Abstract
The cell division protein ZipA has an N-terminal transmembrane domain and a C-terminal globular domain that binds FtsZ. Between them are a charged domain and a P/Q domain rich in proline and glutamine that has been proposed to be an unfolded polypeptide. Here we provide evidence obtained by electron microscopy that the P/Q domain is a flexible tether ranging in length from 8 to 20 nm and invisible in rotary shadowing electron microscopy. We estimated a persistence length of 0.66 nm, which is similar to the persistence lengths of other unfolded and unstructured polypeptides.
The protein FtsZ assembles into a ring at the center of bacterial cells, and this Z ring constricts to effect cell division. A number of accessory proteins localize to the Z ring after FtsZ and are essential for cell division in Escherichia coli. One accessory protein is ZipA (Z-interacting protein A), which has been shown to bind directly to FtsZ (3, 5, 6, 8-11, 13). ZipA is a membrane-anchored protein and consists of five parts: amino acids (aa) 1 to 6 are a charged periplasmic domain; aa 7 to 28 are a transmembrane segment; aa 29 to 85 are a charged domain that is rich in charged amino acids; aa 86 to 185 are called the P/Q domain, which is rich in proline and glutamine; and aa 186 to 328 are a globular domain. The globular domain of ZipA has been shown to bind the conserved C-terminal peptide of FtsZ (5, 6, 8-11), and its atomic structure has been solved (10, 11). The P/Q domain has been proposed, on the basis of its sequence, to be largely an unfolded polypeptide and to serve as a flexible linker tethering FtsZ protofilaments to the membrane (1, 3). RayChaudhuri (13), using the program NNPREDICT, found no evidence for secondary structure in the P/Q domain and only a small stretch of beta sheet and alpha helix at the end of the charged domain. Finally, we performed a sequence alignment of ZipA from six sequenced genomes and found substantial sequence identity for N-terminal residues 1 to 36 (of the E. coli sequence) and for the C-terminal domain (aa 188 to 328). There was very low identity for the intervening segment (aa 38 to 187), with the exception of a few modestly conserved islands. This is consistent with this segment not having a defined structure.
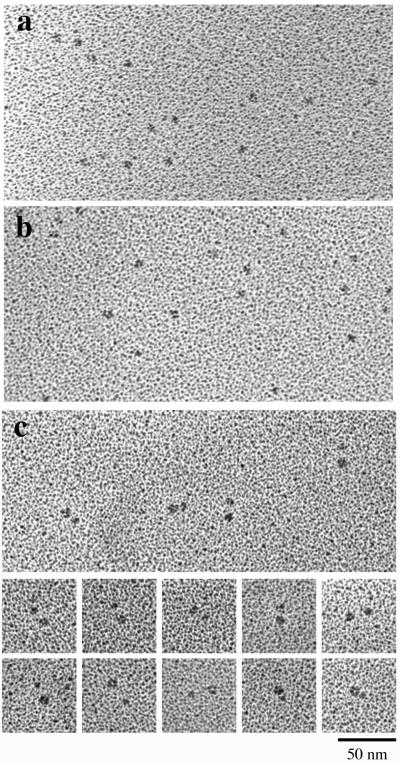
Purified ZipA derivatives (5) were subjected to sedimentation through a glycerol gradient and then examined by rotary shadowing electron microscopy (EM) as previously described (2). Figure 1a shows a micrograph of ZipA(23-328), which contains the complete charged domain, the P/Q domain, and the C-terminal globular domain of native ZipA (5). Figure 1b shows ZipA(186-328), which comprises only the C-terminal globular domain. These two constructs were described previously (4). The C-terminal domain appears as small globular particles about 3.5 to 4 nm in diameter (the diameter was estimated after subtracting 2 nm for the thickness of the platinum shell), consistent with the size (2.5 by 3.5 by 5 nm) of the C-terminal domain determined by X-ray crystallography (10) and nuclear magnetic resonance analysis (11). ZipA(23-328) showed globular particles that were indistinguishable from the C-terminal domain alone. Occasionally, a very small globular particle or a thin extension could be seen close to the main globular particle but, for the most part, there was no visible structure corresponding to the charged and P/Q domains in ZipA(23-328). This suggested that the P/Q domain may be a largely unfolded polypeptide and invisible in the rotary shadowed specimen, consistent with the previous prediction from the sequence (3). We suggest that the charged domain may be unstructured as well because, in our experience, a 60-aa globular domain should be visible in rotary shadowing. This is supported by the lack of predicted secondary structure in the charged domain (13).
FIG. 1.
Rotary shadowing EM of ZipA(23-328) (a) and ZipA(186-328) (b) (the C-terminal domain alone). Both samples show small globular particles of similar sizes. The charged-plus-P/Q domain of ZipA(23-328) is mostly invisible, although occasionally a faint projection or small domain may be discerned. (c) EM of GFP-ZipA. The GFP-ZipA(39-328) fusion protein is visible as pairs of globular domains; presumably, these domains are connected by the charged-plus-P/Q domain, which is not visible. The middle panels show selected pairs with two globular domains of similar sizes, and the bottom panels show pairs with different-sized globular domains.
To investigate the structure in more detail, we constructed a ZipA variant [green fluorescent protein (GFP)-ZipA(39-328)] in which a GFP domain was fused to the N terminus of ZipA(39-328). EM of GFP-ZipA(39-328) showed globular particles in closely spaced pairs (Fig. 1c). The larger globular domain should correspond to the 238-aa GFP, and the smaller one should correspond to the 140-aa ZipA C-terminal domain. The charged-plus-P/Q domain is interpreted to form an invisible link connecting the two globular domains.
An unfolded polypeptide can be modeled as a worm-like chain characterized by a contour length L and a persistence length p. Rivetti et al. (15) provided an elegant analysis of DNA on mica, modeled as a worm-like chain, and concluded that the molecules rearranged by diffusion on the two-dimensional mica substrate. For our analysis, we assume that the flexible polypeptide can also assume a random two-dimensional distribution on mica (see reference 15 for a detailed discussion of the two-dimensional structure versus the three-dimensional structure of the worm-like chain). The distribution of end-to-end lengths of the worm-like chains is given by the formula <R2> = 4pL (for the case in which L >> p), where R is the distance between the ends of the chain, <R2> is the average value of R2, L is the contour length, and p is the persistence length.
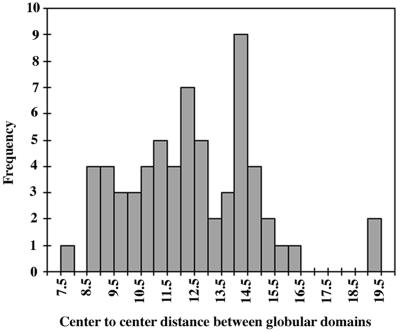
The R value needed for the above formula is the distance between the ends of the flexible chain, but the best estimate we can make is to measure the center-to-center distance between the two globular domains. A histogram of center-to-center distances is shown in Fig. 2. We considered subtracting 4 nm, the sum of the predicted radii of the N- and C-terminal globular domains, from the measured center-to-center distance. This correction would be appropriate if the exit points of the polypeptide from the two globular domains were facing each other (as illustrated in Fig. 3b, c, and e). However, it is also possible that these exit points face away from each other and the polypeptide crosses over or around the domains (as in Fig. 3d), in which case we should add 4 nm. If we assume that the P/Q segment is quite flexible and the orientation of the globular domains is random, the simple center-to-center distance seems to be the best estimate of the end-to-end length of the charged-plus-P/Q segment. Note, however, that each measurement includes an up to ±5-nm error due to the unknown orientation of the globular domains.
FIG. 2.
Histogram of the center-to-center distance between the globular particles in GFP-ZipA(39-328) used as an estimate of the end-to-end length of the charged-plus-P/Q domain (see text for details). The histogram shows a broad distribution of distances, with an average of 12 nm.
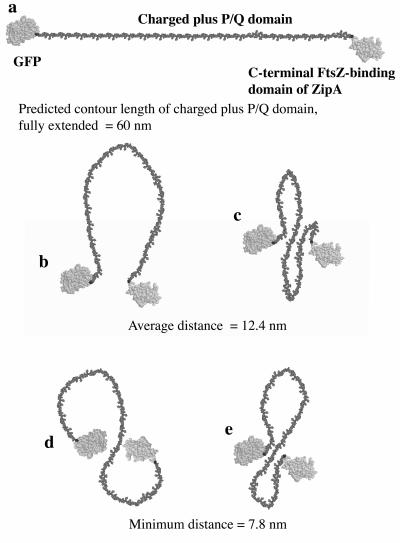
FIG. 3.
Molecular models of conformations of the unstructured charged-plus-P/Q domain of ZipA. The GFP domain is shown at the N terminus, which, in native ZipA, would be replaced by the transmembrane anchor. The maximally extended polypeptide is shown in panel a. A single-loop (b) or winding-chain (c, d, and e) structure can bring the termini closer together. The model was constructed from the atomic structures of GFP and the ZipA C-terminal domain, obtained from the protein database (1ema and 1F47). The extended polypeptide was modeled on segments of beta strands.
A histogram of the center-to-center distances reveals a wide distribution, ranging from 7.8 to 19.5 nm, with an average of 12.4 nm (Fig. 2). The average value of R2 was determined from the data in this histogram, giving <R2> = 159 nm2. The presumed unstructured domain includes the 12-aa spacer followed by the 146-aa charged-plus-P/Q domain (from R39 to M185). This gives a predicted contour length L = 60 nm ([0.38 nm/aa] × 158 aa). The persistence length is now calculated to be p = <R2>/4L = 0.66 nm. This is about twice the length of a peptide bond and close to the values of 0.4 to 0.44 nm estimated from atomic force microscope measurements for unfolded polypeptides of titin and tenascin (12, 14). It is also similar to the p = 0.95 nm calculated by the above equation for the presumably unstructured PEVK domain of titin (7).
We also characterized the ZipA(23-328) and GFP-ZipA(39-328) proteins by sedimentation analysis. Determination of sedimentation coefficients by analytical ultracentrifugation (20,000 rpm in the ANL rotor, Beckman XLA analytical ultracentrifuge) yielded values of 2.2S for ZipA(23-328) and 3.2S for GFP-ZipA(39-328). To estimate the shape of the proteins, we calculated Smax/S, where Smax is the sedimentation coefficient of an unhydrated sphere of protein of the same mass. Smax/S was calculated to be 1.73 for ZipA(23-328) and 1.75 for GFP-ZipA(39-328). These values are indicative of moderately elongated proteins (16) and are consistent with the EM studies described above, suggesting that the P/Q domain of ZipA is largely unfolded.
Figure 3 presents molecular models of the GFP-ZipA structure showing the charged-plus-P/Q domains as unstructured polypeptide. Note that the GFP in our construct is at the position of the transmembrane segment. We propose, on the basis of our EM analyses, that the tether is flexible and can span a maximum distance of 60 nm. In the more relaxed conformations, FtsZ subunits that are bound to the C-terminal FtsZ binding domain of ZipA could be an average of 12 nm from the membrane. Since the FtsZ protofilaments are only 3 to 4 nm thick, the tether is long enough to loop around and bind the C terminus of FtsZ, even if it is facing away from the membrane (see reference 1 for a diagram).
Acknowledgments
We thank Harvey Sage, Biochemistry, Duke University Medical Center, for performing the sedimentation analysis with the XLA analytical ultracentrifuge.
This study was supported by National Institutes of Health grants CA47056 (H.P.E.) and GM-57059 (P.D.B.).
REFERENCES
- 1.Erickson, H. P. 2001. The FtsZ protofilament and attachment of ZipA-structural constraints on the FtsZ power stroke. Curr. Opin. Cell Biol. 13:55-60. [DOI] [PubMed] [Google Scholar]
- 2.Fowler, W. E., and H. P. Erickson. 1979. Trinodular structure of fibrinogen: confirmation by both shadowing and negative stain electron microscopy. J. Mol. Biol. 134:241-249. [DOI] [PubMed] [Google Scholar]
- 3.Hale, C. A., and P. A. J. de Boer. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88:175-185. [DOI] [PubMed] [Google Scholar]
- 4.Hale, C. A., and P. A. J. de Boer. 1999. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J. Bacteriol. 181:167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hale, C. A., A. C. Rhee, and P. A. J. de Boer. 2000. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol. 182:5153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haney, S. A., E. Glasfeld, C. Hale, D. Keeney, Z. He, and P. A. de Boer. 2001. Genetic analysis of the E. coli FtsZ-ZipA interaction in the yeast two-hybrid system: characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J. Biol. Chem. 276:11980-11987. [DOI] [PubMed] [Google Scholar]
- 7.Li, H., A. F. Oberhauser, S. D. Redick, M. Carrion-Vazquez, H. P. Erickson, and J. M. Fernandez. 2001. Multiple conformations of PEVK proteins detected by single-molecule techniques. Proc. Natl. Acad. Sci. USA 98:10682-10686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu, Z., A. Mukherjee, and J. Lutkenhaus. 1999. Recruitment of ZipA to the division site by interaction with FtsZ. Mol. Microbiol. 31:1853-1861. [DOI] [PubMed] [Google Scholar]
- 9.Ma, X., and W. Margolin. 1999. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli ftsZ. J. Bacteriol. 181:7531-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mosyak, L., Y. Zhang, E. Glasfeld, S. Haney, M. Stahl, J. Seehra, and W. S. Somers. 2000. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 19:3179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moy, F. J., E. Glasfeld, L. Mosyak, and R. Powers. 2000. Solution structure of ZipA, a crucial component of Escherichia coli cell division. Biochemistry 39:9146-9156. [DOI] [PubMed] [Google Scholar]
- 12.Oberhauser, A. F., P. E. Marszalek, H. P. Erickson, and J. M. Fernandez. 1998. The molecular elasticity of the extracellular matrix protein tenascin. Nature 393:181-185. [DOI] [PubMed] [Google Scholar]
- 13.RayChaudhuri, D. 1999. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 18:2372-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rief, M., M. Gautel, F. Oesterhelt, J. M. Fernandez, and H. E. Gaub. 1997. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 276:1109-1112. [DOI] [PubMed] [Google Scholar]
- 15.Rivetti, C., M. Guthold, and C. Bustamante. 1996. Scanning force microscopy of DNA deposited onto mica: equilibration versus kinetic trapping studied by statistical polymer chain analysis. J. Mol. Biol. 264:919-932. [DOI] [PubMed] [Google Scholar]
- 16.Schürmann, G., J. Haspel, M. Grumet, and H. P. Erickson. 2001. Cell adhesion molecule L1 in folded (horseshoe) and extended conformations. Mol. Biol. Cell 12:1765-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]