtilis. Mol. Microbiol. 5:3025-3034. [DOI] [PubMed] [Google Scholar]
- 9.Duvall, E. J., N. P. Ambulos, Jr., and P. S. Lovett. 1987. Drug-free induction of a chloramphenicol acetyltransferase gene in Bacillus subtilis by stalling ribosomes in a regulatory leader. J. Bacteriol. 169:4235-4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duvall, E. J., and P. S. Lovett. 1986. Chloramphenicol induces translation of the mRNA for a chloramphenicol resistance gene in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 83:3939-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frisch, D. A., L. W. Harris-Haller, N. T. Yokubaitis, T. L. Thomas, S. H. Hardin, and T. C. Hall. 1995. Complete sequence of the binary vector Bin19. Plant Mol. Biol. 27:405-409. [DOI] [PubMed] [Google Scholar]
- 12.Gaffney, E. F., E. Cundliffe, and T. J. Foster. 1981. Chloramphenicol resistance that does not involve chloramphenicol acetyltransferase encoded by plasmids from gram-negative bacteria. J. Gen. Microbiol. 125:113-121. [DOI] [PubMed] [Google Scholar]
- 13.Goodner, B. W., B. P. Markelz, M. C. Flanagan, C. B. Crowell, Jr., J. L. Racette, B. A. Schilling, L. M. Halfon, J. S. Mellors, and G. Grabowski. 1999. Combined genetic and physical map of the complex genome of Agrobacterium tumefaciens. J. Bacteriol. 181:5160-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lovett, P. S., and E. J. Rogers. 1996. Ribosome regulation by the nascent peptide. Microbiol. Rev. 60:366-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lovett, P. S. 1990. Translation attenuation as the regulator of inducible cat genes. J. Bacteriol. 172:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray, I. A., and W. V. Shaw. 1997. O-acetyltransferase for chloramphenicol and other natural products. Antimicrob. Agents Chemother. 41:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw, W. V. 1967. The enzymatic acetylation of chloramphenicol by extracts of R factor resistant Escherichia coli. J. Biol. Chem. 242:687-693. [PubMed] [Google Scholar]
- 18.Shaw, W. V. 1975. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 43:737-755. [DOI] [PubMed] [Google Scholar]
- 19.Shaw, W. V. 1983. Chloramphenicol acetyltransferase: enzymology and molecular biology. Crit. Rev. Biochem. 14:1-43. [DOI] [PubMed] [Google Scholar]
- 20.Stokes, H. W., and R. M. Hall. 1991. Sequence analysis of the inducible chloramphenicol resistance determinant in the Tn1696 integron suggests regulation by translation attenuation. Plasmid 26:10-19. [DOI] [PubMed] [Google Scholar]
- 21.Suzaki, Y., and S. Okamoto. 1967. The enzymatic acetylation of chloramphenicol by the multiple drug-resistant Escherichia coli. J. Biol. Chem. 242:4722-4728. [PubMed] [Google Scholar]
- 22.Tennigkeit, J., and H. Matzura. 1991. Nucleotide sequence analysis of a chloramphenicol-resistance determinant from Agrobacterium tumefaciens and identification of its gene product. Gene 99:113-116. [DOI] [PubMed] [Google Scholar]
- 23.Turner, S. L., G. C. Ford, A. Mountain, and A. Moir. 1992. Selection of a thermostable variant of chloramphenicol acetyltransferase (Cat-86). Protein Eng. 5:535-541. [DOI] [PubMed] [Google Scholar]
- 24.White, P. A., H. W. Stokes, K. L. Bunny, and R. M. Hall. 1999. Characterisation of a chloramphenicol acetyltransferase determinant found in the chromosome of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 175:27-35. [DOI] [PubMed] [Google Scholar]
- 25.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 26.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. NATO ASI Ser. Ser. A Life Sci. 1999:11-42. [Google Scholar]