Abstract
Dientamoeba fragilis is a protozoan parasite of humans that infects the mucosa of the large intestine and is associated with gastrointestinal disease. We developed a 5′ nuclease (TaqMan)-based real-time PCR assay, targeting the small subunit rRNA gene, for the detection of D. fragilis in human stool specimens and compared its sensitivity and specificity to conventional PCR and microscopic examination by a traditional modified iron-hematoxylin staining procedure. Real-time PCR exhibited 100% sensitivity and specificity.
Dientamoeba fragilis is a pathogenic protozoan parasite, often more prevalent than Giardia intestinalis, which causes gastrointestinal disease in humans (5, 7, 9, 17). Due to the propensity of this organism to cause chronic infection, it is essential that correct diagnosis occur promptly (3, 4, 7, 9, 13, 17). Two genotypes of D. fragilis have been described by analysis of the small subunit rRNA gene (10, 12, 17, 22), with only one predominant in Australia (17).
Diagnosis of D. fragilis relies on direct visualization of the trophozoites in stained fixed fecal smears by light microscopy, as demonstration of the characteristic nuclear structure cannot be achieved in unstained fecal specimens (4). D. fragilis may be difficult to distinguish from nonpathogenic protozoa (9).
The aim of this study was to develop a real-time PCR method that is rapid, highly sensitive, and specific for the detection of D. fragilis in fecal specimens. Results from the real-time assay were compared to those derived by a conventional PCR and microscopic examination using a traditional modified iron-hematoxylin staining procedure in order to determine the usefulness and practicality of this real-time PCR test.
(This research was performed by Damien Stark in partial fulfillment for the degree of Ph.D. at University of Technology Sydney.)
Entamoeba histolytica HM-1:IMSS (ATCC strain 30459) and Trichomonas vaginalis (ATCC strain F1623) were passaged in TYI-S-33 broth: genomic DNA was extracted from them using a QIAamp DNA minikit (QIAGEN, Hilden, Germany).
Stool specimens used in this study were those submitted to St. Vincent's Hospital Department of Microbiology, Sydney, for investigation of diarrhea. Portions of all stool samples were fixed in sodium acetate-acetic acid-formalin and permanently stained using a modified iron-hematoxylin stain (Fronine, Australia) according to the manufacturer's recommendations. DNA was extracted from fresh fecal specimens (<24 h old) using a QIAamp DNA stool minikit (QIAGEN, Hilden, Germany) according to the manufacturer's protocol. To ensure we were using “best practice” for the preparation of DNA from stool specimens using this kit, we also evaluated a modification of the manufacturer's instructions (8) using four samples positive for D. fragilis by microscopy. As a control, these same samples were also extracted following the manufacturer's instructions, and the two sets of DNA were then tested with conventional and real-time PCR.
The small subunit (SSU) rRNA genes from D. fragilis was amplified using the primers TRD3-TRD5 (16), and the 1.8-kb product was cloned into the PCR cloning TA vector, as described by the manufacturer (Invitrogen). After transformation into Escherichia coli (strain DH5α), individual transformants were screened for the presence of cloned DNA by PCR. Plasmid DNA from one of these clones (pDf18S rRNA genes) was purified from bacterial cultures grown in L broth using standard procedures. The purified recombinant DNA was quantified, determined to contain only one insert, and used for the sensitivity testing of the conventional and real-time PCR. Conventional PCR and DNA sequencing, using primers DF 400-DF1250 and DF3-DF4, were performed according to Stark etal. (16). Inhibition controls, comprising patient fecal samples spiked with cloned D. fragilis SSU rRNA genes, were also run to rule out PCR inhibition.
The SSU rRNA gene sequences present in GenBank from enteric protozoa normally associated with clinical signs of gastrointestinal disease in humans were aligned using the computer program Pileup. From this multiple sequence alignment, the following D. fragilis-specific primers and probe were developed: DF3 (5′-GTTGAATACGTCCCTGCCCTTT-3′), DF4 (5′-TGATCCAATGATTTCACCGAGTCA-3′), and dual-labeled TaqMan probe(5′-6-carboxyfluorescein-CACACCGCCCGTCGCTCC TACCG-6-carboxytetramethylrhodamine-3′).
Real-time PCR was performed using a LightCycler (Roche) in a 20-μl reaction volume in a glass capillary tube containing 2 μl of FastStart reaction mix hybridization probes (a component of the FastStart DNA master hybridization probes kit; Roche Diagnostics), 3 mM MgCl2, 0.25 μM forward and reverse primer, 0.2 μM dual-labeled fluorescent probe, and 2 μl of DNA extract.
Reaction conditions were as follows: 10 min at 95°C followed by 35 cycles of 95°C, 10 s at 58°C, and 3 s at 72°C. Temperature change rates were at 20°C/s. Readout was performed in channel F1.
To determine the sensitivity and detection limit of the PCR assay, known concentrations of pDf18S rRNA genes were serially diluted down to approximately 1 plasmid copy. These controls were then run with both the conventional PCR and the real-time PCR.
Optimal extraction of DNA from human feces was achieved by using a commercial kit with no modifications to the extraction procedure. Both the modified and unmodified method produced amplicons in both conventional and real-time PCR. Given that the modified extraction procedure greatly increased the time taken to process the specimens (>1 h), it was deemed that the extra steps taken were not needed. This is in contrast to a study that used the modified technique for extraction of DNA from feline feces (8).
A total of 200 fecal samples were screened by microscopy and conventional and real-time PCR. All 200 fecal samples spiked with pDf18S rRNA genes amplified the correct PCR product, showing that PCR inhibition was not an issue in this study.
Real-time PCR of the samples detected a total of 51 positives (Fig. 1), conventional PCR detected 48 and microscopy 50 D. fragilis-positive while samples. One sample positive by microscopy and negative by both PCR methods was subsequently deemed a false positive when the permanently stained smear was reexamined by an independent experienced microscopist who concluded that the nonpathogenic Endolimax nana was misidentified as D. fragilis. Two samples of the 150 deemed negative for D. fragilis by permanent staining produced amplicons by real-time PCR. One of these samples gave a product with conventional PCR. Upon sequencing of these amplicons, they were confirmed to be derived from D. fragilis DNA by DNA sequence comparisons. One of the 150 samples negative by microscopy was positive by conventional PCR, while 2 of the 150 microscopy samples were positive by real-time PCR. One of these samples contained Blastocystis hominis and E. nana, while the other contained the Entamoeba histolytica/dispar complex, and on subsequent review of each permanent slide, no D. fragilis was detected by microscopy.
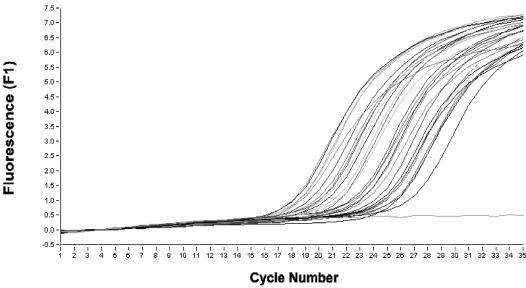
FIG. 1.
Detection of D. fragilis in feces by real-time PCR. One sample is a positive control, one sample is a negative control, and 30 samples are D. fragilis microscopy-positive samples.
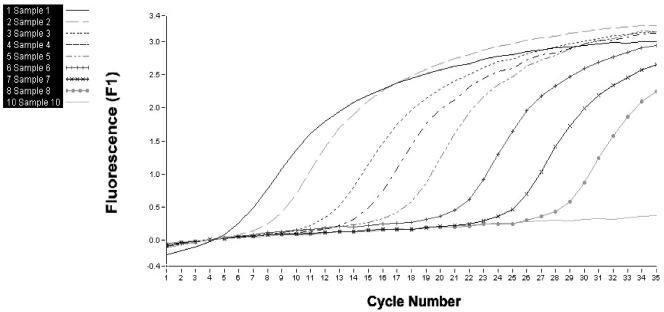
FIG. 2.
Evaluation of sensitivity of real-time PCR using cloned DNA. The results show that the following amounts of target are detectable: sample 1, 10,000,000 rRNA gene copies; sample 2, 1,000,000 rRNA gene copies; sample 3, 100,000 rRNA gene copies; sample 4, 10,000 rRNA gene copies; sample 5, 1,000 rRNA gene copies; sample 6, 100 rRNA gene copies; sample 7, 10 rRNA gene copies; sample 8, 1 rRNA gene copy. Sample 10 is a negative control.
To determine the specificity of the real-time PCR, 29 specimens containing various other protozoan parasites (Table 1) underwent direct DNA extraction from fresh stool samples. No PCR products were obtained from any of these specimens. A further 29 specimens containing no protozoa underwent direct DNA extraction on fresh stool samples. No PCR products were detected in this group either. To further determine any cross-reactivity of the real-time assay, PCR was also performed on genomic DNA from strains of E. histolytica, T. vaginalis, and Trichomonas fetus (ATCC strain 3000). Only T. vaginalis and T. fetus DNA produced a PCR product by this technique.
TABLE 1.
List of specimens used in this study containing various other protozoan parasites
| Specimen no. | Protozoan parasite(s) present |
|---|---|
| 1 | Blastocystis hominis |
| 2 | Entamoeba histolytica |
| 3 | Entamoeba dispar |
| 4 | Entamoeba coli |
| 5 | Entamoeba hartmanni |
| 6 | Giardia intestinalis |
| 7 | Endolimax nana |
| 8 | Iodamoeba butschlii |
| 9 | Cryptosporidium species |
| 10 | Cyclospora species |
| 11 | Chilomastix mesnili |
| 12 | E. histolytica/dispar complex, E. coli, E. nana, B. hominis, and Enteromonas hominis |
| 13 | E. histolytica/dispar complex, E. coli, E. nana, and I. butschlii |
| 14 | E. hartmanni, E. nana, E. hominis, and B. hominis |
| 15 | E. nana, I. butschlii, and B. hominis |
| 16 | Cryptosporidium species and B. hominis |
| 17 | E. nana, I. butschlii, C. mesnili, and B. hominis |
| 18 | G. intestinalis, E. nana, and B. hominis |
| 19 | E. coli, E. nana, and B. hominis |
| 20 | G. intestinalis, E. coli, E. hartmanni, and E. hominis |
| 21 | E. coli, E. nana, I. butschlii, and B. hominis |
| 22 | E. histolytica/dispar complex, E. hartmanni, E. hominis, and B. hominis |
| 23 | E. histolytica/dispar complex, E. nana, E. hominis, and B. hominis |
| 24 | E. coli, E. hartmanni, E. nana, I. butschlii, and B. hominis |
| 25 | E. hartmanni and E. nana |
| 26 | G. intestinalis, E. histolytica/dispar complex, E. hartmanni, E. coli, E. hominis, I. butschlii, and B. hominis |
| 27 | E. histolytica/dispar complex, E. nana, B. hominis, and Cryptosporidium species |
| 28 | C. mesnili, E. hominis, and E. nana |
| 29 | G. intestinalis, E. nana, and B. hominis |
To determine the sensitivity of both conventional and real-time PCR, a known number of copies of pDf18S rRNA genes were then amplified using the same conditions as for the patients' samples. This showed that the detection limit was 100 plasmid copies or an equivalent of approximately 1.0 D. fragilis trophozoite for conventional PCR. The detection limit for the real-time PCR was determined at 1 plasmid copy (a crossing point of 27.87) of the SSU rRNA gene, which is equivalent to approximately 0.01 D. fragilis trophozoite (Fig. 1). It should be noted that we are describing analytical sensitivity and not clinical sensitivity, and these results may differ from those obtained using clinical stool specimens. DNA from stool is complex, containing human, fungal, and plant DNA plus organic material, as well as DNA from thousands of microorganisms. Thus, there is considerably more interaction of primers and probes with this complex fecal DNA than with the purified plasmid.
In summary, based on the analysis of the fecal samples, microscopy showed 92.4% sensitivity and 98.7% specificity and conventional PCR showed 88.9% sensitivity and 100% specificity compared with 100% sensitivity and specificity for real-time PCR (Table 2).
TABLE 2.
Comparison of PCR, conventional PCR, and microscopy for detection of D. fragilis
| Method | No. of samples examined | No. of positive samples detected | Sensitivity (%)a | Specificity (%)b |
|---|---|---|---|---|
| Real-time PCR | 200 | 51 | 100 | 100 |
| Conventional PCR | 200 | 48 | 88.9 | 100 |
| Microscopyc | 200 | 50 | 92.4 | 98.7 |
Calculated as follows: (number of true positives/[number of true positives + number of false negatives]) × 100.
Calculated as follows: (number of true negatives/[number of true negatives + number of false positives]) × 100.
Microscopy was performed using a modified iron-hematoxylin stain (Fronine, Australia).
In this study, we developed a new 5′ nuclease (TaqMan)-based real-time PCR assay, targeting the small subunit rRNA gene, for the detection of D. fragilis in human stool specimens. We then evaluated the ability of real-time PCR to detect D. fragilis in fecal specimens and compared it with conventional PCR and microscopy.
In the comparison, microscopy missed two positive samples and also gave a false positive. The preparation of each slide and the staining procedure using a modified iron-hematoxylin stain are time-consuming, and considerable expertise is also required in the reading and interpretation of the slides by the microscopist. Microscopy showed a sensitivity of 92.4%; this high sensitivity can be attributed to the amount of time spent screening and the highly experienced microscopists who were reading the slides. The conventional PCR detected only 48 positive samples in comparison to the 51 samples detected by the real-time PCR. This sensitivity of 88.9% for the conventional PCR is lower than has been previously reported (16, 17). The real-time PCR was shown to possess a higher level of sensitivity (100%) for the detection of D. fragilis in feces. Real-time PCR methods have been utilized in several areas of clinical parasitology, including the detection of fecal parasites. These real-time PCR assays have been shown to be more sensitive and specific than conventional methods (2, 18, 20, 21). This study represents the first report of the application of a real-time PCR assay for the detection of D. fragilis in human fecal specimens.
Fresh stool samples only were used in this study (<24 h old), as D. fragilis does not produce cysts, and the trophozoites degenerate rapidly once passed (9). Stark et al. (16) demonstrated that aged specimens affected the sensitivity of a conventional PCR assay for detecting D. fragilis in stool specimens.
Of the cultured organisms that were studied by PCR, T. vaginalis and T. fetus DNA produced an amplicon with the D.fragilis primers, and subsequent alignments of T. vaginalis, T. fetus, and D. fragilis SSU rRNA genes showed the priming sites to be highly conserved and almost identical among the three species. This shows the close relationship D. fragilis has with other trichomonads (6, 15). However, T. vaginalis and T. fetus are not found in human stool samples, and the clinical samples containing other enteric protozoa, including trichomonads, used in this study (Chilomastix mesnili, Pentatrichomonas hominis, and Enteromonas hominis) did not produce an amplicon with the primers. This cross-reactivity does not affect the usefulness of this real-time assay in a clinical setting. Inhibition controls were carried out to exclude the possibility of inhibitory substances, and all were negative. Thus, the real-time PCR was shown to have 100% specificity for enteric specimens. On a similar note, we have yet to evaluate the use of this PCR for T. vaginalis in clinical specimens.
Real-time PCR offers several advantages over conventional PCR for a diagnostic laboratory, such as increased sensitivity (1, 11, 14, 19), rapid cycling times, and reduced risk of contamination. The disadvantages of real-time PCR include the cost of the assay, which is substantially higher than that of either microscopy or conventional PCR, and the need for specialized real-time PCR analyzers, which are currently beyond the means of many laboratories.
In summary, this work is the first report of a real-time PCR assay specific for D. fragilis. On fresh stools that had undergone direct DNA extraction promptly (within 24 h), the sensitivity of the PCR was 100% and the specificity was 100%. The PCR method is quick (<2 h) and simple and so offers a diagnostic tool other than light microscopy and conventional PCR for the diagnosis of dientamoebiasis.
Acknowledgments
This research was funded by St. Vincent's Hospital.
REFERENCES
- 1.Bialek, R., N. Binder, K. Dietz, A. Joachin, J. Knobloch, and U. E. Zelck. 2002. Comparison of fluorescence, antigen and PCR assays to detect Cryptosporidium parvum in fecal specimens. Diagn. Microbiol. Infect. Dis. 43:283-288. [DOI] [PubMed] [Google Scholar]
- 2.Blessmann, J., H. Buss, P. A. Nu, B. T. Dinh, Q. T. Ngo, A. L. Van, M. D. Alla, T. F. Jackson, J. I. Ravdin, and E. Tannich. 2002. Real-time PCR for the detection and differentiation of Entamoeba histolytica and Entamoeba dispar in fecal samples. J. Clin. Microbiol. 40:4413-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borody, T. J., E. F. Warren, A. Wettstein, G. Robertson, P. Recabarren, A. Fontella, K. Herdnman, and R. Surace. 2002. Eradication of Dientamoeba fragilis can resolve IBS-like symptoms. J. Gastroenterol. Hepatol. 17(Suppl.):A103. [Google Scholar]
- 4.Butler, W. P. 1996. Dientamoeba fragilis. An unusual intestinal pathogen. Dig. Dis. Sci. 41:1811-1813. [DOI] [PubMed] [Google Scholar]
- 5.Crotti, D., M. L. D'Annibale, G. Fonzo, M. Lalle, S. M. Caccio, and E. Pozio. 2005. Dientamoeba fragilis is more prevalent than Giardia duodenalis in children and adults attending a day care centre in Central Italy. Parasite 12:165-170. [DOI] [PubMed] [Google Scholar]
- 6.Delgado-Viscogliosi, P., E. Viscogliosi, D. Gerbod, J. Kulda, M. L. Sogin, and V. P. Edgcomb. 2000. Molecular phylogeny of parabasalids based on small subunit rRNA sequences, with emphasis on the Trichomonadinae subfamily. J. Eukaryot. Microbiol. 47:70-75. [DOI] [PubMed] [Google Scholar]
- 7.Girginkardesler, N., S. Coskun, I. Cuneyt Balcioglu, P. Ertan, and U. Z. Ok. 2003. Dientamoeba fragilis, a neglected cause of diarrhoea, successfully treated with secnidazole. Clin. Microbiol. Infect. 9:110-113. [DOI] [PubMed] [Google Scholar]
- 8.Gookin, J. L., A. J. Birkenheuer, E. B. Breitschwerdt, and M. G. Levy. 2002. Single-nested PCR for the detection of Tritichomonas foetus in feline feces. J. Clin. Microbiol. 40:4126-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson, E. H., J. J. Windsor, and G. C. Clark. 2004. Emerging from obscurity: biological, clinical, and diagnostic aspects of Dientamoeba fragilis. Clin. Microbiol. Rev. 17:553-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, J. A., and C. G. Clarke. 2000. Cryptic diversity in Dientamoeba fragilis. J. Clin. Microbiol. 38:4653-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pang, X., B. Lee, L. Chui, J. K. Preiksaitis, and S. S. Monroe. 2004. Evaluation and validation of real-time reverse transcription-PCR assay using the LightCycler system for detection and quantitation of norovirus. J. Clin. Microbiol. 42:4679-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peek, R., F. R. Reedeker, and T. van Gool. 2004. Direct amplification and genotyping of Dientamoeba fragilis from human stool specimens. J. Clin. Microbiol. 42:631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Priess, U., G. Ockert, S. Broemme, and A. Otto. 1991. On the clinical importance of Dientamoeba fragilis infections in children. J. Hyg. Epidemiol. Microbiol. Immunol. 35:27-34. [PubMed] [Google Scholar]
- 14.Roy, S., M. Kabir, D. Mondal, I. K. M. Ali, W. A. Petri, Jr., and R. Haque. 2005. Real-time-PCR assay for diagnosis of Entamoeba histolytica infection. J. Clin. Microbiol. 43:2168-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silberman, J. D., C. G. Clark, and M. L. Sogin. 1996. Dientamoeba fragilis shares a recent common evolutionary history with the trichomonads. Mol. Biochem. Parasitol. 76:311-314. [DOI] [PubMed] [Google Scholar]
- 16.Stark, D., N. Beebe, D. Marriott, J. T. Ellis, and J. Harkness. 2005. Detection of Dientamoeba fragilis in fresh stool specimens using PCR. Int. J.Parasitol. 35:57-62. [DOI] [PubMed] [Google Scholar]
- 17.Stark, D., N. Beebe, D. Marriott, J. T. Ellis, and J. Harkness. 2005. A prospective study on the prevalence, genotyping, and clinical relevance of Dientamoeba fragilis infections in an Australian population. J. Clin. Microbiol. 43:2718-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanriverdi, S., A. Tanyeli, F. Baslamisli, F. Koksal, Y. Kilinc, X. Feng, G. Batzer, S. Tzipori, and G. Widmer. 2002. Detection and genotyping of oocysts of Cryptosporidium parvum by real-time PCR and melting curve analysis. J. Clin. Microbiol. 40:3231-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Templeton, K. E., S. A. Scheltinga, A. van der Zee, B. M. W. Diederen, A. M. Kruijssen, H. Goossens, E. Kuijper, and E. C. J. Claas. 2003. Evaluation of real-time PCR for detection of and discrimination between Bordetella pertussis, Bordetella parapertussis, and Bordetella holmesii for clinical diagnosis. J. Clin. Microbiol. 41:4121-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varma, M., J. D. Hester, F. W. Schaefer, M. W. Ware, and H. D. Lindquist. 2003. Detection of Cyclospora cayetanensis using a qualitative real-time PCR assay. J. Microbiol. Methods 53:27-36. [DOI] [PubMed] [Google Scholar]
- 21.Verweij, J. J., J. Blotkamp, E. A. Brienen, A. Aguirre, and A. M. Polderman. 2000. Differentiation of Entamoeba histolytica and Entamoeba dispar cysts using polymerase chain reaction on DNA isolated from faeces with spin columns. Eur. J. Clin. Microbiol. Infect. Dis. 19:358-361. [DOI] [PubMed] [Google Scholar]
- 22.Windsor, J. J., C. G. Clarke, and L. Macfarlane. 2004. Molecular typing of Dientamoeba fragilis. Br. J. Biomed. Sci. 61:153-155. [DOI] [PubMed] [Google Scholar]