Abstract
Escherichia coli grows over a wide range of pHs (pH 4.4 to 9.2), and its own metabolism shifts the external pH toward either extreme, depending on available nutrients and electron acceptors. Responses to pH values across the growth range were examined through two-dimensional electrophoresis (2-D gels) of the proteome and through lac gene fusions. Strain W3110 was grown to early log phase in complex broth buffered at pH 4.9, 6.0, 8.0, or 9.1. 2-D gel analysis revealed the pH dependence of 19 proteins not previously known to be pH dependent. At low pH, several acetate-induced proteins were elevated (LuxS, Tpx, and YfiD), whereas acetate-repressed proteins were lowered (Pta, TnaA, DksA, AroK, and MalE). These responses could be mediated by the reuptake of acetate driven by changes in pH. The amplified proton gradient could also be responsible for the acid induction of the tricarboxylic acid (TCA) enzymes SucB and SucC. In addition to the autoinducer LuxS, low pH induced another potential autoinducer component, the LuxH homolog RibB. pH modulated the expression of several periplasmic and outer membrane proteins: acid induced YcdO and YdiY; base induced OmpA, MalE, and YceI; and either acid or base induced OmpX relative to pH 7. Two pH-dependent periplasmic proteins were redox modulators: Tpx (acid-induced) and DsbA (base-induced). The locus alx, induced in extreme base, was identified as ygjT, whose product is a putative membrane-bound redox modulator. The cytoplasmic superoxide stress protein SodB was induced by acid, possibly in response to increased iron solubility. High pH induced amino acid metabolic enzymes (TnaA and CysK) as well as lac fusions to the genes encoding AstD and GabT. These enzymes participate in arginine and glutamate catabolic pathways that channel carbon into acids instead of producing alkaline amines. Overall, these data are consistent with a model in which E. coli modulates multiple transporters and pathways of amino acid consumption so as to minimize the shift of its external pH toward either acidic or alkaline extreme.
For bacteria to grow within the human body or in the external environment, they need to withstand extreme changes in external pH (20, 56). Resistance to acid contributes to the pathogenesis of enteric pathogens such as Escherichia coli O157:H7 (21, 35, 54, 72) and induces virulence factors such as ToxR in Vibrio cholerae (40). Resistance to base may enhance survival of bacteria as they pass through the pylorus and enter the upper intestine, where they encounter alkaline pancreatic secretion, a phenomenon which has received relatively little study compared to acid stress. External pH can be shifted substantially by bacterial metabolism, either through fermentative generation of acids (9) or through aerobic consumption of acids (36). The effective alkalinization of cultures grown to stationary phase in complex media such as Luria broth is often overlooked (34, 59).
Acid and base have multiple effects on gene expression in E. coli (for a review, see references 20, 45, and 57). Several enzymes of amino acid catabolism are induced at the pH extremes. At low external pH, amino acid decarboxylases generate amines, which are exported and thereby reverse acidification (22, 35, 37, 41, 64). At high pH, catabolism of amino acids could release ammonia, which deprotonates and volatilizes, while channeling the carbon skeleton into acids. There is limited evidence for increased deaminase activity at high pH (8, 22). The best-understood response to high pH is the Na+/H+ antiporter NhaA, which helps maintain internal pH and protects cells from excess sodium at high pH (29). Many questions remain regarding the regulation of internal pH, which is maintained near pH 7.6 during growth over a wide range of external pHs (pH 4.4 to 9.2) (60, 75).
The effects of pH are complex because they intersect with other environmental factors such as oxygenation, growth phase, and various metabolites. External acid amplifies the uptake of membrane-permeant fermentation acids such as acetate or formate (50). Osmolarity and pH coregulate the porins OmpC, OmpF, and MalE (16, 28, 47, 51). Anaerobiosis amplifies the acidic induction of amino acid decarboxylases (37, 59). The alkali-inducible tryptophanase operon is induced by tryptophan (10, 43, 63) or by excreted indole (70), but it is repressed by acetate (31). Oxidative stress is likely to be intensified by acid, which accelerates production of oxygen radicals and increases the concentration of ferrous iron. The alkyl hydroperoxide reductase AhpC is induced at low pH and by membrane-permeant organic acids (8, 32). Membrane-permeant acids also induce the Mar multiple drug resistance regulon, which is coregulated by the SoxRS superoxide stress system (49).
Several genetic systems are coregulated by pH and growth phase. Acetate and other acidic fermentation products induce stationary-phase stress proteins (3, 31) as well as the autoinducer synthesis protein LuxS (31, 52). The growth-phase sigma factor RpoS (33) regulates several components of resistance to both acid and base (27, 34, 61, 71). The overall complexity of the stress response in enteric bacteria is probably greatly underestimated, since intracellular growth produces global patterns substantially different from those predicted by stress experiments under defined conditions (1).
Current proteomic and lac reporter technologies enable us to survey a substantial subset of E. coli proteins over a range of growth conditions (31). In this report, we extended our previous proteomic study of pH (8) to observe proteins grown over a range of pHs from acid through base by using a gel system with increased resolution and sensitivity. Our new analysis revealed pH effects on expression of 22 identified proteins, only 3 of which were previously known to respond to pH. We also demonstrated high-pH induction of three additional genes by using promoter-lacZ fusions. Our study reveals patterns of pH-dependent expression of periplasmic and outer membrane proteins as well as of amino acid catabolism.
MATERIALS AND METHODS
Strains and growth conditions.
The E. coli K-12 strains used in this study are listed in Table 1. Growth media contained LBK broth (10 g of tryptone, 5 g of yeast extract, 7.45 g of KCl) with 100 mM buffer, adjusted for pH using KOH in order to avoid inhibition of growth by Na+ at high pH (29). The buffers used at each pH were as follows: pH 4.9, homopiperazine-N,N′-bis-2(ethanesulfonic acid) (HOMOPIPES); pH 6.0, 2-(N-morpholino)ethanesulfonic acid (MES); pH 6.5, piperazine-N,N′-bis-(2-ethanesulfonic acid) (PIPES); pH 7.0, PIPES or 3-(N-morpholino)propanesulfonic acid (MOPS); pH 8.0, 3-[N-tris(hydroxymethyl)methylamino]propanesulfonic acid (TAPS); pH 9.1, 3-[(1,1-dimethyl-2-hydroxyethyl)amino]-2-hydroxypropanesulfonic acid (AMPSO) (7, 8, 57, 59). Cultures in LBK were grown overnight, diluted 500-fold in fresh medium, and grown to an optical density at 600 nm (OD600) of 0.2 at 37°C in baffled flasks with rotary aeration.
TABLE 1.
Strains of E. coli K-12 used in this study
Optical density readings (OD600) were obtained in a Spectramax Plus microtiter plate spectrophotometer (Molecular Devices). Effects of growth on pH of the culture and growth rates as a function of pH are shown in Fig. 1. Samples from cultures grown to an OD600 greater than 0.4 were diluted 10-fold in fresh medium before measurement of optical density in order to avoid nonlinear readings.
FIG. 1.
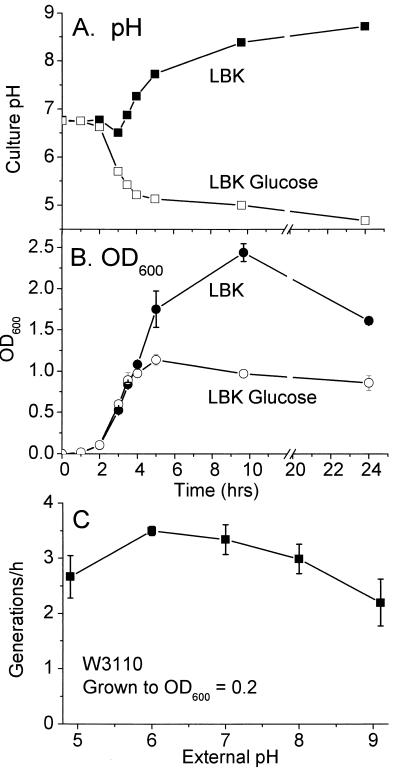
Growth of E. coli W3110 and effects of culture pH. (A) Culture pH as a function of growth in unbuffered LBK (▪) or LBK with 0.5% glucose (□). (B) Optical density as a function of growth in unbuffered LBK (•) or LBK with 0.5% glucose (○). (C) Generation time as a function of pH. Strain W3110 was grown overnight in LBK and then inoculated 1:500 in fresh LBK buffered at 100 mM as described in Materials and Methods. A 20-ml aliquot of medium was rotated at 200 rpm in a 250-ml baffled flask at 37°C. OD600 readings were obtained at 20-min intervals until the OD reached 0.2. Error bars represent standard error of the mean; n = 4.
2-D gel electrophoresis.
All two-dimensional electrophoresis gels (2-D gels) were performed using strain W3110 (42), largely as described elsewhere (31, 58); our most current methods are also maintained online at the website http://www2.kenyon.edu/depts/biology/slonc/labtools/2d_method.html. Protein samples were solubilized in sodium dodecyl sulfate and urea as described previously, and first-dimension IPGphor gel strips were run according to the manufacturer (AP Biotech). For each gel, 50 μg of protein was loaded onto an IPGstrip (pH 4 to 7). 2-D slab gels were performed on the ESA Investigator 2-D Electrophoresis System (Genomic Solutions) as described elsewhere (31). A total of 15 to 18 ml of each culture was chilled, pelleted, and washed with LBK. Cell pellets were then treated with urea-sodium dodecyl sulfate sample buffer and DNase-RNase and applied to an 18-cm polyacrylamide gel strip with an immobilized pH gradient of 4 to 7 for isoelectric focusing. Gel strips were applied to 2-D electrophoretic gels containing 11.5% acrylamide and then silver stained by a procedure compatible with matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis, with omission of glutaraldehyde.
To compare spot quantities between different growth conditions, gels were scanned and digitized and spot densities were quantified using Compugen Z3 version 2.0 software (http://www.2dgels.com/). The Z3 algorithm for pairwise comparisons matches each pair of spots pixel by pixel, discarding values of saturated pixel density as well as values from the edge of the background. A histogram of all spot ratios normalizes them to one, on the assumption that 90% of all proteins should have the same concentration under the two growth conditions; this assumption corrects for loading differences without requiring summation of all spots and subtraction of background, which are procedures that introduce error.
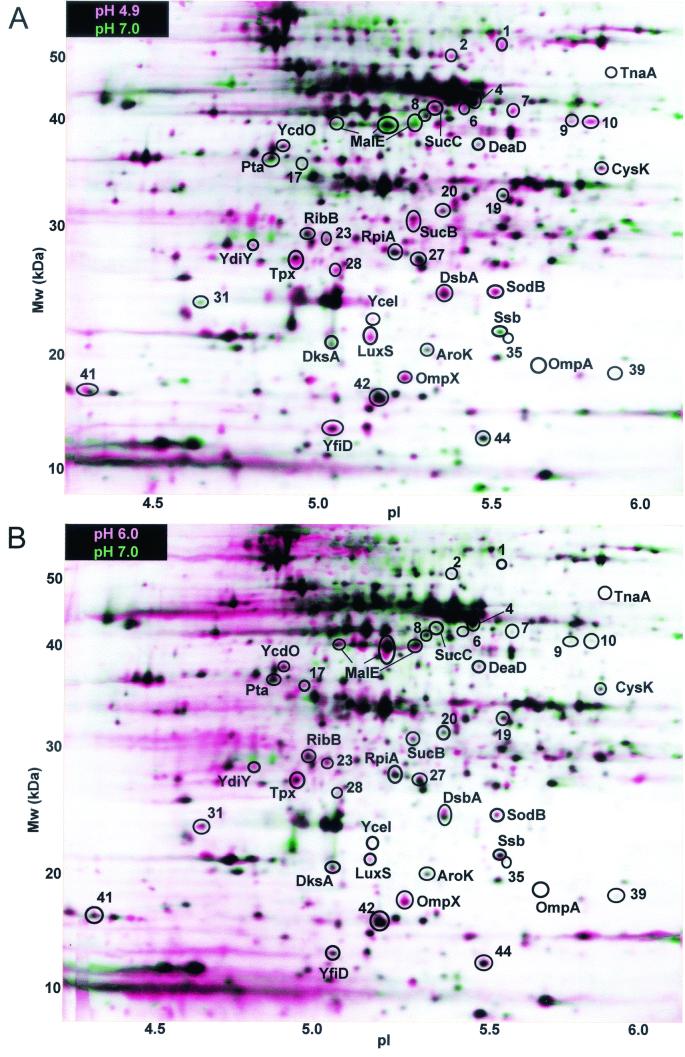
The reference condition was defined as growth at pH 7.0, while four different pH conditions were defined as comparative (pH 4.9, 6.0, 8.0, or 9.1). For each growth condition, spot densities were obtained from three gel images from independently grown cultures. Qualitative visual comparison was performed between composite images from each growth condition, using Z3 software (see Fig. 2). For each growth condition, a set of three replicate gel images was combined digitally into a composite image. Composite images were obtained for the reference condition (growth at pH 7.0) as well as for each comparative condition (growth at pH 4.9, 6.0, 8.0, or 9.1). For each comparative condition, the composite was overlaid digitally onto the reference composite (pH 7.0). In a layered view of experimental and reference images, a spot showing greater density in the comparative image than the reference appears pink, or as a pink border around a dark center, whereas a spot showing greater density in the reference gel appears green (see Fig. 2). Spots that appear pink or green on the layered view usually generate significantly positive or negative expression ratios in the quantitative analysis (described below). In some cases, however, a spot which appears pink or green in a layered view is actually present in only one of the original three gels of a composite; in such cases, the spot does not show a significant overall expression ratio.
FIG. 2.
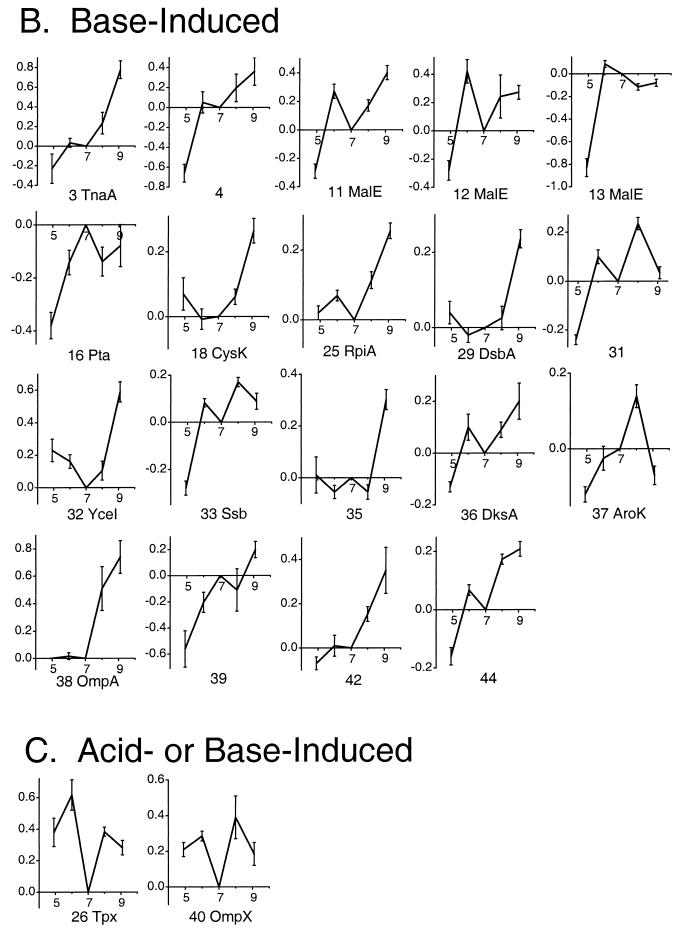
Proteins differentially expressed at each growth pH, relative to expression at pH 7.0. All cultures of E. coli W3110 were grown at 37°C to an OD600 of 0.2 in LBK with 100 mM buffer, as described in Materials and Methods. The horizontal axis represents the approximate pH range of the isoelectric focusing first dimension, and the vertical axis represents the molecular mass in kilodaltons. Each layered view superimposes two composite images representing growth in medium at the comparative pH and growth at the reference pH (pH 7.0). For each pH, the composite image is based on three 2-D gels from replicate cultures. In the layered view, spots that are partly or completely pink indicate proteins expressed to a higher level at the comparative pH than at pH 7.0, whereas spots that are green indicate proteins repressed (spot density greater at pH 7.0). In each layered view, the experimental pH is as follows: A, 4.9; B, 6.0; C, 8.0; D, 9.1.
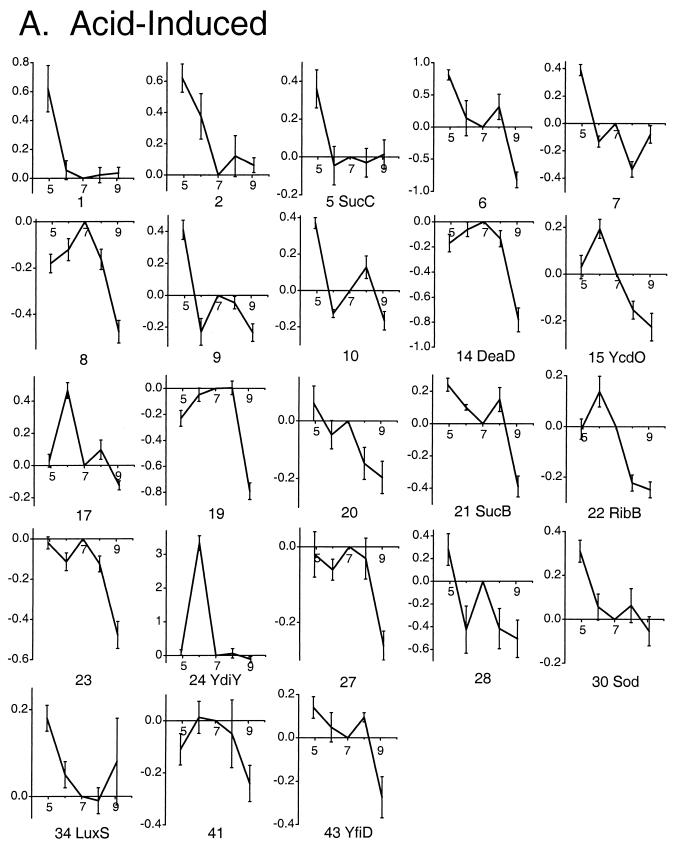
Quantitative analysis was performed using the original individual gel images. For each comparative growth pH, pairwise comparisons were performed between a set of three comparative gels (grown at the comparative pH) and three reference gels (grown at pH 7.0). The differential expression (DE) ratio of the spot densities between comparative and reference images was computed with the Z3 software. A protein spot was considered a candidate for significant induction if seven out of nine pairwise comparisons produced a DE ratio of greater than 1.5 (induced) or less than 0.67 (repressed) (31). Proteins observed under one of the comparative conditions that found no match on gels of the reference condition were scored as having a DE of 10, the expression ratio at which differentially expressed proteins typically start to appear unmatched. This assignment of unmatched proteins results in underestimation of actual expression ratios, but it permits scoring the proteins as induced. The log10 of all nine DE values was obtained, and the mean log10 DE (LDE) was reported as the measure of induction (positive values) or repression (negative values) (see Fig. 3).
FIG. 3.
DE profiles of proteins as a function of growth pH. Serial numbers and protein identification are based on spot position in the gel images (Fig. 2). The horizontal axes represent growth pH, and the vertical axes represent the LDE. For each pH, the LDE values were computed relative to pH 7, using Z3 software as described in Materials and Methods. The LDE values were obtained from pairwise comparison of three individual gel images from the comparative condition and the reference condition (pH 7.0). Error bars represent standard errors of the means. The proteins are grouped by expression categories as follows: A, proteins induced in acid or repressed in base; B, proteins induced in base or repressed in acid; C, proteins induced either in acid or in base relative to pH 7.0.
Identification of proteins.
Proteins were identified by graphical comparison of spot positions from previous studies or by MALDI-TOF analysis of protein spots excised from the silver-stained gel (see Table 2). MALDI-TOF analyses were performed by the Proteomic Mass Spectrometry Laboratory at the University of Massachusetts (http://www.umassmed.edu/proteomic/). A Kratos Axima CFR MALD-TOF mass spectrometer was used to obtain mass spectrometry (MS) data from tryptic peptide mixtures as well as post-source decay analysis of individual peptides. For database searching of MALDI-TOF masses, the Protein Prospector site was used (http://prospector.ucsf.edu).
TABLE 2.
Identified proteins induced by acid or basea
| Protein no. | Protein | Protein name and/or description |
|---|---|---|
| Acid-induced proteins | ||
| 5 | SucC | Succinyl-CoA synthetase, β-chain |
| 14 | DeaD | ATP-dependent RNA helicase |
| 15 | YcdO | Envelope protein |
| 21 | SucB | Dihydrolipoamide succinyltransferase of 2-oxogluterate dehydrogenase complex |
| 22 | RibB | LuxH homolog; 3,4-dihydroxy-2-butanone 4-phosphate synthase (riboflavin biosynthesis) |
| 24 | YdiY | Salt-induced outer membrane protein |
| 30 | SodB | Iron(III) superoxide dismutase, chain A |
| 34 | LuxSc | Autoinducer-2 production protein |
| 43 | YfiDc,d | Low-oxygen pyruvate-formate lyase |
| Base-induced proteins | ||
| 3 | TnaAd | Tryptophanase |
| 11, 12, 13 | MalEd | High-affinity maltodextrin binding protein |
| 16 | Ptac | Phosphotransferase |
| 18 | CysK | Cysteine synthase A; o-acetylserine sulfhydrylase A |
| 25 | RpiA | Ribose 5-phosphate isomerase A |
| 29 | DsbA | Periplasmic thiol:disulfide interchange protein |
| 32 | YceI | Periplasmic protein |
| 33 | Ssb | Single-strand binding protein (helix destabilizing protein) |
| 36 | DksA | DnaK suppressor protein |
| 37 | AroKc | Shikimate kinase I; second activity confers mecillinam resistance |
| 38 | OmpA | Outer membrane protein A |
| Acid- or base-induced proteins, compared to pH 7.0 | ||
| 26 | Tpx | Periplasmic thiol peroxidase |
| 40 | OmpX | Outer membrane protein X |
Overall induction pattern for each identified protein, based on Fig. 3. Fig. 3 shows the full pH profiles of expression for all numbered proteins.
Proteins were identified by MALDI-TOF MS unless noted otherwise. The position of proteins in gels is shown in Fig. 2.
Identified by position, based on reference 31. The effect of pH is new to this paper.
The effect of pH confirms a previous report (8).
Localization of lac fusion insertion.
In order to map the insertion joint of a Mu dI1734 lac fusion strain, a primer sequence homologous to the left end of Mu was designed: AGATGCATTACCTGAAGAAAAACG. Genomic DNA from strain JLS9707 was prepared by using an Epicentre Masterpure kit (Ecogen). PCR sequence analysis was performed on an Applied Biosystems instrument at the Ohio State Plant—Microbe Genomics Facility, under the following cycling conditions: 95°C for 300 s, followed by 45 cycles of 95°C for 30 s, 55°C for 20 s, and 60°C for 240 s.
β-Galactosidase assays.
Overnight cultures were inoculated 1:500 into 2 ml of LBK buffered as above and rotated in tubes at 37°C until growth reached an OD600 of 0.2. For cultures grown to an OD600 of 0.5, the initial dilution was 1:200 or 1:500; no difference in assay result was observed. A microtiter assay of β-galactosidase activity was developed based on references 55 and 38, using a microtiter plate spectrophotometer. For each culture condition, four samples were read at OD600 and two duplicates from each condition were assayed for β-galactosidase activity. Each assay sample (200 μl) was diluted into assay buffer and incubated with chloroform in a polypropylene microtiter plate with shaking for 5 min. Samples were diluted into fresh assay buffer containing o-nitrophenyl-β-d-galactopyranoside (without chloroform) in a transparent polycarbonate microtiter plate. The β-galactosidase assay slope (OD420) was determined over 30 min. Standard activity units were calculated as described elsewhere (55).
RESULTS
Culture growth and protein expression.
In order to obtain reliable expression patterns as a function of pH, it was important to assess the effect of cell metabolism on pH of the culture fluid and to define growth conditions that minimize pH change. The pH of LBK growth medium lacking buffer was measured over a range of culture growth levels (Fig. 1B). Cultures grown in unbuffered LBK acidify slightly in mid-log phase as acetate builds up (18), but the pH steadily increases in stationary phase, rising to well above pH 9. Inclusion of glucose, by contrast, results in steady acidification without recovery (Fig. 1B). For our experiments, we avoided glucose supplementation and included buffers with an appropriate pKa which maintained the culture pH within 0.1 unit of the starting pH (grown to an OD600 of 0.2) or 0.2 unit (grown to an OD600 of 0.5). In LBK media buffered at all pH values, the rate of growth up to an OD600 of 0.2 was reasonably high (Fig. 1C).
For 2-D gels, cells were cultured for approximately five doublings until the OD600 was 0.2. The silver-stained 2-D gel protein patterns were compared digitally by pairing three independently grown cultures from pH 7 with three cultures from each of the four other pH conditions (Fig. 2 ); similarly, patterns from cultures at pH 4.9 were compared with those from pH 9.1, and those from pH 6.0 cultures were compared with those from pH 8.0 (data not shown). For each protein showing significant DE between any of the paired conditions, the mean log ratios were determined for all four pH conditions normalized to pH 7 (Fig. 3). DE was considered significant if the expression ratio was at least 1.5 (mean log ratio, ±0.18). In a control experiment, three independent cultures at pH 7.0 compared with three different cultures grown at the same pH showed virtually no significant protein differences.
The Z3 software detected approximately 560 protein spots in each digitized gel image, of which 44 showed significant DE between at least two different pH conditions as defined above. The layered views depicting expression at each pH compared to pH 7.0 are shown in Fig. 2. The individual gels used to create the composites in Fig. 2 were subjected to pairwise comparison and quantitative analysis (Fig. 3). In Fig. 3, proteins were classified based on whether they showed (i) greater expression under one or both acid conditions (pH 4.9 or 6.0) than at high pH (acid induced); (ii) greater expression at pH 8.0 or 9.1 than at low pH (base induced); or (iii) greater expression at both extremes than at pH 7.0.
The pH-dependent proteins identified by MALDI-TOF MS or by gel position are summarized in Table 2. Only three of the identified proteins (TnaA, MalE, and YfiD) have been shown previously to be regulated by pH (8, 16, 28). The pH dependence of the 19 other identified proteins, as well as the remaining 20 proteins not yet identified, is reported here for the first time.
Proteins with pH-dependent expression profiles.
Several interesting patterns of pH dependence emerged in the induction of porins and exported proteins, metabolic enzymes, and oxidative stress components. Periplasmic or envelope proteins showed diverse responses to pH. Acid induced the periplasmic or exported protein YcdO, as well as the salt-induced outer membrane protein YdiY. High pH induced the periplasmic protein YceI as well as the porins MalE and OmpA (23) (Fig. 3B). MalE appeared in three different isoforms, a common effect of posttranslational modification (24). Each isoform showed generally constant expression at pH 6.0 and above but dipped at pH 4.9. In addition to the acid- and base-specific porins, the virulence-associated porin OmpX (69) showed higher expression in either acid or base than at pH 7.0 (Fig. 3C).
Since membrane-permeant fermentation acids such as acetate accumulate in proportion to the transmembrane pH difference (ΔpH), we expected to find some overlap between induction at low pH (increased ΔpH) and induction by acetate (31). Two acetate-inducible proteins, LuxS and YfiD, were induced at pH 4.9. The acetate-inducible Tpx (protein number 53 in reference 31) was induced in either acid or base compared to pH 7.0 (Fig. 3C). At high pH, on the other hand, several proteins were induced that acetate represses (Pta, TnaA, DksA, AroK, and MalE). This observation is consistent with the fact that the inverted pH gradient at high external pH effectively excludes acid from the cell.
The stress proteins induced by acetate include several members of the RpoS regulon (3, 26, 31). At low pH, however, the proteins we found did not include the RpoS regulon; for example, Dps was virtually absent from our gels, though it is strongly induced by acetate (3, 31). The absence of induction of stationary-phase proteins is a sign that our cultures across the pH range were indeed maintained in early log phase. This is consistent with the high growth rate maintained by E. coli cultures grown in complex media in moderate acid or base (Fig. 1C).
The increased ΔpH also contributes to the cell's proton potential (75), which may have been responsible for the elevation of the TCA cycle components SucB and SucC (2) at low pH. Other pH-modulated proteins were involved in redox reactions or oxidative stress response (Tpx, DsbA, and SodB) (15, 17, 26). Another oxidative stress protein known to be acid inducible, AhpC (8), did not show a pH effect in this study. However, the highly abundant AhpC was largely overexposed in the silver stain, and differential expression may have gone undetected.
High-pH-dependent expression of enzymes of amino acid metabolism.
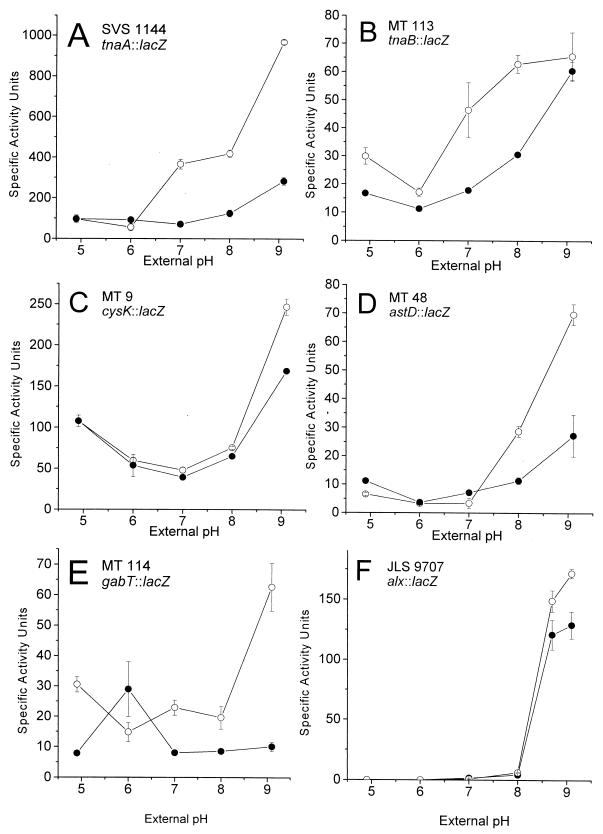
Several enzymes of amino acid catabolism show pH regulation, particularly the decarboxylases at low pH, as well as tryptophan deaminase at high pH (8). We identified a second base-inducible enzyme, CysK (o-acetylserine sulfhydrylase A). The effects of pH on expression of TnaA and CysK, as well as the tryptophan transporter TnaB, were confirmed by lac fusion assays (Fig. 4). These strains contained in-frame gene fusions expected to represent transcriptional as well as translational regulation (4, 63). Both tnaA::lac and cysK::lac showed expression levels highly consistent with their protein profiles: tnaA::lac was strongly induced at high pH, whereas cysK::lac was induced in base but also showed a modest increase at pH 4.9. The expression of cysK as well as tnaB (encoding the tryptophan transporter), downstream of tnaA, is known to be inducible in L broth at high cell density (4) mediated by the metabolite indole (70). In these previous reports, however, the pH dependence was not discussed, although the pH of L broth is known to increase with increasing cell density (59; see also Fig. 1).
FIG. 4.
pH dependence of expression of lacZ gene fusions. Cultures were grown in buffered LBK, and β-galactosidase was assayed as described in Materials and Methods. A, tnaA::lac; B, tnaB::lac; C, cysK::lac; D, astD::lac; E, gabT::lac; F, alx::lac. •, OD600 = 0.2; ○, OD600 = 0.5.
Two additional catabolic genes reported by Rather's group to be induced at high cell density are astD (succinylglutamic semialdehyde dehydrogenase) and gabT (γ-amino butyric acid [GABA] transaminase) (4), which are members of the connected AST (53) and Gab (5, 39) pathways of arginine and glutamate catabolism. Both genes showed strong induction at high pH (Fig. 4). The degree of induction at high pH was amplified at higher cell density. A related glutamate catabolic protein, GadA, shows elevation at high pH (8), but primarily under anaerobiosis. In our gels the GadA protein spot was not observed because that region of the gel (pI 5.2; molecular weight, 52,685) contained too many overlapping proteins with silver stain.
The open reading frame ygjT was identified as the alkali-inducible gene alx.
An extreme alkali-inducible lac fusion to an unknown locus (alx) was identified previously and mapped to 68 min (7). The expression of alx::lac shows an exceptionally steep response to high pH, with a modest increase at higher cell densities (Fig. 4F). We determined the sequence of the alx fusion joint by PCR sequencing as described in Materials and Methods. The sequence obtained showed 100% identity to ygjT, with the fusion joint at base 28 of the coding sequence. The locus alx/ygjT is a putative transmembrane protein whose transcription in a microarray study showed 50% repression by paraquat, an agent of superoxide stress (46). Thus, the alx gene product appears to be another pH-dependent protein involved in redox stress.
DISCUSSION
The pH-dependent expression profiles of various periplasmic and metabolic proteins add to a growing picture of regulation of metabolism so as to minimize excess acidification or alkalization while mobilizing available carbon sources. Cells respond to acid stress by consuming acids and excreting volatile CO2 with alkaline amines, whereas under alkali stress cells produce acids while excreting volatile NH3. These responses are accomplished by manipulating transporters and degradative enzymes. We now report 22 additional components of the growing picture of pH stress response.
Periplasmic and outer membrane proteins.
While the pH dependence of OmpF and OmpC is well known (28, 47, 51), our study revealed pH regulation of several additional periplasmic and membrane proteins, including three whose functions have not yet been characterized (YceI, YcdO, and YdiY). Diverse response patterns were observed, including induction by acid (YcdO and YcdY), base (YceI, OmpA, and MalE), or either pH extreme (OmpX). While their functions are poorly understood, these transporters might make available different kinds of substrates under different conditions; for example, base-induced MalE would provide an acid-generating carbohydrate, whereas acid-induced transporters might transport peptides to generate amines.
The periplasm is also the location of two of the three oxidative stress proteins responsive to pH: Tpx (15) and DsbA (74). DsbA is required to maintain disulfide bonds in periplasmic enzymes at both extremes of pH (6); it was more highly expressed in base, but its overall spot density was high across the pH range (Fig. 2). Another putative redox modulator that showed pH-dependent expression as a lac fusion is the product of alx/ygjT (46). The Alx protein, expressed only near the high pH limit for growth, might serve a function similar to that of DsbA, for example, maintaining disulfides in periplasmic proteins at the highest pH. The one cytoplasmic oxidative stress factor induced by acid was SodB (44). SodB may be induced by acid in order to cope with higher levels of soluble iron, a major oxidative stress factor, as internal pH decreases. Regulation of SodB involves Fur, which is involved in both iron response and acid resistance (66).
Metabolic responses to external acid.
Moderate external acidity (between pH 5 and 7) has favorable as well as unfavorable effects on growth of E. coli. A favorable energetic effect is the increased magnitude of the proton potential, which includes ΔpH (60, 75). In our experiments, growth at low pH induced components of the TCA cycle (SucB and SucC), which would be expected to utilize the high proton potential.
An unfavorable effect of low external pH is the reuptake of fermentation acids to a high concentration (18). While only a small amount of acetate would be produced during growth to an OD600 of 0.2, at external pH as low as 4.9 even a small concentration of a membrane-permeant acid becomes magnified by reentry in the protonated form; in theory, an approximately 300-fold increase would occur if internal pH were maintained at pH 7.4. Thus, it is not surprising that at pH 4.9 to 6.0 several acetate-induced proteins were elevated, notably LuxS and YfiD, and several acetate-repressed proteins were lowered. None of the acetate-inducible RpoS regulon members were affected, however; this is consistent with our maintenance of cells in early to mid-log phase (Fig. 1C).
The induction of LuxS by acid at low cell density is interesting because, previously, LuxS was believed to require growth with glucose to mid-log phase (52, 65). Our results, together with the induction of LuxS by exogenous acetate (31), suggest that in glucose-L broth medium the luxS gene responds to the production of acetate, which peaks in mid-log phase and then declines as the acetate is metabolized. A different potential autoinducer component, the LuxH homolog RibB (12, 48), was induced at low pH but not by acetate. The induction of RibB suggests the possibility of a second pH-dependent autoinducer system.
Metabolic responses to external base.
For E. coli, growth at an external pH of >7.8 generates an inverted pH gradient which drains proton potential (60, 75). Under aerobic growth conditions, alkalinization is inevitable, so long as cells supplied with oxygen keep consuming organic substrates and expelling CO2. The stationary phase in aerated L broth is, in effect, alkaline stress. Late stationary phase selects for GASP (growth advantage in stationary phase) mutants (76), in which cells preferentially metabolize serine, threonine, and alanine—amino acids that are attacked by deaminases, which have been predicted to be induced at high pH. Nevertheless, the pH-dependent expression of enzymes in these pathways has yet to be characterized.
In complex peptide-rich medium, as cell density increases and pH rises we propose that cells direct overall amino acid catabolism into pathways that remove ammonia and generate acids, including deaminases as well as other enzymes that channel metabolites toward fermentation acids instead of excreting amines. Tryptophanase is an especially versatile enzyme that deaminates serine and cysteine as well as tryptophan (36); above pH 9, by mid- to late log phase tryptophanase becomes one of the most highly expressed proteins in the cell (8). Besides TnaA, we have now shown that high pH induces three additional cell density-dependent enzymes: AstD, a component of the major NH3-generating arginine breakdown pathway (53); GabT, which directs GABA into succinate (39); and CysK. CysK is generally considered an anabolic enzyme that produces cysteine but it may also catalyze the reverse pathway (19), removing H2S to make serine, which is deaminated to pyruvate.
Aside from CysK, virtually no biosynthetic enzymes have been observed in the pH stress response. A possible exception is the shikimate kinase I (AroK), which functions in the synthesis of aromatic amino acids. AroK is repressed by acetate (31) and was induced in this study at pH 8 (Fig. 3B). This protein, however, has a second activity which confers mecinillam resistance, possibly associated with regulation of cell division (68). The second activity might be the actual target of pH-dependent expression.
The pH dependence of amino acid catabolism.
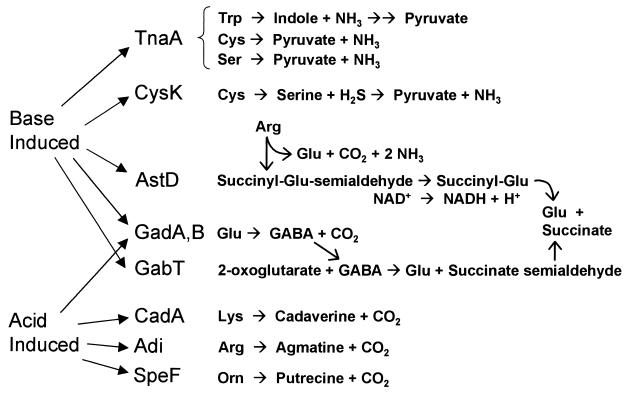
The known pH-regulated catabolic enzymes, including those reported here, are summarized in Fig. 5. This summary emphasizes the connecting catabolic pathways that are used to obtain carbon while reversing acidification or alkalinization. (For further detail of pathways, see reference 36 or the WIT database at http://wit.mcs.anl.gov/WIT2/CGI/.) High pH induces enzymes of pathways generating ammonia, including TnaA and CysK, which consume tryptophan, serine, and cysteine, and AstD, a member of the AST operon for arginine degradation. The AST operon produces glutamate, which could be converted by GadA and GadB (isoforms of glutamate decarboxylase [13]) to GABA. GABA is then directed by GabT into succinate.
FIG. 5.
Summary of proposed pH-dependent pathways of amino acid catabolism. TnaA and CysK generate NH3 and acids. Base-induced AstD, GadA, and GabT participate in arginine and glutamate degradation pathways that avoid generation of amines and direct acids into succinate. Acid-induced CadA, Adi, and SpeF generate CO2 and amines; GadA and GadB decarboxylate glutamate to GABA, which is exported during growth in acid. For further details on pathways, see the WIT database (http://wit.mcs.anl.gov/WIT2/CGI/).
GadA and GadB are known to be induced at pH 5, compared to the response at pH 7 (14); under acid stress, the product GABA is exported by GadC (13, 67). However, this is not necessarily inconsistent with the induction of GadA under anaerobiosis at pH 9 (8). At pH 9, the product GABA would not be exported from the cell but would be converted by GabT into succinate (Fig. 5). This observation could also explain why gad expression is induced in enteropathogenic E. coli as cells leave the acidic stomach to enter the upper intestine, where they encounter alkaline pancreatic secretions (54).
Acid induces the well-studied decarboxylases CadA, Adi, and SpeF, as well as GadA and GadB (reviewed in reference 57). The pH-moderating function of decarboxylases may be enhanced by transporters of amino acids and peptides, such as those induced by acetate: ArtI, FliY, and OppA. OppA transports peptides, either exogenously or through cell wall recycling (30). ArtI transports arginine (73), which might contribute substrate for Adi. FliY transports the cysteine dimer cystine (11); this is interesting, given the observed small degree of acid induction of CysK (Fig. 3A and 4C) as well as possible induction of CysK by acetate (J. Slonczewski, unpublished data). Thus, CysK might participate in some as-yet-undefined acid-consuming pathway.
The growing picture of pH-regulated catabolism has obvious significance for questions of survival of enteric pathogens internally, as well as in amino acid-rich foods (72). Our model points to avenues of investigation for further metabolic components of pH stress response.
Acknowledgments
We thank C. Yanofsky and P. Rather for insightful discussions and for the generous gift of strains. We thank D. Blankenhorn for critical reading of the manuscript.
This work was supported by grant MCB-9982437 from the National Science Foundation.
REFERENCES
- 1.Abshire, K. Z., and F. C. Neidhardt. 1993. Analysis of proteins synthesized by Salmonella typhimurium during growth within a host macrophage. J. Bacteriol. 175:3734-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amarasingham, C. R., and B. D. Davis. 1965. Regulation of α-ketoglutarate dehydrogenase formation in Escherichia coli. J. Biol. Chem. 240:3664-3668. [PubMed] [Google Scholar]
- 3.Arnold, C. N., J. McElhanon, A. Lee, R. Leonhart, and D. A. Siegele. 2001. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J. Bacteriol. 183:2178-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baca-DeLancey, R. R., M. M. South, X. Ding, and P. N. Rather. 1999. Escherichia coli genes regulated by cell-to-cell signaling. Proc. Natl. Acad. Sci. USA 96:4610-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartsch, K., A. von John-Marteville, and A. Schulz. 1990. Molecular analysis of two genes of the Escherichia coli gab cluster: nucleotide sequence of the glutamate:succinic semialdehyde transaminase gene (gabT) and characterization of the succinic semialdehyde dehydrogenase gene (gabD). J. Bacteriol. 172:7035-7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belin, P., E. Quemeneur, and P. L. Boquet. 1994. A pleiotropic acid phosphatase-deficient mutant of Escherichia coli shows premature termination in the dsbA gene. Use of dsbA::phoA fusions to localize a structurally important domain in DsbA. Mol. Gen. Genet. 242:23-32. [DOI] [PubMed] [Google Scholar]
- 7.Bingham, R. J., K. S. Hall, and J. L. Slonczewski. 1990. Alkaline induction of a novel gene locus, alx, in Escherichia coli. J. Bacteriol. 172:2184-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blankenhorn, D., J. Phillips, and J. L. Slonczewski. 1999. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J. Bacteriol. 181:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Böck, A., and G. Sawers. 1996. Fermentation, p. 262-282. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 10.Botsford, J. L. 1975. Metabolism of cyclic adenosine 3′,5′-monophosphate and induction of tryptophanase in Escherichia coli. J. Bacteriol. 124:380-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler, J. D., S. W. Levin, A. Facchiano, L. Miele, and A. B. Mukherjee. 1993. Amino acid composition and N-terminal sequence of purified cystine binding protein of Escherichia coli. Life Sci. 52:1209-1215. [DOI] [PubMed] [Google Scholar]
- 12.Callahan, S. M., and P. V. Dunlap. 2000. LuxR- and acyl-homoserine-lactone-controlled non-lux genes define a quorum-sensing regulon in Vibrio fischeri. J. Bacteriol. 182:2811-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castanie-Cornet, M.-P., and J. W. Foster. 2001. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147:709-715. [DOI] [PubMed] [Google Scholar]
- 14.Castanie-Cornet, M.-P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cha, M. K., H. K. Kim, and I. H. Kim. 1995. Thioredoxin-linked "thiol peroxidase" from periplasmic space of Escherichia coli. J. Biol. Chem. 270:28635-28641. [DOI] [PubMed] [Google Scholar]
- 16.Chagneau, C., M. Heyde, S. Alonso, R. Portalier, and P. Laloi. 2001. External-pH-dependent expression of the maltose regulon and ompF gene in Escherichia coli is affected by the level of glycerol kinase, encoded by glpK. J. Bacteriol. 183:5675-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubrac, S., and D. Touati. 2000. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J. Bacteriol. 182:3802-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.el-Mansi, E. M., and W. H. Holms. 1989. Control of carbon flux to acetate excretion during growth of Escherichia coli in batch and continuous cultures. J. Gen. Microbiol. 135:2875-2883. [DOI] [PubMed] [Google Scholar]
- 19.Flint, D. H., J. F. Tuminello, and T. J. Miller. 1996. Studies on the synthesis of the Fe-S cluster of dihydroxy-acid dehydratase in Escherichia coli crude extract. Isolation of o-acetylserine sulfhydrylases A and B and beta-cystathionase based on their ability to mobilize sulfur from cysteine and to participate in Fe-S cluster synthesis. J. Biol. Chem. 271:16053-16067. [DOI] [PubMed] [Google Scholar]
- 20.Foster, J. W. 2000. Microbial responses to acid stress, p. 99-115. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 21.Foster, J. W., and H. K. Hall. 1990. Adaptive acidification tolerance response of Salmonella typhimurium. J. Bacteriol. 172:771-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gale, E. F., and H. M. R. Epps. 1942. The effect of the pH of the medium during growth on the enzymic activities of bacteria (Escherichia coli and Micrococcus lysodeikticus) and the biological significance of the changes produced. Biochem. J. 36:600-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgellis, D., S. Arvidson, and A. von Gabain. 1992. Decay of ompA mRNA and processing of 9S RNA are immediately affected by shifts in growth rate, but in opposite manners. J. Bacteriol. 174:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gooley, A. A., and N. H. Packer. 1997. The importance of protein co- and post-translational modifications in proteome projects, p. 63-91. In M. R. Wilkins, K. L. Williams, R. D. Appel, and D. F. Hochstrasser (ed.), Proteome research: new frontiers in functional genomics. Springer, Berlin, Germany.
- 25.Green, J., M. L. Baldwin, and J. Richardson. 1998. Downregulation of Escherichia coli yfiD expression by FNR occupying a site at −93.5 involves the AR1-containing face of FNR. Mol. Microbiol. 29:1113-1123. [DOI] [PubMed] [Google Scholar]
- 26.Hengge-Aronis, R. 1996. The general stress response in Escherichia coli, p. 161-178. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 27.Hersh, B. M., F. T. Farooq, D. N. Barstad, D. Blankenhorn, and J. L. Slonczewski. 1996. A glutamate-dependent acid resistance gene in Escherichia coli. J. Bacteriol. 178:3978-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heyde, M., P. Laloi, and R. Portalier. 2000. Involvement of carbon source and acetyl phosphate in the external-pH-dependant expression of porin genes in Escherichia coli. J. Bacteriol. 182:198-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karpel, R., T. Alon, G. Glaser, S. Schuldiner, and E. Padan. 1991. Expression of a sodium proton antiporter (NhaA) in Escherichia coli is induced by Na+ and Li+ ions. J. Biol. Chem. 266:21753-21759. [PubMed] [Google Scholar]
- 30.Kashiwagi, K., Y. Yamaguchi, Y. Sakai, H. Kobayashi, and K. Igarashi. 1990. Identification of the polyamine-induced protein as a periplasmic oligopeptide binding protein. J. Biol. Chem. 265:8387-8391. [PubMed] [Google Scholar]
- 31.Kirkpatrick, C., L. M. Maurer, N. E. Oyelakin, Y. Yontcheva, R. Maurer, and J. L. Slonczewski. 2001. Acetate and formate stress: opposite responses in the proteome of Escherichia coli. J. Bacteriol. 183:6466-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambert, L. A., K. Abshire, D. Blankenhorn, and J. L. Slonczewski. 1997. Proteins induced in Escherichia coli by benzoic acid. J. Bacteriol. 179:7595-7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5:49-59. [DOI] [PubMed] [Google Scholar]
- 34.Lazar, S. W., M. Almirón, A. Tormo, and R. Kolter. 1998. Role of the Escherichia coli SurA protein in stationary-phase survival. J. Bacteriol. 180:5704-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin, J., M. P. Smith, K. C. Chapin, H. S. Baik, G. N. Bennett, and J. W. Foster. 1996. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 62:3094-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McFall, E., and E. B. Newman. 1996. Amino acids as carbon sources, p. 358-379. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 37.Meng, S.-Y., and G. N. Bennett. 1992. Nucleotide sequence of the Escherichia coli cad operon: a system for neutralization of low extracellular pH. J. Bacteriol. 174:2659-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menzel, R. 1989. A microtiter plate-based system for the semiautomated growth and assay of bacterial cells for beta-galactosidase activity. Anal. Biochem. 181:40-50. [DOI] [PubMed] [Google Scholar]
- 39.Metzer, E., R. Levitz, and Y. S. Halpern. 1979. Isolation and properties of Escherichia coli K-12 mutants impaired in the utilization of gamma-aminobutyrate. J. Bacteriol. 137:1111-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, V. L., R. K. Taylor, and J. J. Mekalanos. 1987. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell 48:271-279. [DOI] [PubMed] [Google Scholar]
- 41.Neely, M. N., C. L. Dell, and E. R. Olson. 1994. Roles of LysP and CadC in mediating the lysine requirement for acid induction of the Escherichia coli cad operon. J. Bacteriol. 176:3278-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neidhardt, F. C., and R. A. VanBogelen. 2000. Proteomic analysis of bacterial stress responses, p. 445-452. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 43.Newton, W. A., and E. E. Snell. 1964. Catalytic properties of tryptophanase, a multifunctional pyridoxal phosphate enzyme. Proc. Natl. Acad. Sci. USA 51:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nunoshiba, T., F. Obata, A. C. Boss, S. Oikawa, T. Mori, S. Kawanishi, and K. Yamamoto. 1999. Role of iron and superoxide for generation of hydroxyl radical, oxidative DNA lesions, and mutagenesis in Escherichia coli. J. Biol. Chem. 274:34832-34837. [DOI] [PubMed] [Google Scholar]
- 45.Olson, E. R. 1993. Influence of pH on bacterial gene expression. Mol. Microbiol. 8:5-14. [DOI] [PubMed] [Google Scholar]
- 46.Pomposiello, P. J., M. H. J. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pratt, L. A., W. Hsing, K. E. Gibson, and T. J. Silhavy. 1996. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 20:911-917. [DOI] [PubMed] [Google Scholar]
- 48.Richter, G., R. Volk, C. Krieger, H. W. Lahm, U. Rothlisberger, and A. Bacher. 1992. Biosynthesis of riboflavin: cloning, sequencing, and expression of the gene coding for 3,4-dihydroxy-2-butanone 4-phosphate synthase of Escherichia coli. J. Bacteriol. 174:4050-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosner, J. L., and J. L. Slonczewski. 1994. Dual regulation of inaA by the multiple antibiotic resistance (Mar) and superoxide (SoxRS) stress response systems of Escherichia coli. J. Bacteriol. 176:6262-6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Russell, J. B., and F. Diez-Gonzalez. 1998. The effects of fermentation acids on bacterial growth. Adv. Microb. Physiol. 39:205-234. [DOI] [PubMed] [Google Scholar]
- 51.Sato, M., K. Machida, E. Arikado, H. Saito, T. Kakegawa, and H. Kobayashi. 2000. Expression of outer membrane proteins in Escherichia coli growing at acid pH. Appl. Environ. Microbiol. 66:943-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 53.Schneider, B. L., A. K. Kiupakis, and L. J. Reitzer. 1998. Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. J. Bacteriol. 180:4278-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shin, S., M. P. Castanie-Cornet, J. W. Foster, J. A. Crawford, C. Brinkley, and J. B. Kaper. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microbiol. 41:1133-1150. [DOI] [PubMed] [Google Scholar]
- 55.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 56.Slonczewski, J. L. 2000. pH stress, p. 625-632. In J. Lederberg (ed.), Encyclopedia of microbiology, 2nd ed., vol. 3. Academic Press, London, England.
- 57.Slonczewski, J. L., and J. W. Foster. 1996. pH-regulated genes and survival at extreme pH, p. 1539-1552. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 58.Slonczewski, J. L., and C. Kirkpatrick. Proteomic analysis of Escherichia coli and Helicobacter pylori, using two-dimensional gel electrophoresis. Methods Enzymol., in press. [DOI] [PubMed]
- 59.Slonczewski, J. L., T. N. Gonzalez, F. M. Bartholomew, and N. J. Holt. 1987. Mu d-directed lacZ fusions regulated by low pH in Escherichia coli. J. Bacteriol. 169:3001-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Slonczewski, J. L., B. P. Rosen, J. R. Alger, and R. M. Macnab. 1981. pH homeostasis in Escherichia coli: measurement by 31P nuclear magnetic resonance of methylphosphonate and phosphate. Proc. Natl. Acad. Sci. USA 78:6271-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Small, P., D. Blankenhorn, D. Welty, E. Zinser, and J. L. Slonczewski. 1994. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 176:1729-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith, M. W., and F. C. Neidhardt. 1983. Proteins induced by anaerobiosis in Escherichia coli. J. Bacteriol. 154:336-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stewart, V., R. Landick, and C. Yanofsky. 1986. Rho-dependent transcription termination in the tryptophanase operon leader region of Escherichia coli K-12. J. Bacteriol. 166:217-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stim, K. P., and G. N. Bennett. 1993. Nucleotide sequence of the adi gene, which encodes the biodegradative acid-induced arginine decarboxylase of Escherichia coli. J. Bacteriol. 175:1221-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Touati, D., M. Jacques, B. Tardat, L. Bouchard, and S. Despied. 1995. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J. Bacteriol. 177:2305-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tramonti, A., P. Visca, M. De Canio, M. Falconi, and D. De Biase. 2002. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 184:2603-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vinella, D., B. Gagny, D. Joseleau-Petit, R. D'Ari, and M. Cashel. 1996. Mecillinam resistance in Escherichia coli is conferred by loss of a second activity of the AroK protein. J. Bacteriol. 178:3818-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vogt, J., and G. E. Schulz. 1999. The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. Structure Fold Des. 7:1301-1309. [DOI] [PubMed] [Google Scholar]
- 70.Wang, D., X. Ding, and P. N. Rather. 2001. Indole can act as an extracellular signal in Escherichia coli. J. Bacteriol. 183:4210-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Waterman, S. R., and P. L. Small. 1996. Identification of sigma S-dependent genes associated with the stationary-phase acid-resistance phenotype of Shigella flexneri. Mol. Microbiol. 21:925-940. [DOI] [PubMed] [Google Scholar]
- 72.Waterman, S. R., and P. L. Small. 1998. Acid-sensitive enteric pathogens are protected from killing under extremely acidic conditions of pH 2.5 when they are inoculated onto certain solid food sources. Appl. Environ. Microbiol. 64:3882-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wissenbach, U., S. Six, J. Bongaerts, D. Ternes, S. Steinwachs, and G. Unden. 1995. A third periplasmic transport system for l-arginine in Escherichia coli: molecular characterization of the artPIQMJ genes, arginine binding and transport. Mol. Microbiol. 17:675-686. [DOI] [PubMed] [Google Scholar]
- 74.Wunderlich, M., A. Otto, R. Seckler, and R. Glockshuber. 1993. Bacterial protein disulfide isomerase: efficient catalysis of oxidative protein folding at acidic pH. Biochemistry 32:12251-12256. [DOI] [PubMed] [Google Scholar]
- 75.Zilberstein, D., V. Agmon, S. Schuldiner, and E. Padan. 1984. Escherichia coli intracellular pH, membrane potential, and cell growth. J. Bacteriol. 158:246-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zinser, E. R., and R. Kolter. 1999. Mutations enhancing amino acid catabolism confer a growth advantage in stationary phase. J. Bacteriol. 181:5800-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]