Abstract
Two site-specific shuttle integration vectors were developed with two different chromosomal bacteriophage integration sites to facilitate strain construction in Listeria monocytogenes. The first vector, pPL1, utilizes the listeriophage U153 integrase and attachment site within the comK gene for chromosomal insertion. pPL1 contains a useful polylinker, can be directly conjugated from Escherichia coli into L. monocytogenes, forms stable, single-copy integrants at a frequency of ∼10−4 per donor cell, and can be used in the L. monocytogenes 1/2 and 4b serogroups. Methods for curing endogenous prophages from the comK attachment site in 10403S-derived strains were developed. pPL1 was used to introduce the hly and actA genes at comK-attBB′ in deletion strains derived from 10403S and SLCC-5764. These strains were tested for second-site complementation in hemolysin assays, plaquing assays, and cell extract motility assays. Unlike plasmid-complemented strains, integrated pPL1-complemented strains were fully virulent in the mouse 50% lethal dose assay. Additionally, the PSA phage attachment site on the L. monocytogenes chromosome was characterized, and pPL1 was modified to integrate at this site. The listeriophage PSA integrates in the 3′ end of an arginine tRNA gene. There are 17 bp of DNA identity between the bacterial and phage attachment sites. The PSA prophage DNA sequence reconstitutes a complete tRNAArg gene. The modified vector, pPL2, was integration proficient at the same frequency as pPL1 in common laboratory serotype 1/2 strains as well as serotype 4b strains.
Listeria monocytogenes is a gram-positive facultative intracellular pathogen of humans and many animal species that is capable of causing serious food-borne illness in pregnant and immunocompromised individuals (11). L. monocytogenes has been an important model system that has shed light on the mechanisms of both intracellular parasitism and host cell biology. The study of several of the virulence factors has led to important fundamental insights into the nature of invasion of host cells, escape from the lysosomal compartment into the cytoplasm, intracellular growth, and cell-to-cell spread (8). Additionally, the study of L. monocytogenes has been central in understanding the complex interactions of the eukaryotic actin cytoskeleton and cell motility (7).
Two of the major virulence factors of L. monocytogenes are the pore-forming cytolysin listeriolysin-O (LLO) and the actin-recruiting and -organizing protein ActA. LLO, the product of the hly gene, is responsible for escape from the membrane-bound phagosomal compartment when L. monocytogenes first enters a host cell, allowing the bacterium to quickly gain access to the host cytoplasm (51). LLO is absolutely required for the virulence and pathogenesis of L. monocytogenes (9, 14, 23). Once the bacterium enters the cytoplasm, the ActA protein, a second essential virulence factor (4, 10, 24), enables the bacterium to polymerize host cell actin in a polar manner to propel the bacterium through the cytoplasm and spread the infection from cell to cell without exposure to the extracellular environment (34, 51).
Although tremendous insights have been gained from the study of the intracellular life cycle of L. monocytogenes, there have also been technical limitations to these studies that are related to the genetic manipulation of the bacterium. The current array of tools allow specific genetic alterations to be made, but the process of making these changes is often time-consuming and experimentally cumbersome. For instance, ActA cannot be functionally complemented on a plasmid (39), so the process of evaluating the biological role of ActA is limited to using allelic exchange, which, for L. monocytogenes, can take weeks or months to construct each clone.
With these limitations in mind, we set out to develop a streamlined approach for systematic strain construction. Recently, the complete genome sequence and characterization of the phage attachment site of listeriophage A118 were reported (29), the genome sequence of the PSA prophage was completed (E. Sattelberger, M. Zimmer, R. Calendar, R. B. Inman, S. Scherer, and M. J. Loessner, submitted for publication), and a survey of listeriophage host ranges was conducted (19). These studies facilitated the development of two phage-based integration vectors, the first such vectors for use in L. monocytogenes. Integration vectors have several advantages over plasmids, including single copy number once integrated as well as stability in the absence of selection. Site specificity of the integration vector allows manipulations to be done in an innocuous region of the genome. Furthermore, integration vectors obviate the need for allelic exchange, instead allowing defined molecular constructs to be placed on the chromosome in a single step.
The first of these integration vectors, pPL1, integrates at the comK-attBB′ chromosomal location. We evaluated the role of an intact comK open reading frame on the virulence of L. monocytogenes, determined the prophage status of 25 L. monocytogenes strains at the comK-attBB′ chromosomal position, and used pPL1 to functionally complement LLO and ActA at the comK chromosomal position in the deletion strains. We further characterized the phage attachment site of the PSA prophage within a tRNAArg gene and used this information to modify pPL1 to integrate at the PSA phage attachment site. The second integration vector, pPL2, can be used in a wide array of L. monocytogenes strains independent of serotype and the presence of a prophage at the comK integration site.
MATERIALS AND METHODS
Construction of pPL1 integration vector.
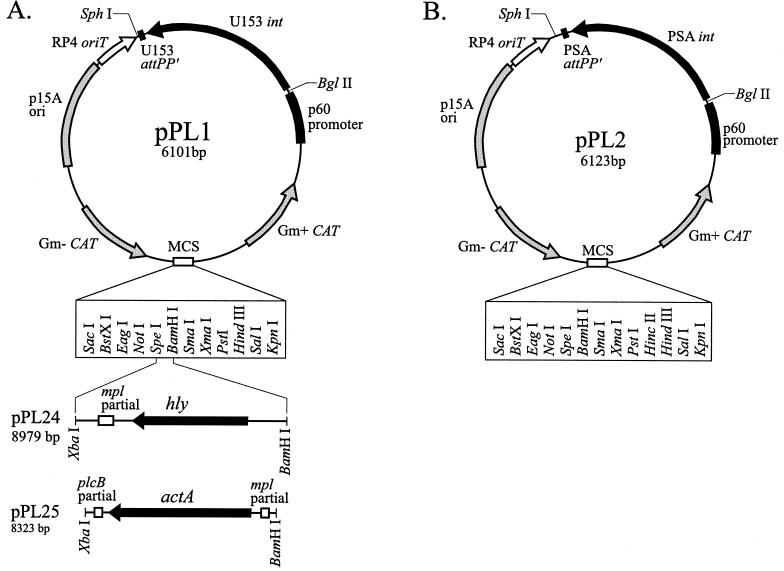
Standard molecular techniques (41) were used in the construction of the 6,101-bp integration vector pPL1 (Fig. 1A). It is a low-copy-number plasmid that replicates autonomously in Escherichia coli and integrates in a site-specific manner in L. monocytogenes and was assembled from six independent DNA sources as follows. Restriction sites in the PCR primers used for construction are underlined. All PCRs used in cloning steps utilized Vent DNA polymerase (New England Biolabs).
FIG. 1.
(A) Plasmid map of the pPL1 integration vector. Chloramphenicol resistance genes and E. coli origin of replication are shown in grey, the RP4 origin of transfer is shown in white, and the U153 integrase gene and L. monocytogenes p60 promoter are shown in black. The multiple cloning site (MCS) is shown at the bottom of the plasmid, with unique restriction sites noted below in a box. pPL24 and pPL25 inserts are shown schematically below the multiple cloning site and were cloned as described in Materials and Methods. Final sizes of the plasmid constructs and the restriction sites used in cloning are noted for each of the inserts. (B) Plasmid map of the integration vector pPL2. The color scheme and genes are the same as in panel A except for the PSA integrase and PSA attPP′ sites, as noted. The multiple cloning site with 13 unique restriction sites is shown below the plasmid.
The multiple cloning site from pBluescript KS− (1) (bp 1 to 171) was cloned after PCR with primers 5′-GGACGTCATTAACCCTCACTAAAGG-3′ and 5′-GGACGTCAATACGACTCACTATAGG-3′. The low-copy-number gram-negative origin of replication and chloramphenicol acetyltransferase (CAT) gene from pACYC184 (5) (bp 172 to 2253) were cloned after PCR with primers 5′-GGACGTCGCTATTTAACGACCCTGC-3′ and 5′-GAGCTGCAGGAGAATTACAACTTATATCGTATGGGG-3′.
For direct conjugation from E. coli to L. monocytogenes, the RP4 origin of transfer (oriT) (37) (bp 2254 to 2624) was cloned from plasmid pCTC3 (53) after PCR with primers 5′-GCACTGCAGCCGCTTGCCCTCATCTGTTACGCC-3′ and 5′-CATGCATGCCTCTCGCCTGTCCCCTCAGTTCAG-3′. The listeriophage U153 integrase gene and attachment site (attPP′) (A. Nolte, P. Lauer, and R. Calendar, unpublished data; bp 2629 to 4127) that direct the site-specific integration of the plasmid were cloned after PCR with primers 5′-GTAGATCTTAACTTTCCATGCGAGAGGAG-3′ and 5′-GGGCATGCGATAAAAAGCAATCTATAGAAAAACAGG-3′.
For expression of the U153 integrase gene, the L. monocytogenes p60 promoter (25) (bp 4134 to 4563) was PCR amplified with primers 5′-CCTAAGCTTTCGATCATCATAATTCTGTC-3′ and and 5′-GGGCATGCAGATCTTTTTTTCAGAAAATCCCAGTACG-3′ and cloned upstream of the integrase gene. From bp 4570 to 6101 is a HindIII-AatII restriction fragment subcloned from pUC18-Cat (a kind gift from Nancy Freitag) and contains the inducible gram-positive CAT gene from pC194 (20) (bp 4788 to 5850).
Cloning of the hly and actA genes into pPL1.
The hly gene was subcloned from plasmid pDP-906 (22) by restriction digestion with BamHI and XbaI, gel purifying a 2.9-kb fragment, and ligating it into pPL1 cut with BamHI and SpeI. The resultant plasmid was designated pPL24 (Fig. 1A). The actA gene was PCR amplified from 10403S genomic DNA with primers 5′-GGTCTAGATCAAGCACATACCTAG-3′ and 5′-CGGGATCCTGAAGCTTGGGAAGCAG-3′. The 2220 bp PCR product was gel purified, cut with BamHI and XbaI, and cloned into pPL1 cut with BamHI and SpeI. The resultant plasmid was designated pPL25 (Fig. 1A).
Phage curing, conjugation, and molecular confirmation of plasmid integration.
Phage curing was accomplished by adapting historical methods (6, 45). L. monocytogenes 10403S derivatives carrying a prophage at comK-attBB′ (integrated in the comK open reading frame as previously described [29]) were grown in BHI at 37°C to 108 CFU/ml and infected with listeriophage U153 (19) at a multiplicity of infection of 20:1 in the presence of 5 mM CaCl2. Cultures were incubated with shaking at 37°C for 75 min, and inhibition of growth was monitored by comparison of the optical density at 600 nm of the infected culture with an uninfected control culture. The infected culture was diluted 10−2 and 10−4 in BHI, and both dilutions were grown at 37°C until the 10−2 dilution culture had increased 100-fold in optical density. The 10−4-fold dilution culture was then diluted 10−2, and 3 μl was plated on BHI.
Fifty colonies were tested for phage release initially by transferring colonies (using a toothpick) into 0.25 ml of LB broth and replica plating at 30°C on a lawn of Mack-4R (DP-L862) (for a list of strains, see Table 1), a nonlysogenic rough strain of L. monocytogenes particularly susceptible to forming plaques. Candidates that did not form plaques were then tested by spotting 10 μl of culture on a lawn of Mack-4R to detect plaque formation. If this second test was negative, whether the candidate could support plaque formation by the phage from the parent 10403S strain (φ10403 [19]) was tested. Curing was confirmed molecularly by PCR with the comK-attBB′-specific primer pair PL60 and PL61 (sequences follow) for the absence of a phage at comK-attBB′. Approximately 10% of colonies were cured by using this procedure.
TABLE 1.
Bacterial strains
| Strain | Relevant genotype or plasmid | Reference |
|---|---|---|
| E. coli | ||
| SM10 | Conjugation donor; F−thi-1 thr-1 leuB6 recA tonA21 lacY1 supE44 (MuC+) λ− [RP4-2(Tc::Mu)] Kmr Tra+ | 44 |
| XL1-Blue | Plasmid manipulations, recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| DP-E4067 | Integration vector, pPL1/SM10 | This study |
| DP-E4068 | hly integration vector, pPL24/SM10 | This study |
| DP-E4069 | actA integration vector, pPL25/SM10 | This study |
| DP-E4190 | Integration vector, pPL2/SM10 | This study |
| L. monocytogenes | ||
| 10403S | Wild type | 3 |
| DP-L4056 | 10403S, phage cured | This study |
| DP-L4027 | DP-L2161, phage cured, Δhly | This study (22) |
| DP-L4029 | DP-L3078, phage cured, ΔactA | This study (46) |
| DP-L4074 | DP-L4056 comK::pPL1 | This study |
| DP-L4075 | DP-L4027 Δhly comK::pPL24 | This study |
| DP-L4076 | DP-L4056 comK::pPL24 | This study |
| DP-L4077 | DP-L4029 ΔactA comK::pPL25 | This study |
| DP-L4078 | DP-L4056 comK::pPL25 | This study |
| SLCC-5764 | Virulence gene overexpresser (Mack, DP-L861) | 27 |
| DP-L862 | Mack-4R (SLCC-5764 rough isolate) | S. Katheriou |
| DP-L4082 | SLCC-5764, Strr derivative | This study |
| DP-L3780 | SLCC-5764 ΔactA (deletion of amino acids 7-633) | This study |
| DP-L4083 | DP-L3780, Strr derivative | This study |
| DP-L4084 | DP-L4082 comK::pPL1 | This study |
| DP-L4085 | DP-L4082 comK::pPL25 | This study |
| DP-L4086 | DP-L4083 comK::pPL1 | This study |
| DP-L4087 | DP-L4083 ΔactA comK::actA | This study |
| DP-L4088 | DP-L1169S 4b strain, Strr | This study |
| DP-L4089 | DP-L1172S 4b strain, Strr | This study |
| DP-L4090 | DP-L4088 comK::pPL1 | This study |
| DP-L4091 | DP-L4089 comK::pPL1 | This study |
| DP-L4199 | EGDe, Strr derivative | This study |
| DP-L4026 | WSLC 1042 (ATCC 23074) | 31 |
| DP-L4061 | WSLC 1042::PSA | This study |
| DP-L4221 | 10403S, tRNAArg::pPL2 | This study |
| DP-L4379 | LO28, Strr derivative | This study |
| L. innocua | ||
| DP-L4391 | CLIP 11262, Strr derivative | 15 |
Recipient strains of L. monocytogenes (SLCC-5764, DP-L1169, and DP-L1172) were made streptomycin resistant for counterselection in conjugation experiments by plate selection on BHI supplemented with 200 μg of antibiotic per ml.
pPL1 plasmid constructs were electroporated into E. coli strain SM10 (44) by standard techniques (41). Bacterial strains were grown to mid-log phase (optical density at 600 nm, ∼0.55) with shaking at 30°C. E. coli donor strains were grown in LB containing 25 μg of chloramphenicol/ml, and L. monocytogenes recipient strains were grown in BHI. Donor culture (2.5 ml) was mixed with 1.5 ml of recipient culture and filtered onto washed 0.45-μm-pore-size HA-type filters (47 mm; Millipore). The filter was washed once with 10 ml of BHI, transferred to a BHI plate with no antibiotics, and incubated for 2 h at 30°C. The bacterial cells were gently resuspended in 2.5 ml of BHI, and 25-μl and 50-μl aliquots were plated in 3 ml of LB top agar on BHI plates supplemented with 7.5 μg of chloramphenicol per ml and 200 μg of streptomycin per ml. Plates were incubated at 30°C overnight and shifted to 37°C for 2 to 3 days.
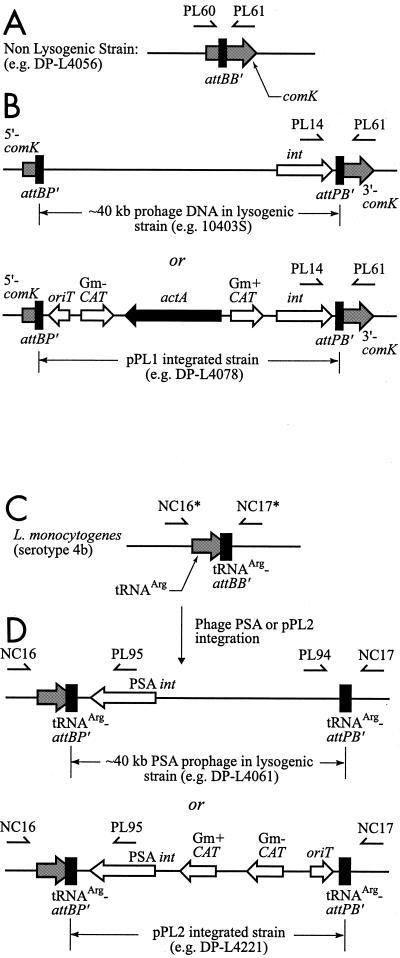
Individual colonies were picked and screened by PCR for integration at the phage attachment site with primers PL14 (5′-CTCATGAACTAGAAAAATGTGG-3′), PL60 (5′-TGAAGTAAACCCGCACACGATG-3′), and PL61 (5′-TGTAACATGGAGGTTCTGGCAATC-3′). PCRs were performed on small portions of individual bacterial colonies picked with sterile P200 pipette tips from BHI plates directly into 20-μl PCR mixtures. The primer pair PL14 and PL61 specifically amplifies attBP′ in a PCR, resulting in a 743-bp product on integrated strains (both prophage and pPL1 derivatives). The primer pair PL60 and PL61 specifically amplifies comK-attBB′ in a PCR, resulting in a 417-bp product only on nonlysogenic strains (i.e., DP-L4056). PCR assays were performed in a Hybaid Omn-E thermocycler with an annealing temperature of 55°C for 30 cycles. Integrants arose at a frequency of approximately 10−4 per donor cell.
Hemolysis on blood plates and hemolytic activity assay.
Hemolysis on blood plates was scored on tryptic soy agar plates supplemented with 5% defimbrinated sheep blood (HemoStat, Davis, Calif.). Hemolytic assays were performed essentially as previously described (40). Hemolytic activity is expressed as the reciprocal of the dilution of culture supernatant required to lyse 50% of sheep erythrocytes.
Plaquing in L2 cells.
Plaque sizes were determined as previously described (26). Each strain was plaqued in six to eight independent experiments and compared with 10403S in each experiment.
SDS-PAGE of surface-expressed ActA.
Surface-expressed ActA protein was prepared from late-log-phase bacterial cultures grown in LB broth (optical density at 600 nm, ∼0.7) by resuspending equivalent amounts in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer and boiling for 5 min, which extracts surface-expressed proteins but does not perturb the cell wall (4, 35). Equivalent amounts were loaded on SDS-7% PAGE gels and visualized with Coomassie blue.
Xenopus laevis cell extract motility assays.
X. laevis egg cytoplasmic extract was prepared as previosuly described (50) and supplemented with tetramethylrhodamine iodoacetamide-labeled actin (49). SLCC-5764-derived strains were grown overnight to stationary phase, washed, added to cell extracts, and incubated for 45 min before microscopic observation.
LD50 determinations.
Limited 50% lethal dose (LD50) determinations were performed in BALB/c mice as previously described (40). Animal experiments were performed in the laboratory of Archie Bouwer at Oregon Health Sciences Center, Portland, Oreg.
Identification of PSA attachment site and construction of pPL2.
The PSA attachment site (tRNAArg-attBB′) DNA sequence was obtained through a combination of inverse PCR and genome walking. Inverse PCR was performed on Sau3AI-digested DP-L4061 DNA (WSLC 1042, lysogenic for PSA) with the divergent primers PL95 (5′-ACATAATCAGTCCAAAGTAGATGC) and PL97 (5′-ACGAATGTAAATATTGAGCGG), which anneal within the PSA int gene. The resultant DNA sequence was used to design further oligonucleotides, and these were used with the Genome Walker kit (Clontech) per the manufacturer's recommendations. DNA sequencing and tRNA analysis were done with MacVector (Accelrys), DNAsis (Hitachi), the BLAST algorithm (2), and tRNAscan-SE (32).
pPL1 was modified to utilize a different attachment site on the L. monocytogenes chromosome by replacing the U153 integrase gene and attachment site in the plasmid. The PSA int and attPP′ were PCR amplified from PSA genomic DNA with primers PL100 (5′-GAAGATCTCCAAAAATAAACAGGTGGTGG) and PL101 (5′-CATGCATGCGTGGAGGGAAAGAAGAACGC) with Vent DNA polymerase, digested with BglII and SphI, and ligated to pPL1 that had been digested with the same enzymes. The resultant plasmid was designated pPL2 (Fig. 1B).
The DNA sequence of the PSA tRNAArg-attBB′ from serotype 1/2 L. monocytogenes strains was obtained by a plasmid trap strategy. DP-L4211 (pPL2 integrated in 10403S) genomic DNA was digested with Nsi I and NheI, which do not cleave in the vector, and ligated under dilute conditions to promote self-ligation. The ligations were transformed into E. coli XL1-Blue, and chloramphenicol-resistant colonies were selected. The plasmids obtained were sequenced with the convergent primers PL94 (5′-GGAGGGAAAGAAGAACGC) and PL95 (sequence above) for attPB′ and attBP′, respectively, which flank attPP′ in the PSA genomic DNA sequence. Further, because of the divergence between the sequences downstream of the tRNAArg gene among serotypes, a serotype 1/2-specific PCR assay across tRNAArg-attBB′ was developed from the 10403S DNA sequence and used to determine the prophage status of various L. monocytogenes strains. Primers PL102 (5′-TATCAGACCTAACCCAAACCTTCC) and PL103 (5′-AATCGCAAAATAAAAATCTTCTCG) specifically amplify a 533-bp PCR product in nonlysogenic serotype 1/2 strains. The primer pair NC16 (5′-GTCAAAACATACGCTCTTATC) and PL95 specifically amplify a 499-bp PCR product in strains that either are lysogenic or contain an integration vector at tRNAArg-attBB′.
Nucleotide sequence accession numbers.
DNA sequences described in this report have been deposited in the EMBL/GenBank/DDBJ databases under accession numbers AJ417488 (6,101-bp pPL1 shuttle integration vector); AJ417489 (3,897-bp HindIII fragment containing the listeriophage U153 integrase gene and attPP′); AJ314913 (2,072 bp surrounding WSLC 1042 tRNAArg-attBB′); AJ417448 (643 bp surrounding 10403S tRNAArg-attBB′); and AJ417449 (6,123-bp pPL2 shuttle integration vector).
RESULTS AND DISCUSSION
pPL1 forms stable, single-copy integrants in various L. monocytogenes strains.
pPL1 is the first shuttle integration vector that we constructed to facilitate strain construction in L. monocytogenes. In order to test the pPL1 vector, we needed an L. monocytogenes strain that lacked a phage at the comK bacterial attachment site. We adapted historical methods to cure L. monocytogenes strains of their prophages and found that after superinfection with phage U153, which has the same attachment site as the endogenous 10403S prophage, we were able to isolate prophage-free strains (see Materials and Methods). The prophage-cured 10403S strain was designated DP-L4056 and used in subsequent experiments.
We attempted both electroporation and conjugation to introduce pPL1 into L. monocytogenes. Despite repeated attempts to electroporate and directly select integrants, these experiments were unsuccessful, probably due to the low efficiency of transformation of L. monocytogenes (typically less than 102 colonies per μg of plasmid DNA for 10403S-derived strains). Much higher electroporation frequencies have been reported for different L. monocytogenes strains (38), and electroporation may be an effective method to introduce pPL1 into these strains. Conjugation of pPL1 from E. coli into L. monocytogenes, on the other hand, was successful. Drug-resistant transconjugants arose at a frequency as high as 1.1 × 10−4 per donor E. coli cell, compared to the frequency of 6.3 × 10−4 observed in our control conjugation with the autonomously replicating plasmid pWM401(oriT) (33). Therefore, the integration frequency is approximately 20% for the bacteria receiving pPL1 by conjugation. All chloramphenicol-resistant pPL1 transconjugants were positive with the PCR assay with primers PL14 and PL61 (Fig. 2B ) and negative with a PCR assay across attPP′ in pPL1 (PL14 paired with a primer in the RP4 oriT), indicating that they were true integrants. In addition, this experiment demonstrated that pPL1 was not maintained as an episomal plasmid and that the vector did not integrate as a concatemer (data not shown).
FIG. 2.
Genomic organization of the attachment sites within the comK gene (A and B) and the tRNAArg gene (C and D). (A) Nonlysogenic L. monocytogenes strain with an intact comK gene. Primers PL60 and PL61 amplify across the bacterial attachment site comK-attBB′. (B) Lysogenic L. monocytogenes strain (with approximately 40 kb of phage DNA inserted into the comK gene) or integrated strain (with pPL1 construct inserted into the comK gene). Primers PL14 and PL61 amplify across the hybrid attachment site comK-attPB′. (C) L. monocytogenes serotype 4b strain nonlysogenic at tRNAArg-attBB′. Primers NC16 and NC17 amplify across the bacterial attachment site tRNAArg-attBB′ in serotype 4b strains. Asterisk indicates that primers NC16 and NC17 are substituted with PL102 and PL103 to amplify across the bacterial attachment site tRNAArg-attBB′ in serotype 1/2 strains. (D) Lysogenic L. monocytogenes strain with approximately 40 kb of phage DNA or 6 kb of pPL2 vector DNA inserted at the 3′ end of the tRNAArg gene. Primers NC16 and PL95 amplify across the hybrid attachment site tRNAArg-attBP′ in both serotype 4b and 1/2 strains.
We tested the stability of the integrants under nonselective growth conditions. Three integrant strains, DP-L4074 and the merodiploid strains DP-L4076 and DP-L4078 (described in the following sections), were passed in liquid BHI medium for 100 generations and plated for single colonies. Ninety-six colonies were then exposed to 0.1 μg of chloramphenicol per ml (to induce CAT gene expression) and patched on plates containing 7.5 μg of antibiotic per ml. All colonies retained drug resistance. Thirty colonies from each nonselective growth experiment were assayed with the PL14/PL61 PCR assay, and all PCRs resulted in the 743-bp product, indicating that all transconjugants had retained the integrated plasmid.
We addressed whether the integration vector would be generally useful for any L. monocytogenes strain with an available attachment site. More than 320 listeriophages have been isolated (summarized in reference 30), and many have restricted host ranges (19). It was unclear whether there was a biological barrier to U153 integrase gene function in host strains that do not support U153 infection. We therefore picked three strains that did not contain a prophage at the comK attachment site, two serotype 4b clinical isolates and SLCC-5764, a serotype 1/2a strain that constitutively expresses the known virulence factors in an unregulated manner and has been useful for studying these virulence factors in vitro. Each of these strains was first made streptomycin resistant for counterselection in conjugation experiments (as described in Materials and Methods). Streptomycin-resistant derivatives were chosen on the basis of having the same growth rate as the parent strain to avoid experimental complications related to viability. The resultant strains, DP-L4088, DP-L4089, and DP-L4082, all proved to be suitable recipients for pPL1 integration at a frequency similar to that in DP-L4056.
A survey of L. monocytogenes isolates was conducted to identify suitable strains that do not harbor a prophage at the comK attachment site by using the PCR assays across comK-attBB′ (primers PL60/PL61) and the hybrid attPB′ (primers PL14/PL61). The results of these experiments (Table 2) indicated that many of the strains commonly used to study the biology and pathogenesis of L. monocytogenes, including 10403S, LO28, and EGDe, had a prophage at comK.
TABLE 2.
Prophage status of various L. monocytogenes strains at comK
| Strain | Description | Source | Serotype | PCR resulta
|
|
|---|---|---|---|---|---|
| PL60/PL61 comK | PL14/PL61 attPB′ | ||||
| 10403S | Wild type | Rabbit pellets | 1/2a | − | + |
| DP-L4056 | 10403S, phage cured | This work | 1/2a | + | − |
| DP-L861 | SLCC-5764 (Mack) | Wild type (overexpresser) | 1/2a | + | − |
| DP-L3818 | WSLC 1118::A118 | Camembert cheese | 4b | − | + |
| DP-L3633 | EGDe | Wild type (1960s, human) | 1/2a | − | + |
| DP-L3293 | LO28 | Wild type (clinical origin) | 1/2c | − | + |
| DP-L185 | F2397 | Los Angeles, Calif.; Jalisco cheese | 4b | + | − |
| DP-L186 | ScottA | Massachusetts outbreak, milk | 4b | + | − |
| DP-L188 | ATTC 19113 | Denmark, human | 3 | + | − |
| DP-L1168 | Clinical | Coleslaw | 4b | + | − |
| DP-L1169 | Clinical | Patient | 4b | + | − |
| DP-L1170 | Clinical | Patient | 4b | + | − |
| DP-L1171 | Clinical | Brie | 1/2b | + | − |
| DP-L1172 | Clinical | Alfalfa tablets | 4b | + | − |
| DP-L1173 | Clinical | Deceased patient | 4b | − | + |
| DP-L1174 | Clinical | Deceased patient | 4b | − | + |
| DP-L3809 | 1981 Halifax | Placenta | 4b | + | − |
| DP-L3810 | 1981 Halifax | Cerebrospinal fluid and brain | 4b | + | − |
| DP-L3812 | 1981 Halifax | Coleslaw | 4b | + | − |
| DP-L3813 | 1996 Halifax | Blood | ? | − | + |
| DP-L3814 | 1981 Halifax | Cerebrospinal fluid | 4b | + | − |
| DP-L3815 | 1993 Halifax | Cerebrospinal fluid | 1/2a | + | − |
| DP-L3816 | 1995 Halifax | Blood | ? | + | − |
| DP-L3817 | 1993 Halifax | Cerebrospinal fluid | 1/2a | + | − |
| DP-L3862 | 1998 Michigan | Patient | 4b | − | + |
−, negative PCR result for primer pair noted at top of column; +, positive PCR result for primer pair noted at top of column. The PL60/PL61 primer pair specifically amplify a 417-bp PCR product in nonlysogenic strains and result in no PCR product in lysogenic strains. The PL14/PL61 primer pair specifically amplify a 743-bp PCR product in lysogenic strains and result in no PCR product in nonlysogenic strains.
Status of comK did not affect the virulence of L. monocytogenes.
We next compared DP-L4056 and DP-L4074 to wild-type 10403S in standard virulence assays to determine if the presence of a prophage at comK, lack of prophage, or the integration vector altered the virulence phenotypes. These three strains were assayed for LLO activity, ability to form plaques in monolayers of L2 cells, and virulence in the mouse LD50 assay (Table 3). Within the confines of these experiments, the data are consistent with the integrity of the comK locus and with the presence of an integrated pPL1 vector not having an effect on the virulence of the bacterium.
TABLE 3.
Complementation of actA and hly at the comK phage attachment site
| Strain | Genotype | Hemolysis on blood plates | Hemolytic activitya (U) | Plaque sizeb % of wt (SD) | LD50c (CFU) |
|---|---|---|---|---|---|
| 10403S | Wild type | + | nd | 100 (na) | ∼2 × 104 |
| DP-L4056 | 10403S phage cured | + | 97 | 101 (1.4) | <1 × 105 |
| DP-L4074 | DP-L4056 comK::pPL1 | + | 98 | 99 (1.4) | <1 × 105 |
| DP-L4027 | DP-L2161 phage cured, Δhly | − | 0 | 0 (0) | 1 × 109d |
| DP-L4075 | DP-L4027 Δhly comK::pPL24 | + | 99 | 97 (3.9) | <1 × 105 |
| DP-L4076 | DP-L4056 comK::pPL24 | + | 198 | 96 (2) | nd |
| DP-L4029 | DP-L3078 phage cured, ΔactA | nd | nd | 0 (0) | 2 × 107d |
| DP-L4077 | DP-L4029 ΔactA comK::pPL25 | nd | nd | 86 (4) | <1 × 105 |
| DP-L4078 | DP-L4056 comK::pPL25 | nd | nd | 72 (6.8) | nd |
Hemolytic activity data shown are from one representative experiment. nd, not determined.
Plaque size is the average of 8 to 10 independent experiments and shown as a percentage of wild-type (wt) size (defined as 100%). Standard deviations are shown in parentheses. na, not applicable.
LD50s of 10403S and an hly mutant were determined previously (40) (the Δhly strain DP-L2161 was described previously [22]), and the LD50 of the ΔactA strain (DP-L1942, a smaller deletion within the actA open reading frame that does not support actin nucleation at the bacterial surface) was also determined previously (4).
LD50 data shown are for the non-phage-cured deletion strains (i.e., DP-L2161 and DP-L1942) and not for the phage-cured strains (i.e., DP-4027 and DP-L4029).
Full complementation of hly at the phage attachment site.
LLO, the gene product of hly, is a secreted pore-forming cytolysin that is responsible for escape from the membrane-bound vacuole when L. monocytogenes first enters a host cell (51). LLO is absolutely required for the intracellular life cycle of L. monocytogenes and for virulence (9, 14, 23). LLO activity can be measured by hemolytic activity on red blood cells (40). hly mutants fail to form plaques in monolayers of L2 cells (48) and are 100,000-fold less virulent in the mouse LD50 assay (40).
We cloned the hly structural gene into pPL1 and conjugated this plasmid from E. coli to the phage-cured wild-type strain and Δhly L. monocytogenes derivatives, resulting in DP-L4076 (an hly merodiploid) and DP-L4075 (hly only at the phage comK-att site). These strains were tested for hemolytic activity on blood plates, relative amount of hemolytic units secreted, ability to form a plaque in a monolayer of L2 cells, and virulence in the mouse LD50 assay (Table 3). The quantitative complementation of hly in the deletion strain background and the doubling of hemolytic units produced in the merodiploid strain indicate the following. First, gene expression is not de facto affected by ectopic expression at the comK chromosomal position. Second, the hly promoter is self-contained. Additionally, a twofold increase in the amount of LLO is not deleterious to the virulence and intracellular life cycle of L. monocytogenes, at least as measured by plaque formation.
Complementation of actA at the phage attachment site approaches wild-type expression.
ActA, a second major L. monocytogenes virulence factor, is responsible for commandeering host cell actin-cytoskeletal factors used for intracellular bacterial motility. ActA is also absolutely required for bacterial pathogenesis, as actA mutants are unable to spread from cell to cell or to form plaques in a cell monolayer (24) and are 1,000-fold less virulent than the wild type (4). Additionally, ActA expression appears to be more complex than that of LLO: two promoters drive actA expression. One is immediately upstream of the actA open reading frame, and the second is in front of the mpl gene, upstream of actA (52).
We constructed several strains to evaluate the complementation of actA at the phage attachment site. The first group included second-site-complemented (DP-L4077) and merodiploid (DP-L4078) strains in the 10403S background. These strains were assayed for plaque formation in an L2 monolayer (Table 3). Integrated ActA did not fully complement in this assay (plaque size of 86% of the wild-type size), and the merodiploid strain formed an even smaller plaque (72% of wild type). We interpret these results to indicate that the second promoter upstream of the mpl gene may make a small contribution to optimal actA expression, which is in agreement with a recent study that evaluated the relative contributions of the mpl and actA promoters both in broth culture and in vivo (43).
Previous research has indicated that there is a threshold level of ActA protein on the bacterial surface that is required for the initiation of intracellular motility (47). Although the level of ActA protein on the surface of intracellular bacteria was not quantitated in these experiments, since the level of LLO appears to scale linearly with copy number (Table 3), we presume that ActA will behave in a similar manner. Therefore, because the merodiploid actA strain makes a smaller plaque than the second-site complemented strain, we hypothesize that there is an upper critical concentration of ActA beyond which cell-to-cell spread is impaired, presumably because there is too much ActA on the bacterial surface for optimal motility. Alternatively, as the regulation of ActA appears to be highly complex in vivo, other explanations for the reduced plaque size are possible.
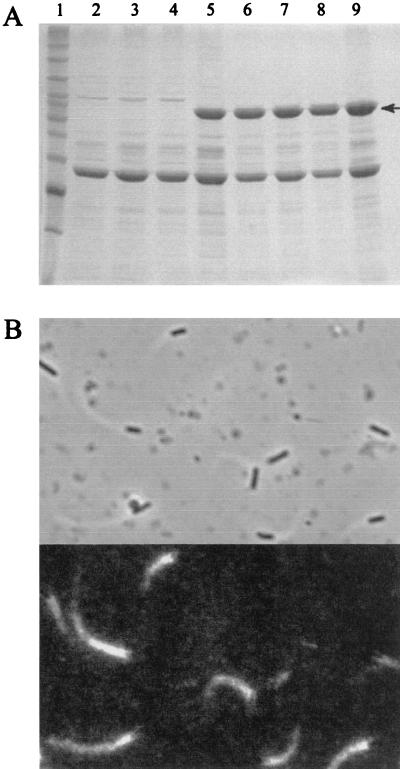
We further tested ActA complementation in the virulence gene-overexpressing strain SLCC-5764, a strain that is suitable for in vitro studies such as actin-based motility in Xenopus cell extracts but not for in vivo studies because the unregulated expression of ActA and other virulence factors causes it to grow poorly inside host cells and form small plaques (D. Portnoy, unpublished observations). ActA is effectively expressed in this strain from the comK-attBB′ site (Fig. 3A, lane 9). Considering the plaque formation data for 10403S-complemented actA strains, it might have been predicted that the merodiploid strain DP-L4085 would make more ActA than the parent strain. However, this was not observed; the parent strain, the complemented strain, and the merodiploid strain all expressed similar levels of ActA (Fig. 3A, lanes 5, 8, and 9). This observation was likely due to the complete lack of regulation and high level of constitutive expression of ActA in SLCC-5764. Additionally, DP-L4087 supported actin nucleation at the bacterial surface, actin tail formation, and bacterial motility in cell extracts (Fig. 3B).
FIG. 3.
Expression and functional complementation of ActA in SLCC-5764. (A) Coomassie blue-stained SDS-PAGE of SLCC-5764-derived strains grown to late log phase. ActA is indicated by an arrow. Lane 1, molecular size markers; lane 2, DP-L3780; lane 3, DP-L4083; lane 4, DP-L4086; lane 5, SLCC-5764; lane 6, DP-L4082; lane 7, DP-L4084; lane 8, DP-L4085; lane 9, DP-L4087. Strains are described in Table 1. (B) Actin tail formation and movement of DP-L4087 in Xenopus cell extracts. The top panel is a phase image; the bottom panel is a fluorescent image of the same field.
The results of these cell extract experiments indicate that the integration vector system for complementation will be useful for in vitro studies of L. monocytogenes motility, facilitating strain construction, and placing various molecular constructs in different host strains for study in a desired set of assays. In particular, several alleles of actA that have unusual motility phenotypes, as described recently (26), have been transferred to the SLCC-5764 ΔactA strain by using pPL1 and are currently being evaluated in cell extracts. The study of these mutants in the simplified cell extract system should yield insights into the activities of poorly understood regions of the ActA protein.
Phage PSA integrates into a tRNAArg gene and pPL2 construction.
pPL1 integration into L. monocytogenes strains that harbor a prophage in the comK attachment site is hindered by the process of first having to cure the prophage from the host strain. To alleviate the need for the phage-curing step, the specificity of pPL1 integration was changed to that of the PSA prophage. PSA (phage from ScottA) is the prophage of L. monocytogenes strain ScottA (30), a serotype 4b strain that was isolated during an epidemic of human listeriosis (12). Using the PSA genomic DNA sequence (E. Sattelberger, M. Zimmer, R. Calendar, R. B. Inman, S. Scherer, and M. J. Loessner, submitted for publication), we identified an integrase-like open reading frame with a contiguous noncoding sequence that we predicted to contain the attPP′ sequences (open reading frame 24 in accession number AJ312240). The PSA integrase sequence was then used to obtain the DNA sequence of PSA-attBB′ from the PSA lysogenic strain DP-L4061 (see Materials and Methods).
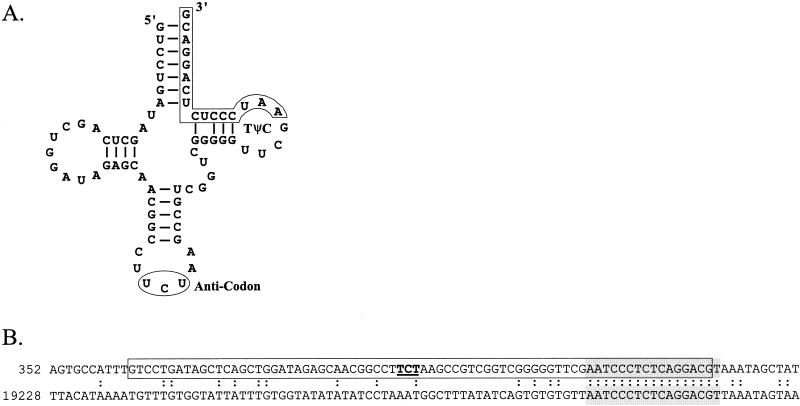
PSA was found to integrate in a tRNAArg gene that is 88% identical to a tRNAArg gene (trnSL-ARG2) from Bacillus subtilis. The anticodon of the tRNAArg gene is 5′UCU, the most commonly used arginine codon in L. monocytogenes. The PSA and bacterial attachment sites share 17 bp of DNA identity, and the tRNAArg-attBB′ contains a short nucleotide sequence that completes the tRNAArg sequence that is interrupted by integration of PSA (Fig. 4). The attachment site of the tRNAArg gene is present only once in the genome of L. monocytogenes strain EGDe (15) and apparently only once in the serotype 4b strain being sequenced by The Institute for Genomic Research and the U.S. Department of Agriculture (http://www.tigr.org/cgi-bin/BlastSearch/blast.cgi?organism=l_monocytogenes-4b). This indicates not only that the PSA integration site unique but also that precise reconstitution of the gene upon integration (or excision) is likely required for survival of the cell.
FIG. 4.
(A) Cloverleaf diagram of tRNAArg used as the PSA attachment site. The arginine anticodon is circled. The region with sequence identity between the tRNA gene and the PSA attPP′ is outlined. The boundaries of the tRNAArg gene and Cove score (82.37) were predicted with tRNAscan-SE (32). (B) Alignment of the tRNAArg-attBB′ region of L. monocytogenes WSLC 1042 (top line) and the attPP′ region of PSA downstream of the integrase gene. The 74-nucleotide tRNAArg gene of L. monocytogenes is boxed, and the 17-bp overlapping region of sequence identity (core integration site) is shaded grey. The tRNAArg gene anticodon is shown in bold and underlined. Identical nucleotide residues are indicated by a colon. The numbers located at the left indicate the nucleotide position in the DNA sequences of the WSLC 1042 attachment site (AJ314913) and PSA genome (AJ312240).
pPL2 was constructed by replacing the U153 listeriophage integrase gene and attachment site in pPL1 with the PSA listeriophage integrase gene and attachment site. pPL2 was transformed into SM10, and the resulting strain was mated into 10403S, EGDe (carrying a streptomycin resistance mutation), and the serotype 4b strain DP-L4088. Chloramphenicol-resistant transconjugants arose from each of these crosses at approximately 10−4 per donor cell, the same rate as pPL1 integration. Two recombinants from each cross were restreaked under drug selection and tested by PCR for the presence of PSA-attBP′ with primers NC16 and PL95 (Fig. 2D). The expected 499-bp PCR product was obtained in each of the colonies tested, indicating that pPL2 integrates into tRNAArg-attBB′ in both serotype 1/2 and 4b strains.
We tested the stability of the integrated pPL2 in both EGDe and DP-L4088 strains with the same nonselective 100-generation experiment described for pPL1. Forty-nine colonies from each of the amplified cultures were tested for chloramphenicol resistance. The EGDe-derived strains retained 100% drug-resistant colonies, indicating complete stability of the integrants. In the case of the DP-L4088 integrant, 2 of the 49 colonies were chloramphenicol sensitive, suggesting that a low level of excision can occur in this serotype 4b strain. In order to test whether precise excision had occurred, we PCR amplified across tRNAArg-attBB′ and sequenced the PCR products. The wild-type DNA sequence was obtained, indicating a precise excision event.
During the course of our PCR experiments, we noted a divergence between the tRNAArg-attPB′ sites from serotype 4b and serotype 1/2 L. monocytogenes strains. To determine nature of this divergence, we isolated and sequenced the tRNAArg-attBB′ site from 10403S (as described in Materials and Methods). We found that the sequence of attPB′ in 10403S (3′ of the tRNAArg gene) is unrelated to that of the serotype 4b strain WSLC 1042. In contrast to this, the sequence of attBP′ (5′ of the tRNAArg gene) in 10403S is 96 to 97% identical to the corresponding regions in L. monocytogenes serotype 4b strain WSLC 1042, the serotype 4b strain sequenced by The Institute for Genomic Research, and the serotype 1/2a strain EGDe sequenced by the European Listeria Consortium. Thus, the bacterial attBB′ sequences recognized by the PSA integrase are likely to lie entirely within the tRNA gene.
Additionally, we tested the availability of the tRNAArg-attBB′ in the common laboratory strains of L. monocytogenes with a PCR assay with primers PL102 and PL103. We found the tRNAArg attachment site to be vacant in strains 10403S and EGEe, indicating that pPL2 may be readily utilized in these backgrounds for strain construction, complementation, and genetic studies without concern for endogenous prophages. Further tests showed that it was not necessary to first check if the tRNAArg site was vacant, because pPL2 could integrate in a strain that already carried a prophage at the tRNAArg attachment site. Primers NC16 and PL95 (Fig. 2D) amplified a 499-bp PCR product from the serotype 1/2c strain LO28, indicating LO28 harbored a prophage at tRNAArg. Nevertheless, when pPL2 was conjugated into the LO28-derived strain DP-L4379, transconjugants arose with a frequency similar to that in strains without a prophage at tRNAArg. This result is quite reasonable: PSA integration into this attachment site reconstitutes a complete tRNAArg gene, allowing the possibility of a second, tandem integration event.
The DP-L4379 integrants were not as stable in the absence of selection as those derived from strains with a vacant attachment site, as described earlier in this report. Only 57 of 96 of the DP-L4379 integrants retained Cmr after 100 generations of growth in nonselective medium. For comparison, strains with a vacant attachment site resulted in 96 of 98 Cmr colonies. However, chloramphenicol selection can maintain the stability of the integrants, allowing strains such as LO28, which have a phage integrated at the tRNAArg-attBB′ attachment site, to be used for strain construction.
The related bacterial species Listeria innocua contains the tRNAArg attachment site DNA sequences (by BLAST search against L. innocua CLIP11262 genome sequence, accession number NC_003212, 99% identical to WSLC 1042 tRNAArg sequence), indicating that it could be a potential recipient for pPL2 integration as well. We have shown that L. innocua can serve as a recipient for pPL2 integration, with approximately fivefold-reduced efficiency of integrants arising from similar experiments already described. This observation may facilitate the study of the L. monocytogenes-specific virulence factor genes in isolation, such as actA and hly, as L. innocua does not harbor these genes (18).
Potential utility of pPL1 and pPL2.
The construction and characterization of the first single-step site-specific integration vectors for use in L. monocytogenes increase the genetic tools available for the study of this pathogen. These vectors will allow more facile strain construction than historical methods and are widely useful for various strains used to study the intracellular life cycle of L. monocytogenes. Additionally, stable merodiploid strains can be constructed to allow refined copy number studies and studies of interactions within a protein through multimerization and testing of the dominance or recessiveness of different alleles of a gene in the same bacterial strain. For instance, the dominant nature of an LLO mutation that makes LLO activity pH independent was recently evaluated in a merodiploid strain by using pPL1 (16). pPL2 has been used to evaluate the role of secA2 in the secretion of proteins and phase transition from rough to smooth colony morphology in L. monocytogenes (28).
pPL1 and pPL2 may also be useful for vaccine development. Several recombinant L. monocytogenes systems have been used to elicit cell-mediated immune responses to viral and cancer antigens in mice (13, 17, 21, 36, 42). One limitation with plasmid-based expression of recombinant proteins in L. monocytogenes is the stability of the plasmids in vivo (i.e., in the host animal) without selection (17, 21). Additionally, chromosomal construction of strains expressing foreign antigens is time-consuming (13, 42). pPL1 and pPL2 alleviate both of these concerns and hold promise to facilitate the construction of new generations of antiviral and anticancer vaccines.
Acknowledgments
We thank Audrey Nolte, Angéline Serre, Walter DeLaurentis, Susanne Rafelski, Ian Glomski, Justin Skoble, and Laurel Lenz for technical help, methods, reagents, and critical discussions during the course of this work. We are grateful to Sophia Katheriou for the kind gift of DP-L862, James Musser for the Halifax clinical isolates, Walter Schlech for serotype information on the Halifax strains, and Archie Bower for LD50 determinations of phage-cured and complemented strains. We thank Philippe Glaser and the European Listeria Consortium for providing sequences from L. monocytogenes strain EGDe prior to publication and The Institute for Genomic Research for making the preliminary sequence of the L. monocytogenes serotype 4b strain available to the public. We are grateful to Neil Fischer for his tremendous help with the preparation of the manuscript.
We are grateful to the Alexander von Humboldt Foundation for a Feodor-Lynen Research Fellowship to M.J.L. This work was supported by National Institutes of Health grants 1 R37 AI29619 and 1 R01 AI27655 to D.A.P.
REFERENCES
- 1.Alting-Mees, M. A., and J. M. Short. 1989. pBluescript II: gene mapping vectors. Nucleic Acids Res. 17:9494.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. [PubMed] [Google Scholar]
- 4.Brundage, R. A., G. A. Smith, A. Camilli, J. A. Theriot, and D. A. Portnoy. 1993. Expression and phosphorylation of the Listeria monocytogenes ActA protein in mammalian cells. Proc. Natl. Acad. Sci. USA 90:11890-11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, D. 1959. A variant of phage P2 originating in Escherichia coli, strain B. Virology 7:112-126. [DOI] [PubMed] [Google Scholar]
- 7.Cossart, P. 2000. Actin-based motility of pathogens: the Arp2/3 complex is a central player. Cell. Microbiol. 2:195-205. [DOI] [PubMed] [Google Scholar]
- 8.Cossart, P., and M. Lecuit. 1998. Interactions of Listeria monocytogenes with mammalian cells during entry and actin-based movement: bacterial factors, cellular ligands and signaling. EMBO J. 17:3797-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cossart, P., M. F. Vicente, J. Mengaud, F. Baquero, J. C. Perez-Diaz, and P. Berche. 1989. Listeriolysin O is essential for virulence of Listeria monocytogenes: direct evidence obtained by gene complementation. Infect. Immun. 57:3629-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domann, E., J. Wehland, M. Rohde, S. Pistor, M. Hartl, W. Goebel, M. Leimeister-Wachter, M. Wuenscher, and T. Chakraborty. 1992. A novel bacterial virulence gene in Listeria monocytogenes required for host cell microfilament interaction with homology to the proline-rich region of vinculin. EMBO J. 11:1981-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleming, D. W., S. L. Cochi, K. L. MacDonald, J. Brondum, P. S. Hayes, B. D. Plikaytis, M. B. Holmes, A. Audurier, C. V. Broome, and A. L. Reingold. 1985. Pasteurized milk as a vehicle of infection in an outbreak of listeriosis. N. Engl. J. Med. 312:404-407. [DOI] [PubMed] [Google Scholar]
- 13.Frankel, F. R., S. Hegde, J. Lieberman, and Y. Paterson. 1995. Induction of cell-mediated immune responses to human immunodeficiency virus type 1 Gag protein by using Listeria monocytogenes as a live vaccine vector. J. Immunol. 155:4775-4782. [PubMed] [Google Scholar]
- 14.Gaillard, J. L., P. Berche, and P. Sansonetti. 1986. Transposon mutagenesis as a tool to study the role of hemolysin in the virulence of Listeria monocytogenes. Infect. Immun. 52:50-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 16.Glomski, I. J., M. M. Gedde, A. W. Tsang, J. A. Swanson, and D. A. Portnoy. 2002. The L. monocytogenes hemolysin has an acidic pH optimum to compartmentalize activity and prevent damage to infected host cells. J. Cell Biol. 156:1029-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goossens, P. L., G. Milon, P. Cossart, and M. F. Saron. 1995. Attenuated Listeria monocytogenes as a live vector for induction of CD8+ T cells in vivo: a study with the nucleoprotein of the lymphocytic choriomeningitis virus. Int. Immunol. 7:797-805. [DOI] [PubMed] [Google Scholar]
- 18.Gouin, E., J. Mengaud, and P. Cossart. 1994. The virulence gene cluster of Listeria monocytogenes is also present in Listeria ivanovii, an animal pathogen, and Listeria seeligeri, a nonpathogenic species. Infect. Immun. 62:3550-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodgson, D. A. 2000. Generalized transduction of serotype 1/2 and serotype 4b strains of Listeria monocytogenes. Mol. Microbiol. 35:312-323. [DOI] [PubMed] [Google Scholar]
- 20.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol. 150:815-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikonomidis, G., Y. Paterson, F. J. Kos, and D. A. Portnoy. 1994. Delivery of a viral antigen to the class I processing and presentation pathway by Listeria monocytogenes. J. Exp. Med. 180:2209-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, S., and D. A. Portnoy. 1994. Characterization of Listeria monocytogenes pathogenesis in a strain expressing perfringolysin O in place of listeriolysin O. Infect. Immun. 62:5608-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kathariou, S., P. Metz, H. Hof, and W. Goebel. 1987. Tn916-induced mutations in the hemolysin determinant affecting virulence of Listeria monocytogenes. J. Bacteriol. 169:1291-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kocks, C., E. Gouin, M. Tabouret, P. Berche, H. Ohayon, and P. Cossart. 1992. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell 68:521-531. [DOI] [PubMed] [Google Scholar]
- 25.Kohler, S., M. Leimeister-Wachter, T. Chakraborty, F. Lottspeich, and W. Goebel. 1990. The gene coding for protein p60 of Listeria monocytogenes and its use as a specific probe for Listeria monocytogenes. Infect. Immun. 58:1943-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lauer, P., J. A. Theriot, J. Skoble, M. D. Welch, and D. A. Portnoy. 2001. Systematic mutational analysis of the amino-terminal domain of the Listeria monocytogenes ActA protein reveals novel functions in actin-based motility. Mol. Microbiol. 42:1163-1177. [DOI] [PubMed] [Google Scholar]
- 27.Leimeister-Wachter, M., W. Goebel, and T. Chakraborty. 1989. Mutations affecting hemolysin production in Listeria monocytogenes located outside the listeriolysin gene. FEMS Microbiol. Lett. 53:23-29. [DOI] [PubMed] [Google Scholar]
- 28.Lenz, L. L., and D. A. Portnoy. Identification of a second Listeria secA gene that promotes protein secretion and is associated with the rough phenotype. Mol. Microbiol., in press. [DOI] [PubMed]
- 29.Loessner, M. J., R. B. Inman, P. Lauer, and R. Calendar. 2000. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol. Microbiol. 35:324-340. [DOI] [PubMed] [Google Scholar]
- 30.Loessner, M. J., I. B. Krause, T. Henle, and S. Scherer. 1994. Structural proteins and DNA characteristics of 14 Listeria typing bacteriophages. J. Gen. Virol. 75:701-710. [DOI] [PubMed] [Google Scholar]
- 31.Loessner, M. J., G. Wendlinger, and S. Scherer. 1995. Heterogeneous endolysins in Listeria monocytogenes bacteriophages: a new class of enzymes and evidence for conserved holin genes within the siphoviral lysis cassettes. Mol. Microbiol. 16:1231-1241. [DOI] [PubMed] [Google Scholar]
- 32.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moors, M. A., V. Auerbuch, and D. A. Portnoy. 1999. Stability of the Listeria monocytogenes ActA protein in mammalian cells is regulated by the N-end rule pathway. Cell. Microbiol. 1:249-257. [DOI] [PubMed] [Google Scholar]
- 34.Mounier, J., A. Ryter, M. Coquis-Rondon, and P. J. Sansonetti. 1990. Intracellular and cell-to-cell spread of Listeria monocytogenes involves interaction with F-actin in the enterocytelike cell line Caco-2. Infect. Immun. 58:1048-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mourrain, P., I. Lasa, A. Gautreau, E. Gouin, A. Pugsley, and P. Cossart. 1997. ActA is a dimer. Proc. Natl. Acad. Sci. USA 94:10034-10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan, Z. K., G. Ikonomidis, A. Lazenby, D. Pardoll, and Y. Paterson. 1995. A recombinant Listeria monocytogenes vaccine expressing a model tumour antigen protects mice against lethal tumour cell challenge and causes regression of established tumours. Nat. Med. 1:471-477. [DOI] [PubMed] [Google Scholar]
- 37.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 38.Park, S. F., and G. S. Stewart. 1990. High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene 94:129-132. [DOI] [PubMed] [Google Scholar]
- 39.Pistor, S., L. Grobe, A. S. Sechi, E. Domann, B. Gerstel, L. M. Machesky, T. Chakraborty, and J. Wehland. 2000. Mutations of arginine residues within the 146-KKRRK-150 motif of the ActA protein of Listeria monocytogenes abolish intracellular motility by interfering with the recruitment of the Arp2/3 complex. J. Cell Sci. 113:3277-3287. [DOI] [PubMed] [Google Scholar]
- 40.Portnoy, D. A., P. S. Jacks, and D. J. Hinrichs. 1988. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J. Exp. Med. 167:1459-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., T. Maniatis, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Shen, H., M. K. Slifka, M. Matloubian, E. R. Jensen, R. Ahmed, and J. F. Miller. 1995. Recombinant Listeria monocytogenes as a live vaccine vehicle for the induction of protective antiviral cell-mediated immunity. Proc. Natl. Acad. Sci. USA 92:3987-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shetron-Rama, L. M., H. Marquis, H. G. Bouwer, and N. E. Freitag. 2002. Intracellular induction of Listeria monocytogenes actA expression. Infect. Immun. 70:1087-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 45.Six, E. 1960. Prophage substitution and curing in lysogenic cells superinfected with hetero-immune phage. J. Bacteriol. 80:728-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skoble, J., D. A. Portnoy, and M. D. Welch. 2000. Three regions within ActA promote Arp2/3 complex-mediated actin nucleation and Listeria monocytogenes motility. J. Cell Biol. 150:527-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, G. A., J. A. Theriot, and D. A. Portnoy. 1996. The tandem repeat domain in the Listeria monocytogenes ActA protein controls the rate of actin-based motility, the percentage of moving bacteria, and the localization of vasodilator-stimulated phosphoprotein and profilin. J. Cell Biol. 135:647-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun, A. N., A. Camilli, and D. A. Portnoy. 1990. Isolation of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect. Immun. 58:3770-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Theriot, J. A., and D. C. Fung. 1998. Listeria monocytogenes-based assays for actin assembly factors. Methods Enzymol. 298:114-122. [DOI] [PubMed] [Google Scholar]
- 50.Theriot, J. A., J. Rosenblatt, D. A. Portnoy, P. J. Goldschmidt-Clermont, and T. J. Mitchison. 1994. Involvement of profilin in the actin-based motility of L. monocytogenes in cells and in cell-free extracts. Cell 76:505-517. [DOI] [PubMed] [Google Scholar]
- 51.Tilney, L. G., and D. A. Portnoy. 1989. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite Listeria monocytogenes. J. Cell Biol. 109:1597-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vazquez-Boland, J. A., C. Kocks, S. Dramsi, H. Ohayon, C. Geoffroy, J. Mengaud, and P. Cossart. 1992. Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect. Immun. 60:219-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams, D. R., D. I. Young, and M. Young. 1990. Conjugative plasmid transfer from Escherichia coli to Clostridium acetobutylicum. J. Gen. Microbiol. 136:819-826. [DOI] [PubMed] [Google Scholar]