Abstract
Alanine racemases are ubiquitous prokaryotic enzymes providing the essential peptidoglycan precursor d-alanine. We present evidence that the enzymes from Pseudomonas aeruginosa and Escherichia coli function exclusively as homodimers. Moreover, we demonstrate that expression of a K35A Y235A double mutation of dadX in E. coli suppresses bacterial growth in a dominant negative fashion.
In most bacteria the d-alanine-d-alanyl dipeptide is essential for cross-linking within the peptidoglycan cell wall. It is produced from the natural amino acid l-alanine by the subsequent action of two enzymes, alanine racemases, to create d-alanine and d-alanine-d-alanyl ligase. While some organisms contain only one alanine racemase, others have two isozymes, encoded by the alr and the dadX genes. The latter group includes Escherichia coli (4) and Pseudomonas aeruginosa (12). Little is known about the advantages or disadvantages of having two enzymes or about the interaction between the enzymes. alr expression is constitutive and seems to provide the d-alanine necessary to maintain cell growth, while dadX gene expression is induced by l-alanine to a level much greater than that of alr and thus is most active when l-alanine is used as a carbon and energy source (4).
Alanine racemase is an attractive target for the development of new antibiotics; the enzyme is ubiquitous among bacteria and generally absent in higher eukaryotes, although it has recently been found to function as a means of osmoregulation in some lower marine eukaryotes (7, 10).
There are still open questions regarding the phylogenetic history of the two alanine racemase genes. For instance, the sequence identity between Alr and DadX enzymes from the same organism is not significantly greater than between enzymes from different bacteria (12). Likewise, the structural differences between alanine racemases deserve further attention. While the alanine racemases from Salmonella enterica serovar Typhimurium (1, 15), Thermus thermophilus (8), and Shigella sp. (19) have been described as monomers, the crystal structure of the alanine racemase from Bacillus stearothermophilus reveals a dimer with two identical polypeptide chains (9). The dimer has two active sites, each of which contains residues from both monomers. It is noteworthy that, especially within these active center domains, alanine racemase protein sequences from different bacteria share a high level of conservation. Pyridoxal-5′-phosphate (PLP), the essential cofactor of alanine racemases, is in aldimine linkage to an N-terminal lysine residue as a protonated Schiff's base (6). This residue (K39), together with a tyrosine residue near the C terminus (Y265) from the other subunit in the dimer, has been identified as being critical for the racemization of alanine (16, 17). The contribution of these two residues to enzyme activity was further confirmed through cocrystallization of the B. stearothermophilus alanine racemase with one of its inhibitors, 1-aminoethylphosphonate (11). While the lysine residue is clearly the catalytic base for the racemization of l- to d-alanine (17), the tyrosine ring is necessary to stabilize the hydrogen-bonding network in the active site with d-alanine as the substrate and to catalyze the reverse reaction, i.e., the conversion of d- to l-alanine (16, 18).
For our studies we selected the DadX enzyme from E. coli, which shares an 85% identity with the Salmonella enzyme, and the alanine racemases from P. aeruginosa, an important opportunistic human pathogen. We are particularly interested in whether both species of alanine racemase act as dimers, whether they can form mixed heterodimers, and how this interaction might be exploited for inhibiting the enzyme. If the enzymes are obligate dimers, interrupting the interaction between the monomers might be an attractive means of inhibiting cell growth.
Cloning of the alanine racemases from E. coli and P. aeruginosa.
Gene-specific primers were designed based on the sequences of the respective alanine racemase in GenBank (accession numbers: alrPA, AF165882; dadXPA, AF165881; dadXEC, AE000217; subscripts PA and EC indicate P. aeruginosa and E. coli, respectively). Chromosomal DNA was prepared from P. aeruginosa PAO1 and E. coli MG1655 (5), the genes were amplified by touchdown-PCR (3), and the resulting fragments were ligated into the N-terminal 6-His tag fusion vector pET28 (Table 1). The plasmids were transformed into E. coli MB1457 and verified by DNA sequence analysis. Some members of our group previously reported the cloning of the two Pseudomonas genes (12); the sequence of the E. coli gene was identical to the published sequence for strain MG1655 (GenBank accession no. AE000217). Sequence comparison using BLAST revealed a 49% overall identity between the two DadX proteins and also between the two Pseudomonas alanine racemases, AlrPA and DadXPA. In particular, the two areas around the key lysine residue near the N terminus and the tyrosine residue closer to the C terminus displayed strong sequence identity with the conserved motifs, ([AS]-V-[VI]-K-A-[ND]-A-Y-G-H-G) and (G-E-x-V-G-Y-G-[AG]-[TR]-[WY]), respectively.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| MG1655 | lam rph-1 | Laboratory stock |
| BL21(DE3)/pLysS | F−ompT hsdSB(rB− mB−) gal dcm (DE3) Cmr | Novagen, Madison, Wis. |
| MB414 | recA56 srl::Tn10 | Laboratory stock |
| MB1457 | DH5α recA+ | Laboratory stock |
| MB1547 | supE thi hsdΔ5 Δ(lac-proAB) endA F′[traD36 proAB lacIqlacZΔM15] | Laboratory stock |
| MB2859 | MB1547 alr::FRT | This work |
| MB2946 | MG1655 alr::FRT dadX::FRT recA56 srl::Tn10 | This work |
| P. aeruginosa | ||
| PAO1 | Wild-type strain | Laboratory stock |
| Plasmids | ||
| pET28 | T7lac promoter expression vector, Kmr | Novagen |
| pBAD18 | Apr oriColE1 | 2 |
| pBAD33 | Cmr orip15 | 2 |
| pUC19 | Apr | 14 |
| pMB3051 | pUC19-dadXEC (K35A Y253A) | This work |
Mutagenesis of the alanine racemase variants.
Site-directed mutagenesis was used to replace the two putative key residues, lysine and tyrosine, in the active sites of DadXPA, DadXEC, and AlrPA. Two PCRs were set up using the pET28-cloned wild-type genes as the template, one combining the T7 terminator primer with a mutagenesis primer and the other combining the T7 promoter primer with the complementary mutagenesis primer. The completed reaction mixtures were treated with DpnI and purified. A second, four-cycle PCR containing equimolar amounts of the partially overlapping PCR products created the full-length product through self-priming. T7 primers were added, and the PCR was continued under standard conditions. The PCR products were ligated into pET28, and all mutations were verified by sequencing the entire gene.
Complementation by pairs of mutants.
The alanine racemase-deficient E. coli strain MB2946, which was an alr dadX mutant constructed as described earlier (13), was the host used for the posttranslational complementation analysis of the various alanine racemase genes investigated in this study. In order to prevent intermolecular recombination, we introduced the recA allele from MB414 into MB2946 by P1vir transduction. The various alanine racemase variants of alrPA, dadXPA, and dadXEC were transferred from pET28 into pBAD18 and pBAD33. These vectors can coexist in the same cell and encode resistance to different antibiotics, allowing the easy selection of cotransformation events.
All pair-wise combinations of the various pBAD18 and pBAD33 constructs were cotransformed into E. coli MB2946 and screened for restoration of prototrophy (Table 2). Plasmid selection initially occurred on d-alanine-supplemented Luria-Bertani agar with ampicillin and chloramphenicol. Fifty single colonies were then picked and transferred to drug plates without d-alanine, and the plates were scored for growth after 24 h at 37°C. None of the mutant alleles was able to complement growth of the alanine racemase-deficient double mutant of E. coli, whereas the wild-type alleles could. With respect to the cotransformants, allelic complementation occurred only when both plasmids expressed mutants of the same enzyme; i.e., the DadXPA (K33A) mutant complemented only when cotransformed with the DadXPA (Y253A) mutant, but not with the DadXEC (Y253A) mutant or AlrPA (Y254A) mutant. It is notable that all three enzymes behaved identically in this respect. The fact that complementation does occur demonstrates that all three alanine racemases tested here function as homodimers. The N terminus of one monomer interacts with the C terminus of the other monomer and vice versa. Each of the monomers contributes residues to the active site, i.e., the lysine and the tyrosine residues crucial for the catalytic activity of the enzyme.
TABLE 2.
Complementation of an E. coli alanine racemase double mutant with combinations of mutant alanine racemases from E. coli and P. aeruginosaa
| Plasmid construct | No second plasmid | pBAD18-dadXPA (K33A) | pBAD33-dadXPA (K33A) | pBAD18-dadXPA (Y253A) | pBAD33-dadXPA (Y253A) | pBAD18-dadXEC (K35A) | pBAD33-dadXEC (K35A) | pBAD18-dadXEC (Y253A) | pBAD33-dadXEC (Y253A) | pBAD18-alrPA (K34A) | pBAD33-alrPA (K34A) | pBAD18-alrPA (Y254A) | pBAD33-alrPA (Y254A) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pBAD18-dadXPA | + | ||||||||||||
| pBAD18-dadXPA (K33A) | − | + | − | − | |||||||||
| pBAD33-dadXPA | + | ||||||||||||
| pBAD33-dadXPA (K33A) | − | + | − | − | |||||||||
| pBAD18-dadXPA (Y253A) | − | + | − | − | |||||||||
| pBAD33-dadXPA (Y253A) | − | + | − | − | |||||||||
| pBAD18-dadXEC | + | ||||||||||||
| pBAD18-dadXEC (K35A) | − | − | + | − | |||||||||
| pBAD33-dadXEC | + | ||||||||||||
| pBAD33-dadXEC (K35A) | − | − | + | − | |||||||||
| pBAD18-dadXEC (Y253A) | − | − | + | − | |||||||||
| pBAD33-dadXEC (Y253A) | − | − | + | − | |||||||||
| pBAD18-alrPA | + | ||||||||||||
| pBAD18-alrPA (K34A) | − | − | − | + | |||||||||
| pBAD33-alrPA | + | ||||||||||||
| pBAD33-alrPA (K34A) | − | − | − | + | |||||||||
| pBAD18-alrPA (Y254A) | − | − | − | + | |||||||||
| pBAD33-alrPA (Y254A) | − | − | − | + |
−, d-alanine auxotrophy; +, restoration of prototrophy.
Our inability to observe prototrophy when pairs of mutants of different proteins were used could have a number of possible explanations, among which we cannot distinguish. The monomers may be unable to form heterologous dimers at all, due to subtle structural differences, or dimers might be formed, but the subunits are unable to form a functional active site or they form one with activity too low to be observed.
To rule out the possibility that our results were due to recombination between homologous genes despite the recA host, and not protein complementation, we checked for intermolecular recombination between the two plasmids in MB2946 by retransformation and electrophoretic analysis of the plasmid DNA. There was no evidence for the formation of plasmid multimers, only monomeric plasmids were observed, and no wild-type recombinants were found (data not shown).
Formation of functional heteroalleles in vitro.
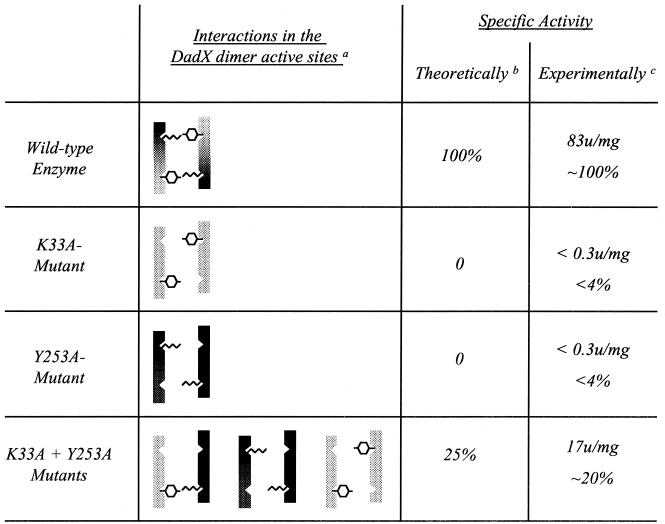
To further verify our observations, we investigated the formation of mixed dimers from purified DadX alanine racemase of P. aeruginosa in vitro. It had previously been observed that there was no significant difference in the specific activities of AlrPA and DadXPA compared to those of their His-tagged derivatives (12); thus, DadXPA (K33A) and DadXPA (Y253A) were purified independently (Fig. 1) by virtue of their His tags and assayed in a spectrophotometric alanine racemase assay in the d→l direction (1). Both proteins were inactive in the assay, as expected from their failure to complement the E. coli alanine racemase mutant. When both protein preparations were simply combined in an equal stoichiometric ratio, we were still unable to measure any enzyme activity. However, when a stoichiometric mixture of both variants was dialyzed against 50 mM Tris-HCl (pH 8.0)-6 M urea to separate the existing inactive dimers and then refolded and redimerized by successive dialysis against 50 mM Tris-HCl (pH 8.0) containing 4, 2, and 0 M urea, significant alanine racemase activity was restored (Fig. 2). About 20% of the specific activity of the wild-type enzyme was seen in this preparation. A loss of 50% of the specific activity would be expected because the denaturation and renaturation process will restore both inactive homodimers and active heterodimers; additionally, the active heterodimers have only a single active site, instead of two in the wild-type enzyme, resulting in another 50% loss of activity, giving a maximum theoretical activity of 25%, provided there is no bias in the affinity of the different monomers for each other.
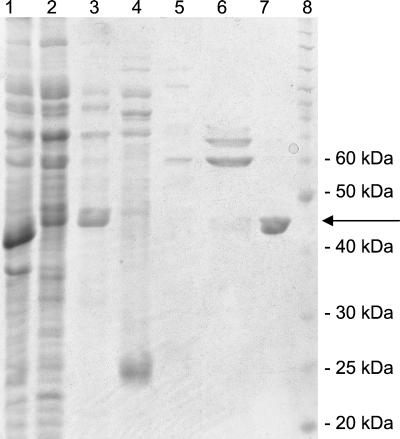
FIG. 1.
Purification of Pseudomonas aeruginosa His6-DadX (K33A) from E. coli BL21(DE3)/pLysS, pET28-dadXPA (K33A). Lane 1, crude extract; lane 2, flowthrough fraction, not binding to Pharmacia's His-Trap matrix; lanes 3 to 6, elution of non-His-tagged proteins with 1, 5, 10, 50, and 100 mM imidazole, respectively; lane 7, elution of His6-DadX (K33A) with 500 mM imidazole; lane 8, molecular weight standard (Invitrogen). The wild-type and the His6-DadX (Y253A) proteins were purified in the same way (not shown). An arrow indicates the His6-DadX protein.
FIG. 2.
Restoration of P. aeruginosa DadX activity through denaturation and renaturation of a mixture of two independently generated mutant proteins. a, in the alanine racemase dimer two catalytic sites are formed with K33 and Y253 as two of their key residues; the possible active and inactive combinations for the mutants studied are listed. b, in determining values for theoretically determined specific activity, it was presumed that there is no difference in mutual affinity between wild-type and mutant monomers. c, purified enzyme variants were used to determine the specific activity in a spectrophotometric assay measuring the conversion of d- to l-alanine; the protein was denatured in 6 M urea and renatured as described; the wild-type specific activity was also measured after denaturation and renaturation (note that since 0.3 U/mg was our detection limit in the assay, no activity was detectable at <0.3 U/mg).
While our findings do not necessarily exclude the possibility that the three alanine racemases studied here exist as monomers, our experiments do conclusively show that they are active as homodimeric units. Given the elaborate structural and mechanistical investigations of the B. stearothermophilus enzyme as described earlier, the level of homology within the active center domains throughout most bacterial alanine racemases, and the recently established X-ray structure of P. aeruginosa DadX protein as a homodimer (P. M. Le Magueres and K. L. Krause, unpublished data), it seems likely that alanine racemases are universally active as dimers. Earlier studies had already shown that the wild-type enzyme follows Michaelis-Menten kinetics (12); cooperation between the two active sites is thus unlikely.
Alanine racemase mutants as dominant negative alleles can act like a peptide antibiotic.
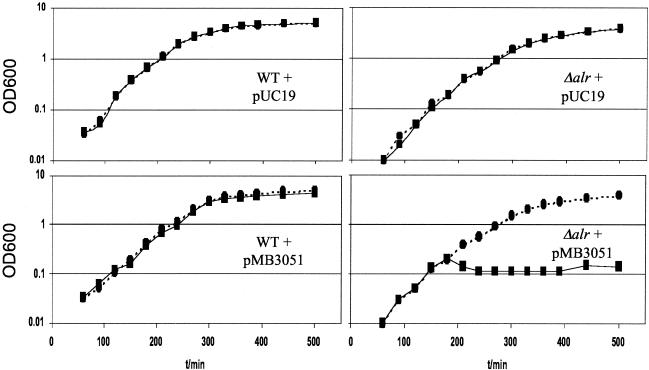
Alanine racemase is a potentially attractive target for the development of novel antibiotics. We investigated whether it is possible to inhibit bacterial growth by expressing a mutant alanine racemase. For this purpose, a dadXEC double mutant (K35A Y253A) was constructed by transferring the insert from pET28-dadXEC (K35A) into pUC19 and replacing the SphI-EcoRI C-terminal fragment in the resulting plasmid with the SphI-EcoRI fragment from pET28-dadXEC (Y253A). Sequencing of the complete gene confirmed the presence of both active-site mutations in the insert of the newly designed plasmid, pMB3051. Expression in E. coli MB2859, an alr::FRT mutant of E. coli MB1547 (lacIq), created as described earlier (13), was studied. The dadXEC (K35A Y253A) allele in pMB3051 is under the control of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible lacUV5 promoter, and when its expression was induced, bacterial growth was effectively inhibited (Fig. 3).
FIG. 3.
Dominant negative effect of a dadXEC double mutant on E. coli alr. The K35A Y253A allele of dadXEC (pMB3051) or a control plasmid (pUC19) was transformed into strains MB2859 (alr) and MB1547 (wild type). Cell growth was monitored with (solid line, square symbols) and without (dotted line, round symbols) the addition of 1 mM IPTG (added 180 min after inoculation) in Luria-Bertani medium at 37°C.
Expression of the double-mutant protein was apparently sufficient to sequester enough of the wild-type protein into inactive dimers to preclude wild-type DadX dimers from being formed. While this is an encouraging result, it has to be considered that our experiment was performed with an alr mutant. In the wild-type strain, the alr gene provides sufficient alanine racemase to support growth. Alr is not significantly affected by the DadX overexpression. Any potential inhibitor protein or inhibitor peptide would have to be engineered to bind sufficiently well to both Alr and DadX to effectively control bacterial growth or be intended for those organisms, such as Mycobacterium tuberculosis and Mycobacterium avium (13), that have only a single alanine racemase gene. However, this demonstrates the potential effectiveness of new alanine racemase inhibitors, peptide based or not, that do not target the catalytic mechanism of the enzyme but instead block dimer formation. This could have the added advantage of being highly strain specific and not interfere with other PLP-containing or alanine-utilizing enzymes.
Acknowledgments
This work was supported by National Institutes of Health grant AI46340 and the Robert A. Welch Foundation.
We are grateful to the members of the Briggs and Krause labs, our collaborators, for helpful discussions.
REFERENCES
- 1.Esaki, N., and C. T. Walsh. 1986. Biosynthetic alanine racemase of Salmonella typhimurium: purification and characterization of the enzyme encoded by the alr gene. Biochemistry 25:3261-3267. [DOI] [PubMed] [Google Scholar]
- 2.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hecker, K. H., and K. H. Roux. 1996. High and low annealing temperatures increase both specificity and yield in touchdown and stepdown PCR. BioTechniques 20:478-485. [DOI] [PubMed] [Google Scholar]
- 4.Lobocka, M., J. Hennig, J. Wild, and T. Klopotowski. 1994. Organization and expression of the Escherichia coli K-12 dad operon encoding the smaller subunit of d-amino acid dehydrogenase and the catabolic alanine racemase. J. Bacteriol. 176:1500-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mak, Y. M., and K. K. Ho. 1992. An improved method for the isolation of chromosomal DNA from various bacteria and cyanobacteria. Nucleic Acids Res. 20:4101-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morollo, A. A., G. A. Petsko, and D. Ringe. 1999. Structure of a Michaelis complex analogue: propionate binds in the substrate carboxylate site of alanine racemase. Biochemistry 38:3293-3301. [DOI] [PubMed] [Google Scholar]
- 7.Nomura, T., I. Yamamoto, F. Morishita, Y. Furukawa, and O. Matsushima. 2001. Purification and some properties of alanine racemase from a bivalve mollusc Corbicula japonica. J. Exp. Zool. 289:1-9. [DOI] [PubMed] [Google Scholar]
- 8.Seow, T. K., K. Inagaki, T. Nakamura, R. Maeda, T. Tamura, and H. Tanaka. 2000. Purification and some characteristics of a monomeric alanine racemase from an extreme thermophile, Thermus thermophilus. J. Biosci. Bioeng. 90:344-346. [DOI] [PubMed] [Google Scholar]
- 9.Shaw, J. P., G. A. Petsko, and D. Ringe. 1997. Determination of the structure of alanine racemase from Bacillus stearothermophilus at 1.9-Å resolution. Biochemistry 36:1329-1342. [DOI] [PubMed] [Google Scholar]
- 10.Shibata, K., K. Shirasuna, K. Motegi, Y. Kera, H. Abe, and R. Yamada. 2000. Purification and properties of alanine racemase from crayfish Procambarus clarkii. Comp. Biochem. Physiol. B 126:599-608. [DOI] [PubMed] [Google Scholar]
- 11.Stamper, G. F., A. A. Morollo, D. Ringe, and C. G. Stamper. 1998. Reaction of alanine racemase with 1-aminoethylphosphonic acid forms a stable external aldimine. Biochemistry 37:10438-10445. [DOI] [PubMed] [Google Scholar]
- 12.Strych, U., H. C. Huang, K. L. Krause, and M. J. Benedik. 2000. Characterization of the alanine racemases from Pseudomonas aeruginosa PAO1. Curr. Microbiol. 41:290-294. [DOI] [PubMed] [Google Scholar]
- 13.Strych, U., R. L. Penland, M. Jimenez, K. L. Krause, and M. J. Benedik. 2001. Characterization of the alanine racemases from two mycobacteria. FEMS Microbiol. Lett. 196:93-98. [DOI] [PubMed] [Google Scholar]
- 14.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 15.Wasserman, S. A., E. Daub, P. Grisafi, D. Botstein, and C. T. Walsh. 1984. Catabolic alanine racemase from Salmonella typhimurium: DNA sequence, enzyme purification, and characterization. Biochemistry 23:5182-5187. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe, A., Y. Kurokawa, T. Yoshimura, and N. Esaki. 1999. Role of tyrosine 265 of alanine racemase from Bacillus stearothermophilus. J. Biochem. (Tokyo) 125:987-990. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe, A., Y. Kurokawa, T. Yoshimura, T. Kurihara, K. Soda, N. Esaki, and A. Watababe. 1999. Role of lysine 39 of alanine racemase from Bacillus stearothermophilus that binds pyridoxal 5′-phosphate. Chemical rescue studies of Lys39→Ala mutant. J. Biol. Chem. 274:4189-4194. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe, A., T. Yoshimura, B. Mikami, and N. Esaki. 1999. Tyrosine 265 of alanine racemase serves as a base abstracting alpha-hydrogen from L-alanine: the counterpart residue to lysine 39 specific to D-alanine. J. Biochem. (Tokyo) 126:781-786. [DOI] [PubMed] [Google Scholar]
- 19.Yokoigawa, K., R. Hirasawa, H. Ueno, Y. Okubo, S. Umesako, and K. Soda. 2001. Gene cloning and characterization of alanine racemases from Shigella dysenteriae, Shigella boydii, Shigella flexneri, and Shigella sonnei. Biochem. Biophys. Res. Commun. 288:676-684. [DOI] [PubMed] [Google Scholar]