Abstract
Antigen 43 (Ag43), a self-recognizing outer membrane protein of Escherichia coli, has been converted into an efficient and versatile tool for surface display of foreign protein segments. Ag43 is an autotransporter protein characterized by the feature that all information required for transport to the outer membrane and secretion through the cell envelope is contained within the protein itself. Ag43 consists of two subunits (α and β), where the β-subunit forms an integral outer membrane translocator to which the α-subunit is noncovalently attached. The simplicity of the Ag43 system makes it ideally suited as a surface display scaffold. Here we demonstrate that the Ag43 α-module can accommodate and display correctly folded inserts and has the ability to display entire functional protein domains, exemplified by the FimH lectin domain. The presence of heterologous cysteine bridges does not interfere with surface display, and Ag43 chimeras are correctly processed into α- and β-modules, offering optional and easy release of the chimeric α-subunits. Furthermore, Ag43 can be displayed in many gram-negative bacteria. This feature is exploited for display of our chimeras in an attenuated Salmonella strain.
The ability of cells to display proteins on the surface is essential in a wide range of biological phenomena, such as adhesion, colonization, biofilm formation, cell-cell recognition, signal transduction, and immunoreactions. In recent years, a variety of expression systems have been developed for display of heterologous peptides on cell surfaces. By grafting foreign proteins to naturally occurring surface proteins, it has been possible to express chimeric proteins on the surfaces of bacteriophages, bacteria, and yeast cells (4, 16, 24). Cells displaying heterologous peptides and proteins have led to the design of recombinant vaccines, reagents for diagnostics, whole-cell biocatalysts and bioadsorbants, and systems for combinatorial library screening.
Many different types of surface proteins of gram-negative bacteria have been used to display foreign peptides; these surface proteins include flagellin (36, 41), fimbriae (reviewed in reference 30), LamB (5, 6, 32), OmpA (13), FhuA (12), PhoE (1), peptidoglycan-associated lipoprotein (10, 14), S-layer protein (2), OprI (8), immunoglobulin Aβ (29, 51, 52), AIDA-I (34, 37), and VirGβ (50). One of the problems usually encountered with such display systems is the limited size of heterologous grafts that can be displayed without detrimental effects on the structure and/or function of the carrier protein and often the low copy number of the carrier protein. Also, expression of many, if not most, surface proteins is often restricted to confined host backgrounds, which can limit the versatility of corresponding display systems. Clearly, an ideal display system should combine the ability to accommodate large inserts with a high copy number and a broad host range.
Antigen 43 (Ag43) is a surface protein of Escherichia coli present at ∼50,000 copies per cell (42). The protein is a member of the autotransporter family, for which all information required for transport to the outer membrane and secretion through the cell envelope is contained in the protein itself (22). Ag43 is encoded by the flu gene, situated at 43 min on the E. coli chromosome (9). The expression is phase variable due to the concerted action of the Dam methylase (positive regulation) and the global regulator OxyR (negative regulation) (18, 23, 53). Ag43 is synthesized as a precursor of 1,039 amino acids, which is processed into a mature form consisting of two subunits, α and β, of 499 and 488 amino acid residues, respectively (19, 23). The β-subunit integrates into the outer membrane, where it forms a pore-like β-barrel structure in analogy with its AIDA-I homologue (23, 38). The α-subunit reaches the cell exterior assisted by the β-subunit and remains attached to the cell surface, presumably through noncovalent interaction with the β-subunit; however, it can easily be detached by brief heating to 60°C.
Ag43 has been found to induce characteristic surface properties on host cells, such as autoaggregation and a frizzy colony morphology (19, 20). Furthermore, we recently demonstrated that Ag43 is involved in bacterial biofilm formation (27, 28). Another important property of Ag43 is that, albeit of E. coli ancestry, it can be expressed in a fully functional form in a broad spectrum of gram-negative bacteria, including Pseudomonas fluorescens, Klebsiella pneumoniae, Enterobacter cloacae, Serratia liquefaciens, and many others (27; unpublished data). These attributes make Ag43 a highly versatile protein and potentially an ideal candidate for display of heterologous sequences on the surface of a broad range of organisms.
In this report we investigate the use of Ag43 for display of different immunogenic epitope tags and entire protein domains. Furthermore, due to the broad host range of Ag43, we examine the ability to express Ag43 chimeras in a well-characterized attenuated Salmonella vaccine strain.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strains and plasmids are listed in Table 1. In this study we used E. coli K-12 strain HEHA16 (leu lysA metE proB purE thi trp str supE fhuA fim::kan Δflu) (27). Salmonella enterica serovar Typhimurium SL5325 is a lipopolysaccharide-deficient (rfaJ) derivative of SL5316 (aroA) (39). SL5316 is of the FIRN (rhamnose negative, inositol negative, and permanently nonfimbriate) biotype and unable to produce type 1 fimbriae (11). Cells were grown at 37°C on solid or in liquid Luria-Bertani (LB) medium supplemented with the appropriate antibiotics unless otherwise stated.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference |
|---|---|---|
| Escherichia coli K-12 HEHA16 | BD1428; fim::kan Δflu | 27 |
| Salmonella enterica SL5325 | aroA rfaJ989 | 39 |
| Plasmids | ||
| pBAD/myc-HisA | 17 | |
| pKKJ143 | flu gene in pBAD/myc-HisA | This study |
| pKKJ135 | pKKJ143 with BglII site in flu gene | This study |
| pKKJ136 | CTP3 epitope in BglII site of pKKJ135 | This study |
| pKKJ138 | Chlam12 epitope in BglII site of pKKJ135 | This study |
| pKKJ146 | fimH lectin domain in XhoI/BglII sites of pKKJ135 | This study |
| pMAS4 | fimH gene in pBR322 | 46 |
Recombinant DNA techniques.
The flu gene was amplified by PCR from a plasmid preparation of pHHA147 (27) with primers KK70 (5′-CGCGCTCGAGATAATAAGGAAAAGCTATGAAC) and KK39 (5′-CGGCGAAGCTTCTGTCAGAAGGTCAC) containing XhoI and HindIII sites, respectively. The resulting PCR products were digested with XhoI and HindIII and inserted into the XhoI and HindIII sites of the pBAD/myc-HisA plasmid to produce pKKJ143 (Fig. 1). In this construct, expression of the flu gene is under control of the arabinose-inducible araBAD promoter.
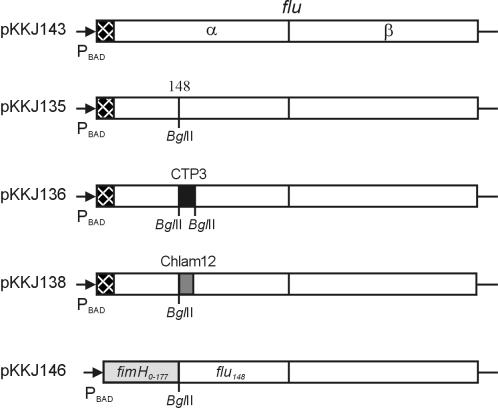
FIG. 1.
Overview of the plasmid constructs used in this study (not drawn to scale). Checkered boxes represent the flu signal peptide, and shaded boxes represent heterologous sequences: CTP3 epitope sequence (black box), Chlam12 epitope sequence (gray box), and FimH1-177 (light gray box).
A BglII linker was inserted at amino acid position 148 in the flu gene according to the following procedure: plasmid pKKJ143 was used as the template for PCR with primers KK70 and HH184 (5′-CCCAGATCTATCCGGTCCCCCTTCCG) and with primers HH185 (5′-GGGAGATCTAACGGTGATACCGGGCAGTT) and KK39. The PCR products were cleaved with XhoI and BglII and with BglII and HindIII. In a triple ligation, both PCR fragments obtained were inserted into the XhoI and HindIII sites of pBAD/myc-HisA to produce pKKJ135 (Fig. 1).
Two oligonucleotides, KK3 (5′-GATCTGTTGAAGTTCCGGGATCCCAGCATATCGATAGTCAGAAAAAAGCTA) and KK4 (5′-GATCTAGCTTTTTTCTGACTATCGATATGCTGGGATCCCGGAACTTCAACA), encoding amino acids 50 to 64 of the cholera toxin B chain (CTB), were designed so that they contained an internal BamHI site at amino acid position 54 and were flanked with BglII overhangs. These oligonucleotides were annealed, phosphorylated, and ligated into pKKJ135 digested with BglII. The resultant plasmid (pKKJ136) was checked by BamHI digestion and sequencing. In a similar procedure, two oligonucleotides, KK12 (5′-GATCTAAAGAACCGAACAAAGGCGTTAATCCGGATGAAGTTG) and KK13 (5′-GATCCAACTTCATCCGGATTAACGCCTTTGTTCGGTTCTTTA), encoding a 12-amino-acid linear epitope sequence of Chlamydia trachomatis DnaK, Chlam12 (3), flanked by BglII and BamHI sites, were introduced into the BglII site of pKKJ135 to produce pKKJ138 (Fig. 1).
Construction of fimH-flu fusion.
The sector encoding the signal peptide and the first 156 amino acids of fimH was amplified with primers KK85 (5′-CGCGCTCGAGATAAGAAGAGAGGATTGTAATGAAAC) and HH190 (5′-CCCAGATCTGCCGCCAGTAGGCACCACCA). The resulting fragment was cut with XhoI and BglII and inserted directly into the XhoI and BglII sites of pKKJ135 to produce plasmid pKKJ146. In this construct, the initial 199 codons of the flu gene are replaced with the first 177 codons of fimH, i.e., the signal peptide and lectin domain.
DNA manipulations.
Isolation of plasmid DNA was carried out with the QIAprep Spin miniprep kit (Qiagen). Restriction endonucleases were used according to the manufacturer's specifications (Biolabs). PCRs were done as previously described (47). Amplified products were sequenced to ensure fidelity of the PCR with the ABI Prism BigDye Terminator cycle sequencing ready reaction kit (PE Applied Biosystems). Samples were run on a Perkin-Elmer ABI Prism 310 genetic analyzer (PE Applied Biosystems) as described in the manufacturer's specifications.
Colony morphology.
Colony morphology was examined by with a Carl Zeiss Axioplan epifluorescence microscope, and digital images were captured with a 12-bit cooled slow-scan charge-coupled device camera (KAF 1400 chip; Photometrics, Tucson, Ariz.) controlled by the PMIS software (Photometrics).
Immunofluorescence microscopy.
Surface presentation of Ag43, Ag43-CTP3, Ag43-Chlam12, and Ag43-FimH was assessed by immunofluorescence microscopy with antibodies directed against the α-subunit of Ag43 (raised by T. Chakraborty, University of Giessen, Giessen, Germany), CTB (47), Chlam12 (kindly donated by S. Birkelund, University of Aarhus, Aarhus, Denmark) (3), or FimH (kindly provided by E. Sokurenko, University of Washington, Seattle). Cell fixation, immunolabeling, and microscopy were carried out as previously described (43) with a fluorescein isothiocyanate (FITC)-labeled secondary antibody. Surface-associated FimH receptor activity was assessed by receptor fluorescence microscopy with d-mannose-bovine serum albumin (BSA)-biotin and FITC-conjugated streptavidin.
Western immunoblotting.
Samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride microporous membrane filters as described previously (47). Reconstituted dried skimmed milk (10% [wt/vol]) was used as the blocking reagent, and serum raised against the α-subunit of Ag43 or FimH was used as the primary serum. Peroxidase-conjugated anti-rabbit immunoglobulin serum was used as the secondary antibody.
Receptor blot.
Receptor blots of FimH-Ag43 fusions were carried out essentially as described previously (45). d-mannose coupled to BSA was used as the primary reagent. Specific d-mannose binding was visualized by incubating the filter with anti-BSA rabbit serum and subsequently with horseradish peroxidase-conjugated anti-rabbit immunoglobulin serum. Tetramethylbenzidine was used as the substrate.
Release and purification of Ag43 chimeric fusion proteins.
Overnight cultures of the strains were inoculated in fresh LB supplemented with 0.2% arabinose and appropriate antibiotics and grown to identical optical densities at 600 nm (OD600; 2.5). Cells were harvested by centrifugation and gently washed in cold phosphate-buffered saline, and the centrifugation tube was placed in a 60°C water bath for 10 min in order to release the Ag43 α-fragment. Cells were immediately removed by centrifugation, and the proteins in the supernatant were precipitated with acetone (75%, vol/vol). The pellet was dried and resuspended in 0.1 M Tris buffer (pH 8).
RESULTS
Construction of recombinant gene fusions.
Recently we subjected the flu gene to linker insertion mutagenesis in connection with structure-function studies of Ag43 (unpublished data). One of the findings from these studies was that in-frame insertion of a BglII linker (encoding Arg-Ser) in front of the codon corresponding to amino acid 148 in the α-subunit of Ag43 did not affect the production, surface location, or autoaggregation of Ag43. This observation led us to believe that position 148 in Ag43 could be suitable for insertion and display of heterologous sequences on the surface of E. coli.
In order to investigate this tenet, two different well-characterized epitope sequences, CTP3 and Chlam12, were chosen as heterologous passenger sequences and introduced into Ag43. The CTP3 epitope, comprising amino acids 50 to 64 of the CTB chain, is known to comprise a conformational loop on the surface of CTB and has previously been shown by us to be authentically displayed on bacterial surfaces after fusion to different surface proteins (43, 47). A synthetic DNA segment encoding the CTP3 loop was made by annealing two complementary 51-base oligonucleotides which were designed to contain BglII overhangs in order to allow insertion into the BglII linker in the flu gene (Fig. 1). In a similar manner, a 12-amino-acid linear epitope, KEPNKGVNPDEV (presently called Chlam12), of Chlamydia trachomatis DnaK protein was introduced in front of position 148 in Ag43 (Fig. 1) The Chlam12 epitope is specifically recognized by the monoclonal antibody MAb35.2 (3). The tightly regulated arabinose-inducible araBAD promoter was used for expression of the gene fusions (17).
We have previously shown that the physical presence of type 1 fimbriae on the E. coli cell surface blocks Ag43-mediated autoaggregation (19). In order to abolish any interference from type 1 fimbriae, the plasmid constructs were transformed into a fim flu double mutant (HEHA16) of E. coli BD1428 (27).
Ag43 function is unaffected by insertion of heterologous sequences.
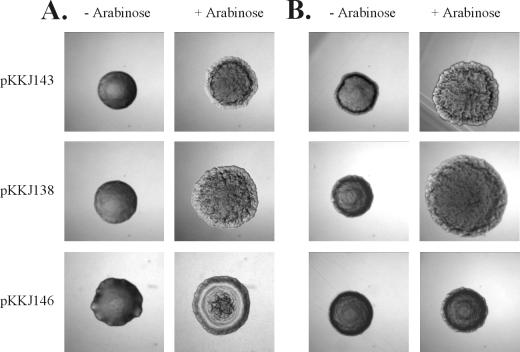
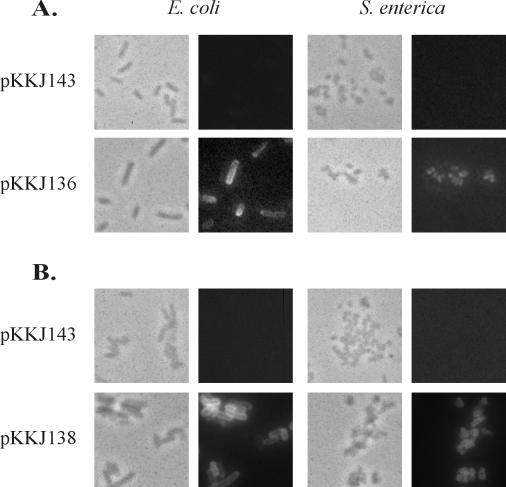
The phenotypic hallmarks of Ag43 expression are induction of a frizzy colony morphology and autoaggregation of cells (19, 27). To investigate whether insertion of heterologous peptides influenced the functionality of Ag43, we examined these parameters. When cells containing plasmids pKKJ136 (Ag43-CTP3) and pKKJ138 (Ag43-Chlam12) were grown on arabinose-containing solid medium, distinct frizzy colony morphotypes were observed, similar to that of cells expressing wild-type Ag43 (pKKJ143) (Fig. 2). No change in colony morphology was observed in control experiments where the cells were grown in the absence of arabinose. Also, when cells harboring pKKJ136 or pKKJ138 were left standing after growth in liquid medium in the presence of arabinose, they were observed to autoaggregate and settle exactly like the wild-type control (data not shown). Taken together, the results indicated that the fusion proteins were presented on the cell surface and that introduction of the heterologous sequences did not interfere with the functionality of Ag43.
FIG. 2.
Colony morphology of cells examined by phase contrast microscopy after overnight growth on solid medium with (+) and without (−) 0.2% arabinose. (A) Recombinant E. coli strain HEHA16; (B) recombinant S. enterica strain SL5325. Strains expressing both wild-type and chimeric versions of Ag43 have frizzy colony morphologies except in the case of plasmid pKKJ146. No change in morphology was observed for a strain containing the control plasmid, pBAD/myc-HisA. Colonies were photographed at the same magnification.
Expression and processing of Ag43-CTP3 and Ag43-Chlam12 fusion proteins.
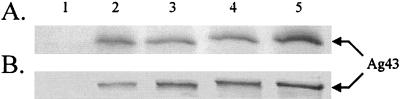
The amount of Ag43 protein produced by the clones was examined by SDS-PAGE and Western immunoblotting after arabinose induction. A band migrating at the expected apparent molecular mass for the Ag43 α-fragment (∼50 kDa) was observed from hosts carrying plasmid pKKJ143 or pKKJ135, and bands migrating with slightly higher apparent molecular masses were seen in the case of plasmids encoding the fusion proteins, i.e., pKKJ136 (Ag43-CTP3) and pKKJ138 (Ag43-Chlam12) (Fig. 3A). Also, the presence of foreign inserts seemed to have little influence on the level of Ag43 produced and on the correct processing of Ag43 into α- and β-subunits (Fig. 3A).
FIG. 3.
Western immunoblot of total cell lysates of (A) recombinant E. coli strain HEHA16 and (B) recombinant S. enterica strain SL5325 containing plasmids pBAD/myc-HisA, pKKJ143, pKKJ135, pKKJ136, and pKKJ138 (lanes 1 to 5, respectively). The position of Ag43α protein is indicated.
Passenger epitopes are authentically presented on the cell surface in the context of the Ag43 fusion proteins.
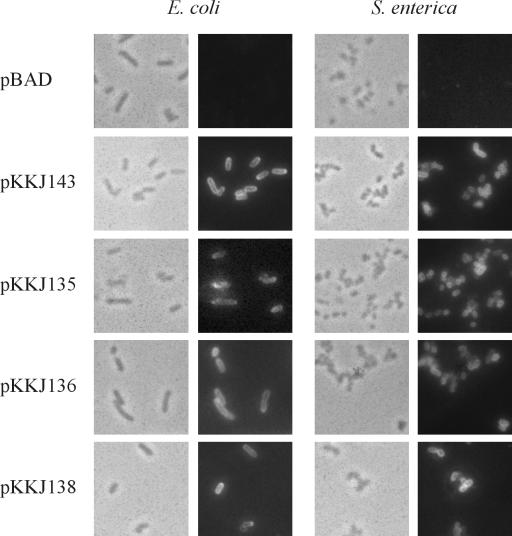
The results obtained regarding the colony morphology and autoaggregation of the strains expressing the Ag43-CTP3 and Ag43-Chlam12 chimeras prompted us to investigate more directly the surface display of the chimeras and, notably, whether the passenger epitopes were accessible and exposed in authentic conformations. This was carried out by immunofluorescence microscopy with antibodies directed against Ag43 and the CTP3 and Chlam12 epitopes. Ag43 antiserum readily reacted with cells harboring plasmids pKKJ143, pKKJ135, pKKJ136, and pKKJ138, again demonstrating that the Ag43 chimeras were translocated to the cell surface (Fig. 4). Furthermore, antibodies directed against the passenger epitopes were seen to specifically recognize cells expressing the corresponding fusion proteins, indicating that the CTP3 and Chlam12 epitopes were indeed not only accessible on the cell surface but also authentically folded in the context of the Ag43 scaffold (Fig. 5). No reactions against the passenger epitope sera were seen in control experiments with either wild-type Ag43 or vector controls.
FIG. 4.
Immunofluorescence microscopy showing surface presentation of Ag43 on recombinant E. coli HEHA16 cells and S. enterica SL5325 cells. Phase contrast microscopy (the left panels on each side) and fluorescence microscopy (right panels on each side) were performed. To detect the presence of Ag43 on the cell surface, a polyclonal rabbit serum raised against the α-subunit of Ag43 was used, and this was detected by FITC-labeled anti-rabbit immunoglobulin serum.
FIG. 5.
Immunofluorescence microscopy showing surface presentation of CTP3 epitope sequence (A) and Chlam12 epitope sequence (B) on recombinant E. coli HEHA16 and S. enterica SL5325 cells. Phase contrast microscopy (the left panels on each side) and fluorescence microscopy (right panels on each side) were performed. To detect the presence of CTP3 on the cell surface, a polyclonal rabbit serum raised against CTP3 was used, and this was detected by FITC-labeled anti-rabbit immunoglobulin serum. To detect the presence of Chlam12 on the cell surface, a monoclonal antibody raised against Chlam12 was used, and this was detected by FITC-labeled anti-mouse immunoglobulin serum.
Expression of Ag43 chimeras in Salmonella strains.
Attenuated Salmonella strains have been shown to be effective live vaccine strains and candidates for recombinant vaccine delivery systems (31, 40, 48). We have previously shown that Ag43 can be transported to and displayed on the surface of a wide range of gram-negative bacteria (27). This prompted us to investigate the possibility of expressing our Ag43 chimeras in the aroA-attenuated S. enterica serovar Typhimurium reference strain SL5325 with a view to demonstrating Ag43-mediated foreign antigen display in a bona fide live vaccine strain. Strain SL5325 forms nonfrizzy colonies and does not autoaggregate (Fig. 2B). Transformation of SL5325 with pKKJ143, pKKJ135, pKKJ136, and pKKJ138 gave rise to colonies with the characteristic frizzy morphology associated with Ag43 surface presentation after induction (Fig. 2B). Furthermore, arabinose-induced cultures of the transformed SL5325 clones readily autoaggregated in line with the corresponding E. coli clones (data not shown).
The amount of Ag43 produced and the processing of Ag43 (both wild-type and chimeric versions), as assessed by SDS-PAGE and Western immunoblotting, were similar in SL5325 and E. coli (Fig. 3). Furthermore, immunofluorescence microscopy revealed that Ag43 and the passenger epitopes (CTP3 and Chlam12) were displayed in an accessible and conformationally authentic form on the surface of SL5325 (Fig. 4 and Fig. 5A and B). The results suggest that Ag43 can be used to display foreign immunogenic determinants on the surface not only of E. coli but also of a relevant live vaccine strain.
Ag43 display of a foreign functional protein domain, the lectin moiety of FimH.
Having demonstrated Ag43-mediated display of small heterologous passenger peptides of 12 to 15 amino acids, we decided to investigate whether Ag43 was capable of accommodating and displaying entire protein domains. For this purpose, we selected the N-terminal domain of a well-characterized bacterial adhesin, FimH, as a model. Immunization with purified FimH has shown promise for prevention of E. coli urinary tract infections (33). FimH is the 279-amino-acid mannose-recognizing adhesin of type 1 fimbriae. It consists of two domains, a lectin domain encompassing the N-terminal 156 amino acid residues, linked to the C-terminal organelle integration domain via a short tetrapeptide loop. We have previously manipulated the fimH gene and shown that the FimH lectin domain can be expressed and purified as a stable, functional entity from the periplasm of E. coli (45).
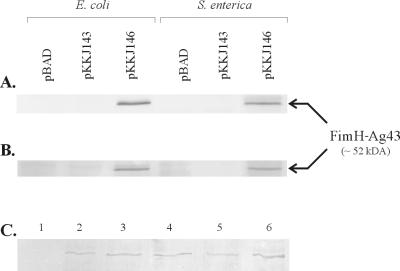
In order to create a FimH-Ag43 chimera, the first 199 codons of flu, i.e., the sector upstream of position 148, were replaced with the first 177 codons of fimH, i.e., signal peptide plus lectin domain, and placed downstream of the pBAD promoter (Fig. 1). E. coli host cells harboring this construct (pKKJ146) were seen to exhibit a peculiar nonfrizzy colony morphology after induction (Fig. 2), but the cells were unable to autoaggregate. Analysis of lysates of cells harboring pKKJ146 by SDS-PAGE and Western immunoblotting with antisera raised against FimH revealed a protein band with an apparent molecular mass of ∼52 kDa, in accordance with the expected size of the FimH-Ag43α fusion product, i.e., 514 amino acid residues (Fig. 6A). The ability of the fusion protein to recognize d-mannose was assayed by receptor blotting of whole-cell lysates (see Materials and Methods). The FimH-Ag43α chimera was found to recognize d-mannose, indicating that the fusion protein retained the FimH receptor recognition faculty (Fig. 6B).
FIG. 6.
Western immunoblot and receptor blot of total whole-cell lysate of recombinant cells with anti-FimH serum (A) and mannose-BSA (B). The apparent molecular size of the FimH-Ag43 fusion protein is indicated. (C) Coomassie brilliant blue-stained SDS-PAGE gel of wild-type and chimeric Ag43 α-modules released from the cell surface of E. coli strain HEHA16 containing plasmid pBAD/myc-HisA, pKKJ143, pKKJ135, pKKJ136, pKKJ138, or pKKJ146 (lanes 1 to 6, respectively).
Ag43-assisted surface display of the lectin moiety of FimH.
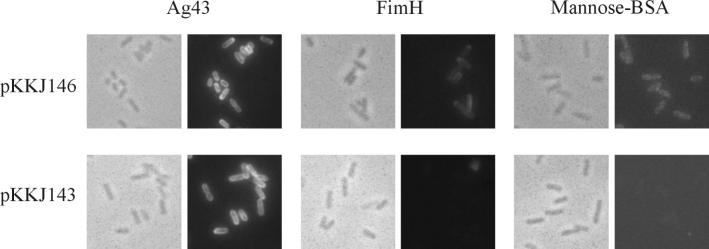
Fluorescence microscopy was used to establish surface presentation of the FimH-Ag43 fusion product. Cells containing plasmid pKKJ146 were seen to react with both anti-FimH serum and anti-Ag43 serum (Fig. 7). Additionally, the functionality of the FimH lectin moiety was assessed by receptor fluorescence microscopy with d-mannose-coupled BSA as the receptor target (Fig. 7). The results indicate that the lectin domain of FimH is presented on the bacterial surface in a functional form in the context of the FimH-Ag43α fusion.
FIG. 7.
Immunofluorescence microscopy showing surface presentation and receptor functionality of the FimH-Ag43 fusion protein on recombinant E. coli HEHA16 cells. Cells expressing plasmids pKKJ146 and pKKJ143 were reacted with primary antibodies directed against Ag43 and FimH, respectively, and FITC-labeled secondary antibody was used for detection. Additionally, receptor functionality was probed by using mannose-BSA-biotin, which was subsequently detected by FITC-conjugated streptavidin.
Ag43 chimeras can be liberated from carrier cells and purified.
The α-subunit of wild-type Ag43 is attached to the cell surface via noncovalent interaction with the β-subunit but can be released by brief heating to 60°C. In order to test whether this faculty was conserved in the chimeric versions of Ag43, identical aliquots of host cells expressing our chimeric Ag43 as well as adequate controls were processed to release the Ag43 α-subunits and subsequent protein isolation (see Materials and Methods). SDS-PAGE revealed that the Ag43α chimeras in all cases were released and could be purified from the supernatants in yields comparable to that of wild-type Ag43α (Fig. 6C)
DISCUSSION
The double membrane system of gram-negative bacteria constitutes a formidable barrier for proteins destined for the cell exterior. However, the bacteria have over eons developed various systems to route proteins through this obstacle. Five major systems can be distinguished: type I, of which E. coli hemolysin is a key example; type II, exemplified by the pullulanase system of Klebsiella; type III, required for translocation of some of the components of the secretion machinery; type IV, exemplified by the Agrobacterium tumefaciens T-DNA delivery system; and finally the type V system, comprising the autotransporter protein family (reviewed in references 21 and 35). By these systems, a range of proteins can reach the cell exterior, where they are key players in a number of natural processes, such as adhesion, biofilm formation, motility, and cell aggregation.
However, many proteins of interest in medicine and biotechnology are neither surface displayed nor of bacterial origin. An attractive way to display such proteins or segments thereof on the bacterial surface is to graft them into permissible positions on naturally occurring surface proteins. Autotransporter proteins, of which Ag43 is a paradigm, do not require auxiliary proteins (e.g., periplasmic chaperones), as many other systems do. This inherent simplicity makes them obvious candidates for scaffold-assisted surface display of foreign proteins or protein segments.
Our data indicate that Ag43 can act as a versatile scaffold for surface display of heterologous inserts and that the system offers a number of interesting features. (i) Ag43 can accommodate and display correctly folded foreign inserts. (ii) Ag43 is able to display entire and functional protein domains. (iii) Ag43 is a high-valency display system, i.e., the high copy number of wild-type Ag43 (∼50,000 copies per cell) does not seem to be significantly affected by foreign inserts or change of host strain. (iv) The carriage of passenger peptides does not interfere with correct processing of Ag43 into α- and β-moieties. (v) Ag43 can be displayed in a wide range of gram-negative bacteria.
Two immunogenic reporter epitopes, CTP3 and Chlam12, were displayed on the bacterial surface in the context of Ag43 and shown by immunofluorescence microscopy to be authentically folded. The presence of the foreign inserts did not interfere with either the yield or functionality of the carrier protein. This prompted us to investigate the possibility of displaying an entire protein domain with the Ag43 system. The d-mannose-specific lectin moiety of FimH was chosen as a candidate domain for this purpose due to its well-characterized binding features and its promise as a candidate vaccine against urinary tract-infectious E. coli. The FimH lectin moiety was surface displayed in the context of the FimH-Ag43 chimera and presented in a functional configuration, as evidenced by its affinity for a cognate receptor target, d-mannose-BSA. Unlike other Ag43 constructs, FimH-Ag43 did not give rise to a frizzy colony morphology, and cells were unable to autoaggregate. It might be that the frizzy colony morphology is dependent on cell-to-cell aggregation and that this faculty depends on structural information residing in the segment encompassing the N-terminal 148 amino acids of the Ag43 α-subunit.
FimH contains two disulfide bridges, one of which is located in the lectin moiety. Disulfide bridge formation of fimbrial structural proteins has been demonstrated to take place in the periplasm (15, 26). During biogenesis, the structural components are to a large extent folded before final translocation to the cell exterior (25, 49). Accordingly, the FimH lectin moiety in the FimH-Ag43 fusion is probably extensively folded before the final Ag43 β-subunit-assisted transit to the cell exterior takes place. The FimH lectin domain forms an ellipsoid structure with a width of ∼2.5 nm (7). Although Ag43 does not possess cysteine bridges itself, the presence of such features in a heterologous passenger domain is apparently not incompatible with traverse of the membrane system and surface display. A similar observation was noted with another autotransporter, the immunoglobulin A protease of Neisseria gonorrhoeae (52).
Wild-type Ag43 is processed into an α- and a β-subunit, presumably by autocatalysis. The α-subunit contains a sequence similar to that of the consensus sequence for the active site of aspartyl proteases, LLADSGAAVSGT (44), at amino acid positions 335 to 346. The present constructs were engineered to contain foreign inserts in front of amino acid position 148, leaving the potential protease site intact. All of our engineered Ag43 versions were processed into α- and β-subunits of expected sizes, lending further credence to the tenet of autocatalytic processing of Ag43. The fact that the Ag43 chimeras are not only surface displayed on bacteria but can additionally be released by simple heat treatment and purified offers several advantages. In a vaccine context, this offers capabilities for both live and component vaccine development approaches. In the current FimH-based candidate vaccine, FimH is purified as a complex with the FimC chaperone from the periplasm of E. coli hosts (33), a rather tedious procedure in comparison. Here we suggest two easy modes: an easy-to-purify component vaccine and a live vaccine candidate strain.
We have previously demonstrated that Ag43 can be surface exposed in a functional form in many gram-negative bacteria (27). This faculty is here demonstrated in a Salmonella strain, the potential vaccine candidate strain SL5325. Aromatic-dependent Salmonella strains have been demonstrated to be highly efficient live vaccine strains in many contexts (31, 40, 48). The apparent efficiency with which the Ag43 chimeras were displayed in the aroA-attenuated SL5325 strain bodes well for the development of new efficient live vaccine strains combining the excellent features of both systems.
Acknowledgments
This work was supported by the BIOPRO Center, part of the Danish National Strategic Environmental Research Program, and the Danish Medical (9802358) and Natural Sciences (21-01-0296) Research Councils.
REFERENCES
- 1.Agterberg, M., H. Adriaanse, A. van Bruggen, M. Karperien, and J. Tommassen. 1990. Outer-membrane PhoE protein of Escherichia coli K-12 as an exposure vector: possibilities and limitations. Gene 88:37-45. [DOI] [PubMed] [Google Scholar]
- 2.Bingle, W. H., J. F. Nomellini, and J. Smit. 1997. Cell surface display of a Pseudomonas aeruginosa strain K pilin peptide within the paracrystalline S-layer of Caulobacter crescentus. Mol. Microbiol. 26:277-288. [DOI] [PubMed] [Google Scholar]
- 3.Birkelund, S., A. G. Lundemose, and G. Christiansen. 1989. Characterization of native and recombinant 75-kilodalton immunogens from Chlamydia trachomatis serovar L2. Infect. Immun. 57:2683-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boder, E. T., and K. D. Wittrup. 1997. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15:553-557. [DOI] [PubMed] [Google Scholar]
- 5.Brown, S. 1992. Engineered iron oxide-adhesion mutants of the Escherichia coli phage lambda receptor. Proc. Natl. Acad. Sci. USA 89:8651-8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charbit, A., A. Molla, W. Saurin, and M. Hofnung. 1988. Versatility of a vector for expressing foreign polypeptides at the surface of gram-negative bacteria. Gene 70:181-189. [DOI] [PubMed] [Google Scholar]
- 7.Choudhury, D., A. Thompson, V. Stojanoff, S. Langermann, J. Pinkner, S. J. Hultgren, and S. D. Knight. 1999. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285:1061-1066. [DOI] [PubMed] [Google Scholar]
- 8.Cornelis, P., J. C. Sierra, A. Lim, Jr., A. Malur, S. Tungpradabkul, H. Tazka, A. Leitao, C. V. Martins, C. di Perna, L. Brys, P. De Baetseller, and R. Hamers. 1996. Development of new cloning vectors for the production of immunogenic outer membrane fusion proteins in Escherichia coli. Bio/Technology 14:203-208. [DOI] [PubMed] [Google Scholar]
- 9.Diderichsen, B. 1980. flu, a metastable gene controlling surface properties of Escherichia coli. J. Bacteriol. 141:858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubel, S., F. Breitling, P. Fuchs, M. Braunagel, I. Klewinghaus, and M. Little. 1993. A family of vectors for surface display and production of antibodies. Gene 128:97-101. [DOI] [PubMed] [Google Scholar]
- 11.Duguid, J. P., E. S. Anderson, G. A. Alfredsson, R. Barker, and D. C. Old. 1975. A new biotyping scheme for Salmonella typhimurium and its phylogenetic significance. J. Med. Microbiol 8:149-166. [DOI] [PubMed] [Google Scholar]
- 12.Etz, H., D. B. Minh, C. Schellack, E. Nagy, and A. Meinke. 2001. Bacterial phage receptors, versatile tools for display of polypeptides on the cell surface. J. Bacteriol. 183:6924-6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freudl, R. 1989. Insertion of peptides into cell surface-exposed areas of the Escherichia coli OmpA protein does not interfere with export and membrane assembly. Gene 82:229-236. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs, P., F. Breitling, S. Dubel, T. Seehaus, and M. Little. 1991. Targeting recombinant antibodies to the surface of Escherichia coli: fusion to a peptidoglycan associated lipoprotein. Bio/Technology 9:1369-1372. [DOI] [PubMed] [Google Scholar]
- 15.Genevaux, P., P. Bauda, M. S. DuBow, and B. Oudega. 1999. Identification of Tn10 insertions in the dsbA gene affecting Escherichia coli biofilm formation. FEMS Microbiol. Lett. 173:403-409. [DOI] [PubMed] [Google Scholar]
- 16.Georgiou, G., C. Stathopoulos, P. S. Daugherty, A. R. Nayak, B. L. Iverson, and R. Curtiss III. 1997. Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat. Biotechnol. 15:29-34. [DOI] [PubMed] [Google Scholar]
- 17.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haagmans, W., and M. van der Woude. 2000. Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol. Microbiol. 35:877-887. [DOI] [PubMed] [Google Scholar]
- 19.Hasman, H., T. Chakraborty, and P. Klemm. 1999. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J. Bacteriol. 181:4834-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hasman, H., M. A. Schembri, and P. Klemm. 2000. Antigen 43 and type 1 fimbriae determine colony morphology of Escherichia coli K-12. J. Bacteriol. 182:1089-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson, I. R., J. P. Nataro, J. B. Kaper, T. F. Meyer, S. K. Farrand, D. L. Burns, B. B. Finlay, and J. W. St. Geme III. 2000. Renaming protein secretion in the gram-negative bacteria. Trends Microbiol. 8:352.. [PubMed] [Google Scholar]
- 22.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370-378. [DOI] [PubMed] [Google Scholar]
- 23.Henderson, I. R., and P. Owen. 1999. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and OxyR. J. Bacteriol. 181:2132-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill, H. R., and P. G. Stockley. 1996. Phage presentation. Mol. Microbiol. 20:685-692. [DOI] [PubMed] [Google Scholar]
- 25.Hultgren, S. J., C. H. Jones, and S. Normark. 1996. Bacterial adhesins and their assembly, p. 2730-2756. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 26.Jacob-Dubuisson, F., R. Striker, and S. J. Hultgren. 1994. Chaperone-assisted self-assembly of pili independent of cellular energy. J. Biol. Chem. 269:12447-12455. [PubMed] [Google Scholar]
- 27.Kjaergaard, K., M. A. Schembri, H. Hasman, and P. Klemm. 2000. Antigen 43 from Escherichia coli induces inter- and intraspecies cell aggregation and changes in colony morphology of Pseudomonas fluorescens. J. Bacteriol. 182:4789-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kjaergaard, K., M. A. Schembri, C. Ramos, S. Molin, and P. Klemm. 2000. Antigen 43 facilitates formation of multispecies biofilms. Environ. Microbiol. 2:695-702. [DOI] [PubMed] [Google Scholar]
- 29.Klauser, T., J. Pohlner, and T. F. Meyer. 1990. Extracellular transport of cholera toxin B subunit with Neisseria IgA protease beta-domain: conformation-dependent outer membrane translocation. EMBO J. 9:1991-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klemm, P., and M. A. Schembri. 2000. Fimbrial surface display systems in bacteria: from vaccines to random libraries. Microbiology 12:3025-3032. [DOI] [PubMed] [Google Scholar]
- 31.Koesling, J., B. Lucas, L. Develioglou, T. Aebischer, and T. F. Meyer. 2001. Vaccination of mice with live recombinant Salmonella typhimurium aroA against H. pylori: parameters associated with prophylactic and therapeutic vaccine efficacy. Vaccine 20:413-420. [DOI] [PubMed] [Google Scholar]
- 32.Kotrba, P., L. Doleckova, V. de Lorenzo, and T. Ruml. 1999. Enhanced bioaccumulation of heavy metal ions by bacterial cells due to surface display of short metal binding peptides. Appl. Environ. Microbiol. 65:1092-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langermann, S., R. Mollby, J. E. Burlein, S. R. Palaszynski, C. G. Auguste, A. DeFusco, R. Strouse, M. A. Schenerman, S. J. Hultgren, J. S. Pinkner, J. Winberg, L. Guldevall, M. Soderhall, K. Ishikawa, S. Normark, and S. Koenig. 2000. Vaccination with FimH adhesin protects Cynomolgus monkeys from colonization and infection by uropathogenic Escherichia coli. J. Infect. Dis. 181:774-778. [DOI] [PubMed] [Google Scholar]
- 34.Lattemann, C. T., J. Maurer, E. Gerland, and T. F. Meyer. 2000. Autodisplay: functional display of active beta-lactamase on the surface of Escherichia coli by the AIDA-I autotransporter. J. Bacteriol. 182:3726-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lory, S. 1998. Secretion of proteins and assembly of bacterial surface organelles: shared pathways of extracellular protein targeting. Curr. Opin. Microbiol. 1:27-35. [DOI] [PubMed] [Google Scholar]
- 36.Lu, Z., K. S. Murray, V. van Cleve, E. R. LaVallie, M. L. Stahl, and J. M. McCoy. 1995. Expression of thioredoxin random peptide libraries on the Escherichia coli cell surface as functional fusions to flagellin: a system designed for exploring protein-protein interactions. Bio/Technology 13:366-372. [DOI] [PubMed] [Google Scholar]
- 37.Maurer, J., J. Jose, and T. F. Meyer. 1997. Autodisplay: one-component system for efficient surface display and release of soluble recombinant proteins from Escherichia coli. J. Bacteriol. 179:794-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maurer, J., J. Jose, and T. F. Meyer. 1999. Characterization of the essential transport function of the AIDA-I autotransporter and evidence supporting structural predictions. J. Bacteriol. 181:7014-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nevola, J. J., B. A. D. Stocker, D. C. Laux, and P. S. Cohen. 1985. Colonization of the mouse intestine by an avirulent Salmonella typhimurium strain and its lipopolysaccharide-defective mutants. Infect. Immun. 50:152-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newton, S. M., C. O. Jacob, and B. A. D. Stocker. 1989. Immune response to cholera toxin epitope inserted in Salmonella flagellin. Science 244:70-72. [DOI] [PubMed] [Google Scholar]
- 41.Newton, S. M., T. M. Joys, S. A. Anderson, R. C. Kennedy, M. E. Hovi, and B. A. D. Stocker. 1995. Expression and immunogenicity of an 18-residue epitope of HIV1 gp41 inserted in the flagellar protein of a Salmonella live vaccine. Res. Microbiol. 146:203-216. [DOI] [PubMed] [Google Scholar]
- 42.Owen, P. 1992. The gram-negative outer membrane: structure, biochemistry and vaccine potential. Biochem. Soc. Trans. 20:1-6. [DOI] [PubMed] [Google Scholar]
- 43.Pallesen, L., L. K. Poulsen, G. Christiansen, and P. Klemm. 1995. Chimeric FimH adhesin of type 1 fimbriae: a bacterial surface display system for heterologous sequences. Microbiology 141:2839-2848. [DOI] [PubMed] [Google Scholar]
- 44.Rao, J. K., J. W. Erickson, and A. Wlodawer. 1991. Structural and evolutionary relationships between retroviral and eucaryotic aspartic proteinases. Biochemistry 30:4663-4671. [DOI] [PubMed] [Google Scholar]
- 45.Schembri, M. A., H. Hasman, and P. Klemm. 2000. Expression and purification of the mannose recognition domain of the FimH adhesin. FEMS Microbiol. Lett. 188:147-151. [DOI] [PubMed] [Google Scholar]
- 46.Schembri, M. A., L. Pallesen, H. Connell, D. L. Hasty, and P. Klemm. 1996. Linker insertion analysis of the FimH adhesin of type 1 fimbriae in an Escherichia coli fimH-null background. FEMS Microbiol. Lett. 137:257-263. [DOI] [PubMed] [Google Scholar]
- 47.Stentebjerg-Olesen, B., L. Pallesen, L. B. Jensen, G. Christiansen, and P. Klemm. 1997. Authentic display of a cholera toxin epitope by chimeric type 1 fimbriae: effects of insert position and host background. Microbiology 143:2027-2038. [DOI] [PubMed] [Google Scholar]
- 48.Stocker, B. A. D. 2000. Aromatic-dependent salmonella as antibacterial vaccines and as presenters of heterologous antigens or of DNA encoding them. J. Biotechnol. 83:45-50. [DOI] [PubMed] [Google Scholar]
- 49.Strom, M. S., and S. Lory. 1993. Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47:565-596. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki, T., M. C. Lett, and C. Sasakawa. 1995. Extracellular transport of VirG protein in Shigella. J. Biol. Chem. 270:30874-30880. [DOI] [PubMed] [Google Scholar]
- 51.Valls, M., S. Atrian, V. de Lorenzo, and L. A. Fernandez. 2000. Engineering a mouse metallothionein on the cell surface of Ralstonia eutropha CH34 for immobilization of heavy metals in soil. Nat. Biotechnol. 18:661-665. [DOI] [PubMed] [Google Scholar]
- 52.Veiga, E., V. de Lorenzo, and L. A. Fernandez. 1999. Probing secretion and translocation of a beta-autotransporter with a reporter single-chain Fv as a cognate passenger domain. Mol. Microbiol. 33:1232-1243. [DOI] [PubMed] [Google Scholar]
- 53.Warne, S. R., J. M. Varley, G. J. Boulnois, and M. G. Norton. 1990. Identification and characterization of a gene that controls colony morphology and auto-aggregation in Escherichia coli K12. J. Gen. Microbiol. 136:455-462. [DOI] [PubMed] [Google Scholar]