Abstract
Screening of random fragments of Escherichia coli genomic DNA for their ability to increase the novobiocin resistance of a hypersusceptible ΔacrAB mutant resulted in the isolation of a plasmid containing baeR, which codes for the response regulator of the two-component regulatory system BaeSR. When induced for expression, baeR cloned in multicopy plasmid pTrc99A significantly increased the resistance of the ΔacrAB host strain to novobiocin (16-fold) and to deoxycholate (8-fold). Incubation of cells with novobiocin followed by a chromatographic assay for intracellular drug showed that overproduced BaeR decreased drastically the drug accumulation, presumably via increased active efflux. The genes baeSR are part of a putative operon, yegMNOB baeSR. Direct binding of BaeR to the yegM promoter was demonstrated in vitro by gel retardation assay. The gene yegB, which codes for a major facilitator superfamily transporter, was not necessary for increased resistance, but deletion of yegO or an in-frame deletion of yegN, both of which code for resistance-nodulation-cell division-type multidrug transporters, abolished the BaeR-induced increase in resistance. It is likely that both YegN and YegO produce a complex(es) with the membrane fusion protein family member YegM and pump out novobiocin and deoxycholate. We accordingly propose to rename yegMNOB as mdtABCD (mdt for multidrug transporter). Finally, the expression of two other genes, yicO and ygcL, was shown to be regulated by BaeR, but it is not known if they play any roles in resistance.
Multidrug transporters are a large and diverse group of proteins capable of protecting cells against a wide variety of environmental toxins by active extrusion of noxious compounds. Multidrug transporters in bacteria are classified into five families based on sequence similarity. These families are the major facilitator (MFS), resistance-nodulation-cell division (RND), small multidrug resistance, multidrug and toxic compound extrusion, and ATP-binding cassette families (5, 30). Many putative and proven drug transporters of all five families exist in the Escherichia coli genome (31), and a large fraction of them are multidrug transporters (30). Since the spectra of substrates of many of these multidrug transporters overlap (25), it is intriguing why bacteria, with their economically organized genomes, harbor such a large set of multidrug efflux genes.
The key to understanding how bacteria utilize these multiple transporters lies in the analysis of the regulation of transporter expression. The data available today show that multidrug transporters are often expressed under precise and elaborate control at the level of transcription. Expression of many transporters increases following exposure to various environmental toxins and inducing compounds (1, 14, 17). Thus, in E. coli, the repressor EmrR derepresses transcription of the multidrug efflux genes emrAB in response to a variety of chemicals, including EmrAB substrates (4, 17, 42). Expression of the major E. coli efflux pump AcrAB is subject to multiple levels of regulation. First, it is modulated by the local repressor AcrR (20). At the level of global regulation, acrAB transcription is activated by MarA (3, 29) in response to exposure to various chemicals, such as salicylate (2, 7). Another global regulator, SoxS, also activates expression of acrAB and a group of other genes in response to oxidative stress (10, 41). Both MarA and SoxS belong to the XylS/AraC family of transcription regulators (6), and yet another member of this family, RobA, also increases acrAB expression (37, 41). Interestingly, expression of acrAB and three other E. coli loci implicated in multidrug resistance, tolC, acrEF, and acrD, was recently found to be activated by SdiA (39), an E. coli homologue of the LuxR quorum-sensing regulator. SdiA acts as a positive regulator of genes involved in septation, motility, and chemotaxis (36, 38, 39), suggesting a link between these cellular processes and multidrug resistance.
However, not much data is available on the regulation of E. coli multidrug transporter genes other than acrAB and emrAB. In this study, we report positive regulation of the recently identified multidrug resistance locus yegMNOB (26) by a response regulator, BaeR, of the putative BaeSR two-component signal transduction system (24), a phenomenon that leads to increased resistance to novobiocin and deoxycholate.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains used were derivatives of E. coli K-12 (Table 1). Cells were grown at 37°C unless specified otherwise, with shaking at 200 rpm in Luria-Bertani (LB) broth containing 10 g of Bacto Tryptone, 10 g of Bacto Yeast Extract, and 5 g of NaCl per liter. The antibiotics chloramphenicol (25 μg/ml) and ampicillin (100 μg/ml) were used for the selection of plasmid-containing cells.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic or genotype | Reference(s) or source | |
|---|---|---|---|
| E. coli strains | |||
| AG100 | K-12 argE3 thi-1 rpsL xyl mtl Δ(gal-uvrB) supE44 | 13, 29 | |
| AG100A | Same as AG100; ΔacrAB::Tn903 Kanr | 29 | |
| N8453 | F− ΔlacU169 rpsL Δmar rob::kan sox::cam | J. L. Rosner | |
| WZM120 | F− ΔlacZ39 rpsL45 or rpsL110 melB4 ΔacrAB::Tn903 Kanr | 21 | |
| LMG194 | F− ΔlacX74 galE thi rpsL ΔphoA (PvuII) Δara714 leu::Tn10 | Invitrogen | |
| AG100A ΔbaeSR::cat | This study | ||
| AG100A ΔyegB::cat | This study | ||
| AG100A ΔyegOB::cat | This study | ||
| AG100A ΔyegN ΔyegB::cat | This study | ||
| AG100A ΔyegMNOB::cat | This study | ||
| Plasmids | |||
| pTrc99A | Ampr, multicopy cloning vector for gene expression under IPTG-inducible Ptrc promoter | Pharmacia Biotech | |
| pNN387 | Cmr, single-copy vector containing promoterless lacZY | 12 | |
| pBAD/Myc-His A | Ampr, cloning vector for dosage-dependent expression under arabinose-inducible PBAD promoter | Invitrogen |
Enzymes.
Restriction endonucleases and DNA-modifying enzymes were purchased from commercial sources and used as recommended by the suppliers. PCR was performed by using the PLATINUM Taq DNA polymerase High Fidelity (Invitrogen/Life Technologies, Carlsbad, Calif.), which allows for high-fidelity amplification of long PCR products.
Screening for positive regulators of multidrug resistance.
DNA manipulation generally followed standard practice (35). A genomic library was made by partial DNase I digestion of the chromosomal DNA. Fragments approximately 0.5 to 3 kb in size were purified from low-melting-point agarose and treated with T4 DNA polymerase and Klenow fragment to create blunt ends. The fragments were ligated into the SmaI site of vector pTrc99A, which contains an IPTG (isopropyl-β-d-thiogalactoside)-inducible Ptrc promoter. The ligation products were used to transform the acrAB deletion strain AG100A. Cells were plated on LB agar medium containing 100 μg of ampicillin/ml, 100 μM IPTG, and inhibitory concentrations of various drugs.
DNA sequencing.
Automated sequencing of the samples was performed at the DNA Sequencing Facility at the University of California, Berkeley.
Drug susceptibility measurements.
The MICs of toxic compounds were determined by serial twofold dilutions in LB medium containing 100 μg of ampicillin/ml and, when appropriate, 0.02% l-arabinose or 100 to 200 μM IPTG. Exponential-phase cells (5 × 105 per ml) were used as inocula. After overnight incubation at 37°C, cell growth was examined visually.
Assay of novobiocin accumulation.
Cells were grown at 37°C in LB medium supplemented with 100 μg of ampicillin/ml to an optical density at 600 nm (OD600) of approximately 0.4, at which time IPTG was added to a final concentration of 1 mM and incubation was continued for 1.5 h longer. Cells were collected by centrifugation and resuspended in a buffer containing 50 mM potassium phosphate buffer (pH 7.0), 1 mM MgSO4, and 0.2% glucose, so that the cell density was eight times that of the original culture. The cell suspension was divided into two portions. Novobiocin was added to one of them to a final concentration of 10 μg/ml; the other one served as a control. The cells were incubated at 37°C for 20 min; 2-ml samples were then taken, and cells were collected by filtration through 0.22-μm-pore-size Millipore filters and washed with 1 ml of the buffer used for resuspension. Collected cells were resuspended in 0.4 ml of H2O and disrupted by sonication. Total protein content in the samples was determined with a BCA protein assay kit (Pierce, Rockford, Ill.) by using the protocol supplied by the manufacturer. The concentration of the samples was adjusted to 7.5 mg of total protein/ml with water, and then 3 volumes of 0.2 M NH4H2PO4 and 4 volumes of methanol were added. The samples were incubated at room temperature for 5 min with occasional mixing and were centrifuged in a tabletop centrifuge at 13,000 rpm. One milliliter of the supernatants was used to determine the novobiocin content by reversed-phase chromatography (33) by using a Supercosil LC-18-DB column (Supelco, Inc., Bellefonte, Pa.) with a Shimadzu model SPD-M6A high-performance liquid chromatography (HPLC) apparatus. The initial mobile phase was 5 mM phosphoric acid and methanol at a ratio of 50:50 (vol/vol). Novobiocin was eluted with a linear gradient to 5 mM phosphoric acid, methanol, and acetonitrile at a ratio of 20:0:80 (vol/vol/vol), with detection at 340 nm and authentic novobiocin samples used as a control.
Gene disruption with the phage λ Red system.
Gene disruption was performed according to the method of Datsenko and Wanner (9), with recombination between short homologous DNA regions catalyzed by the phage λ Red recombinase. A curable expression plasmid encoding Red recombinase (pKD46) was introduced into strain AG100A. The chloramphenicol resistance gene cat, flanked by Flp recognition target sites, was amplified by PCR with primers with 40-nucleotide extensions that were homologous to the beginning and end of the coding sequence of the gene to be disrupted. Plasmid pKD3 was used as a template. This PCR product was used to transform the recipient AG100A strain expressing Red recombinase, and recombinant clones were isolated as chloramphenicol-resistant colonies. pKD46 vector was eliminated by incubation at a nonpermissive temperature of 37°C, as confirmed by the loss of resistance to ampicillin. Chromosomal DNA was isolated from the mutants obtained, and the structure of the rearranged loci was confirmed by using a series of PCRs with primers complementary to cat and to the regions adjacent to the site of its insertion.
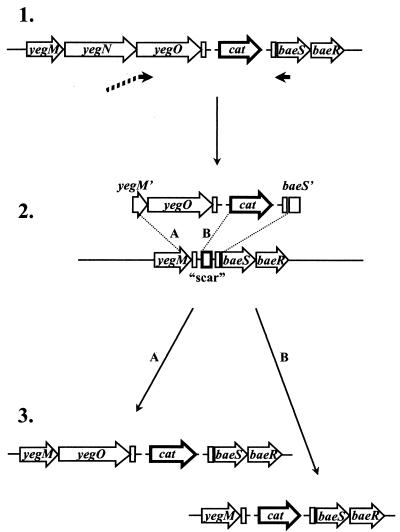
Construction of the AG100A ΔyegN ΔyegB::cat strain.
First, the strains AG100A ΔyegB::cat and AG100A ΔyegNOB::cat (Fig. 1), in which cat is substituted for the coding sequences of the deleted genes, were created by using the Red recombinase-based approach outlined above. The cat gene was eliminated from the AG100A ΔyegNOB::cat strain according to the method of Datsenko and Wanner (9), by introducing a helper plasmid expressing Flp recombinase, which acts on the Flp recognition target sites flanking cat in this strain. The Flp helper plasmid was cured by incubation at a nonpermissive temperature of 43°C. The loss of cat and of the Flp helper plasmid was confirmed by the loss of ampicillin and chloramphenicol resistance. The expected genomic rearrangements in the resulting strain, AG100A ΔyegNOB, were confirmed by using a series of PCRs with primers surrounding the site of cat excision. The strain retained a short “scar” sequence originating from pKD3 vector at the site of cat excision (for the predicted scar sequence, see reference 9). A hybrid primer, which contained a sequence corresponding to the last 45 nucleotides of yegM and the ATG start codon of yegN (which overlaps, by one nucleotide, the yegM stop codon TGA), followed by the sequence corresponding to nucleotides 4 to 21 of yegO, was used with a primer annealing within the baeS gene in a PCR with chromosomal DNA from the AG100A ΔyegB::cat strain as a template (Fig. 2). The resulting PCR product was introduced into the AG100A ΔyegNOB strain transformed with pKD46 as a substrate for Red-assisted recombination, and chloramphenicol-resistant mutants were selected. Two alternative recombination events can lead to the insertion of cat into the chromosome of the AG100A ΔyegNOB strain, one within yegM sequences and the other within pKD3-derived sequences surrounding cat and retained within the scar region in AG100A ΔyegNOB. Chloramphenicol-resistant mutants were screened by using a series of PCRs to select clones in which recombination occurred within the yegM gene. In the isolated strain, AG100A ΔyegN ΔyegB::cat, the coding sequence of yegN gene is deleted starting with nucleotide 4, and the coding sequence of yegO starting with the second codon is merged with the remaining ATG start codon of yegN. To confirm the structure of this junction, the region was amplified by PCR and sequenced.
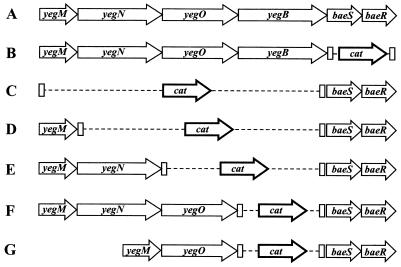
FIG. 1.
Organization of the yegMNOB baeSR locus in wild-type E. coli (A) and in the strains AG100A ΔbaeSR::cat (B), AG100A ΔyegMNOB::cat (C), AG100A ΔyegMNOB::cat (D), AG100A ΔyegOB::cat (E), AG100A ΔyegB::cat (F), and AG100A ΔyegN ΔyegB::cat (G). Note that we propose to rename the locus yegMNOB as mdtABCD (see Results).
FIG. 2.
Construction of the strain AG100A ΔyegN ΔyegB::cat. (1) Genomic DNA from the strain AG100A ΔyegOB::cat is used as a template in a PCR with a hybrid primer whose sequence at the 5′ end corresponds to the last 45 nucleotides of yegM followed by nucleotides 2 to 21 of yegO. (2) The product of the PCR described above is introduced into strain AG100A ΔyegNOB expressing Red recombinase. Two possible recombination events, labeled A and B, can lead to insertion of the cat gene into the genome. (3) Chloramphenicol-resistant clones are screened by PCR to select mutants in which recombination occurred according to mechanism A, resulting in the insertion of the yegO gene next to the yegM gene. Note that we propose to rename the locus yegMNOB as mdtABCD (see Results).
β-Galactosidase assay.
To test the effect of overexpressed BaeR on the transcription of various reporter constructs, cells were grown at 37°C in LB medium supplemented with 100 μg of ampicillin/ml and 25 μg of chloramphenicol/ml. When cultures reached an OD600 of 0.4 to 0.6, they were divided into two halves and l-arabinose was added to one of them to 0.2%, leaving the other half as a control. Incubation was continued for 3 h longer, and β-galactosidase activity in the cells was assayed by using o-nitrophenyl-β-d-galactopyranoside (ONPG) as a substrate essentially as described by Miller (22) with the following modifications. Cells from 0.25- or 0.5-ml samples were collected by centrifugation, resuspended in 0.8 ml of Z-buffer (22), and permeabilized in the presence of chloroform and sodium dodecyl sulfate (SDS) as recommended by Miller (22). The samples were preincubated at 30°C for 15 min, and 160 μl of a solution containing 4 mg of ONPG/ml in Z-buffer was added. Reactions were allowed to proceed at 30°C for 15 to 60 min and were stopped by the addition of 0.4 ml of 1 M Na2CO3. After centrifugation, the absorbance of the supernatants at 420 nm was measured, and the specific activity of β-galactosidase was calculated as follows: 1,000 × A420/(time of reaction × OD600 of cells × sample volume), with the time of reaction measured in minutes and the sample volume measured in milliliters.
Expression and purification of His6-tagged BaeR.
The baeR gene was amplified from K-12 genomic DNA by using primers that introduced BspHI and SalI sites at the ends of the amplified fragment. This PCR product was cloned between the NcoI and SalI sites of the expression vector pBAD/Myc-HisA (Invitrogen Corporation, Carlsbad, Calif.), resulting in the merging of the initiator ATG of the baeR gene with the initiator ATG of the vector and an in-frame addition of eight extra codons to the last codon of the cloned fragment: GTC GAC (CAT)6, which encodes the peptide Val Asp His6. The insertion in the resulting plasmid, pBADBaeR-His, was sequenced to confirm the fidelity of the PCR. In the pBAD/Myc-HisA vector, heterologous gene expression is directed by the araBAD promoter, which is subject to positive control by l-arabinose and negative control by d-glucose. Resulting constructs were transformed into E. coli LMG194 (Invitrogen), and BaeR-His6 was purified on Ni2+-nitrilotriacetic acid Superflow resin (Qiagen, Valencia, Calif.) as follows. Cells were grown at 30°C in 80 ml of LB medium containing 100 μg of ampicillin/ml to an OD600 of 0.7, at which time l-arabinose was added to 0.0002% and incubation continued for 6 h. Cells were harvested by centrifugation, resuspended in 1 ml of the binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-Cl [pH 7.9]) containing 1 mg of lysozyme/ml, left on ice for 30 min, and disrupted by sonication. The lysate was clarified by centrifugation at 17,000 × g for 30 min at 4°C, and 2.5 ml of the 50% slurry of the Superflow resin in the binding buffer was added. The mixture was left for 1 h at 4°C with slow stirring and then poured into a column, which was washed with 7 ml of binding buffer followed by 10 ml of wash buffer (60 mM imidazole, 0.5 M NaCl, 20 mM Tris-Cl [pH 7.9]). BaeR-His6 was eluted from the column with 8 ml of a linear 60 mM-to-2 M gradient of imidazole in 0.5 M NaCl-20 mM Tris-Cl (pH 7.9). Fractions were analyzed by SDS-polyacrylamide gel electrophoresis, and those containing at least 95% pure protein were combined and dialyzed overnight against two changes of 20 mM Tris-HCl-200 mM NaCl (pH 7.4). The concentration of the protein was determined with a BCA protein assay kit. Glycerol was added to 15%, and the protein was stored in aliquots at −70°C.
Electrophoretic mobility shift assay.
DNA probes were generated by PCR by using K-12 genomic DNA as a template. Fragments were end labeled with T4 polynucleotide kinase and [γ-32P]ATP (5,000 Ci/mmol; Amersham Pharmacia Biotech, Piscataway, N.J.) and purified by using a Qiaquick nucleotide removal kit (Qiagen). Labeled fragments (20 fmol) were combined with 0 to 26.4 pmol of purified BaeR-His6 in 20-μl reaction volumes containing 10 mM Tris-Cl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 2.5% glycerol, 0.05% NP-40, and 50 ng of poly(dI-dC)/μl. After incubation for 25 min at room temperature, 5 μl of a 5× sample loading solution (50% glycerol, 0.025% bromophenol blue) was added and the mixtures were loaded directly onto a prerun 4% polyacrylamide slab gel. Electrophoresis was conducted at 4°C in 0.04 M Tris-0.02 M acetic acid buffer. The gel was dried and exposed with X-ray film to visualize bands.
Search for BaeR-regulated genes in the E. coli genome.
The chromosomal DNA of the K-12 strain of E. coli was digested with either RsaI or HaeIII. The fragments were ligated with pNN387 reporter vector that was digested with HindIII and end filled by using Klenow fragment. The ligation products were used to transform WZM120 cells harboring pTrcBaeR. Transformants were plated onto LB agar plates containing 100 μg of ampicillin/ml, 25 μg of chloramphenicol/ml, 60 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)/ml, and 0.2 mM IPTG to induce the expression of BaeR. Blue colonies were restreaked on duplicate plates with the same medium, one containing IPTG and the other not containing IPTG. Plasmids were isolated from the colonies in which the production of β-galactosidase was either enhanced or decreased in the presence of IPTG. To retest the selected reporter library plasmids in a fresh genetic background, the obtained plasmid preparations were used to transform LMG194 cells carrying pBADBaeR expression vector or pBAD/Myc-HisA as a control. The transformants obtained were used to test the changes in β-galactosidase activity in response to the induction of BaeR synthesis with 0.2% arabinose.
RESULTS
Overexpression of BaeR increases resistance to novobiocin and deoxycholate.
Expression of multidrug efflux genes in E. coli is often regulated in a complex manner, as described in the introduction. We therefore carried out screens of the E. coli genomic library for genes that increased multidrug resistance levels of this organism. Our preliminary effort with an acrAB+ host strain, however, predominantly yielded already known transcriptional activators of the AcrAB transporter, such as marA, soxS, and sdiA (data not shown).
We therefore used a host strain lacking functional acrAB genes in the screening in order to identify regulatory elements involved in the expression of other multidrug resistance systems. Furthermore, the library was made from the chromosomal DNA of strain N8453, in which the well-known global regulatory genes marA, rob, and soxS were deleted or incapacitated. Random fragments of the chromosomal DNA were cloned into the SmaI site of vector pTrc99A, which contains an IPTG-inducible Ptrc promoter, as described in Materials and Methods. The recombinant plasmids were transformed into the acrAB deletion strain AG100A, and the transformants were plated on LB agar containing 100 μg of ampicillin/ml, 100 μM IPTG, and inhibitory concentrations of various drugs. In one experiment, the medium contained 5 μg of novobiocin/ml, which had an MIC of 1.25 μg/ml against AG100A. When one of the transformant colonies that grew up on this medium was purified and reexamined in the presence and absence of IPTG, we indeed found 8- to 16-fold increased novobiocin MICs against it in the presence of IPTG (data not shown). Introduction of the plasmid isolated from this strain (the plasmid is called pTrcBaeR on the basis of results described below) into fresh AG100A cells resulted in the same phenotype of novobiocin resistance: MICs increased 4- and 16-fold over those against the recipient strain in the absence and presence of IPTG, respectively (Table 2). However, when pTrcBaeR was used to transform AG100 cells, which express the AcrAB pump, no effect on novobiocin resistance was observed (Table 2), presumably because AG100 already was highly resistant to this drug, whose MIC against it was 160 μg/ml. pTrcBaeR was also found to increase the resistance of both AG100A and AG100 cells to deoxycholate about eightfold. A variety of other drugs, including common substrates of multidrug transporters, such as norfloxacin, chloramphenicol, tetracycline, erythromycin, acriflavin, gentamicin, amikacin, carbonyl cyanide m-chlorophenylhydrazone, nalidixic acid, rifampin, SDS, and ethidium bromide, were tested, but no change in resistance levels was detected.
TABLE 2.
Effect of pTrcBaeR on novobiocin and deoxycholate resistance of wild-type, acrAB, and acrAB yegMNOB strains
| Strain/plasmid | Novobiocin MIC (μg/ml)
|
Deoxycholate MIC (mg/ml)
|
||
|---|---|---|---|---|
| No IPTG | 200 μM IPTG | No IPTG | 200 μM IPTG | |
| AG100/pTrc99A | 160 | 160 | 1.5 | 1.5 |
| AG100/pTrcBaeR | 160 | 160 | 1.5 | 12.5 |
| AG100A/pTrc99A | 1.25 | 1.25 | 1.5 | 1.5 |
| AG100A/pTrcBaeR | 5 | 20 | 1.5 | 12.5 |
| AG100A ΔyegMNOB::cat/ pTrc99A | 0.63-1.25 | 0.63-1.25 | 1.5 | 1.5 |
| AG100A ΔyegMNOB::cat/ pTrcBaeR | 0.63-1.25 | 0.63-1.25 | 1.5 | 1.5 |
Sequencing of the plasmid revealed an insertion containing most of the coding sequence of the gene baeR, along with the 113-bp upstream sequence from the initiation codon ATG. The last 8 codons of baeR (Glu-Ala-Asp-Ala-Cys-Arg-Ile-Val) were replaced by 17 vector-derived codons (Gly-Ser-Ser-Arg-Val-Asp-Leu-Gln-Ala-Cys-Lys-Leu-Gly-Cys-Phe-Gly-Gly) inthis construct. BaeR is the response regulator of a putative two-component system, which also includes the sensor kinase BaeS, encoded by a gene located upstream of baeR (24). It seemed likely that in cells carrying pTrcBaeR, overexpressed BaeR was causing the transcriptional activation of genes involved in multidrug resistance.
To test whether the alteration of the carboxyl-terminal sequence of baeR affects the regulatory activity of the protein, the full-length wild-type baeR was amplified by PCR and then cloned in pBAD/Myc-His A vector under control of the arabinose-inducible PBAD promoter and vector-encoded translational signals to obtain pBADBaeR, which was then introduced into the ΔacrAB strain WZM120. Novobiocin MICs for these cells were 8 times higher (16 versus 2 μg/ml) and deoxycholate MICs were 8 to 16 times higher for cells grown in the presence of 0.2% arabinose than for those grown without arabinose or for control cells carrying empty pBAD/Myc-His A vector (with or without arabinose), a result suggesting that the intact BaeR produced by this plasmid and the C-terminally-altered BaeR produced by pTrcBaeR had comparable activities. Furthermore, pBADBaeR did not increase the MICs of various other agents, including nalidixic acid, norfloxacin, and SDS, against WZM120, even in the presence of arabinose (data not shown). Many experiments in this study were therefore carried out with pTrcBaeR.
Effect of BaeR on novobiocin accumulation.
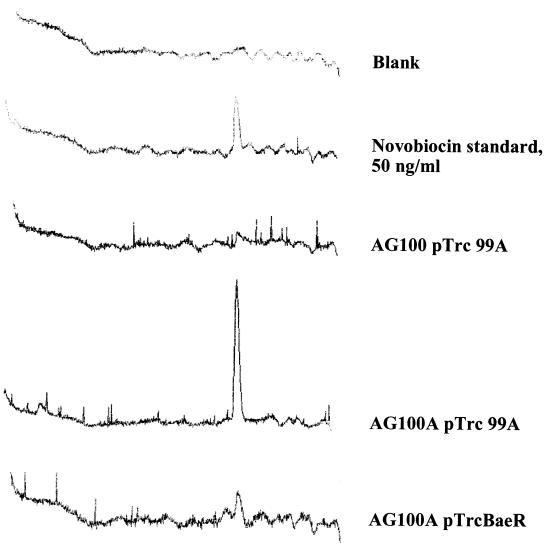
Two major mechanisms of resistance to novobiocin are modifications of the novobiocin target enzyme DNA gyrase and drug efflux. To test whether efflux is responsible for resistance conferred by pTrcBaeR, we assayed the levels of novobiocin accumulation in the cells, as described in Materials and Methods. In brief, cells were exposed to 10 μg of novobiocin/ml for 20 min and cell extracts were analyzed for novobiocin by HPLC (Fig. 3). Novobiocin levels in wild-type AG100 cells were almost undetectable (Fig. 3). In contrast, AG100A cells, which do not express the AcrAB pump, showed much higher accumulation of the drug (Fig. 3), as expected from our knowledge (21) that novobiocin is a good substrate for the AcrAB efflux transporter. The introduction of pTrcBaeR into AG100A cells significantly reduced the accumulation of novobiocin (Fig. 3), indicating that BaeR-induced resistance is caused by drug efflux. The plasmid pTrcBaeR, however, did not decrease the novobiocin accumulation level down to the levels observed for the acrAB+ AG100 cells, a finding consistent with the intermediate levels of novobiocin resistance seen for AG100A containing this plasmid (Table 2).
FIG. 3.
Novobiocin accumulation is reduced in acrAB cells overexpressing BaeR. Mid-exponential-phase cells were induced with 1 mM IPTG for 1.5 h, concentrated 8 times in a buffer containing 50 mM potassium phosphate buffer (pH 7.0), 1 mM MgSO4, and 0.2% glucose, and exposed to 10 μg of novobiocin/ml for 20 min. Cells were disrupted, and novobiocin levels were determined by HPLC. Samples contained 940 μg of total protein in a 1-ml volume. A novobiocin standard (50 ng) was used for reference.
Putative sensor kinase BaeS is not essential for the effect of BaeR.
Bacterial response regulators are often inactive or have low activity in their basal, nonphosphorylated state and are activated via phosphorylation of a specific Asp residue by their corresponding sensor kinases, which are activated by a specific signal. To test whether the putative sensor kinase BaeS is required for the activity of the overexpressed BaeR, we disrupted the baeS and baeR genes in strain AG100A by using the λ Red system (8), as described in Materials and Methods. The structure of the altered baeS-baeR loci in the mutant was confirmed by using a series of PCRs with primers complementary to cat and to the adjacent regions. In the mutant strain AG100A ΔbaeSR::cat, the cat gene substituted for the majority of the coding sequences of the two genes, leaving the first 53 bp of baeS and the last 81 bp of baeR (Fig. 1). The orientation of the cat gene coincided with the direction of transcription of the baeS and baeR genes. Introduction of pTrcBaeR into the AG100A ΔbaeSR::cat strain and the AG100A strain resulted in identical increases in novobiocin and deoxycholate MICs after IPTG induction (Table 3). Thus, the presence of baeS does not seem to be required for the activity of overexpressed BaeR in these cells.
TABLE 3.
Effects of deletions within the yegMNOB baeSR locus on the drug resistance conferred by pTrcBaeRa
| Strain/plasmid | Novobiocin MIC (μg/ml) | Deoxycholate MIC (mg/ml) |
|---|---|---|
| AG100A/pTrc99A | 3.1 | 3.1 |
| AG100A/pTrcBaeR | 25 | 25 |
| AG100A ΔbaeSR::cat/pTrc99A | 3.1 | 3.1 |
| AG100A ΔbaeSR::cat/pTrcBaeR | 25 | 25 |
| AG100A ΔyegB::cat/pTrc99A | 3.1 | 3.1 |
| AG100A ΔyegB::cat/pTrcBaeR | 25 | 25 |
| AG100A ΔyegOB::cat/pTrc99A | 1.6 | 1.6 |
| AG100A ΔyegOB::cat/pTrcBaeR | 1.6 | 1.6 |
| AG100A ΔyegN ΔyegB::cat/pTrc99A | 1.6 | 1.6 |
| AG100A ΔyegN ΔyegB::cat/pTrcBaeR | 1.6 | 1.6 |
All MIC tests were performed in the presence of 100 μM IPTG.
Increase in novobiocin and deoxycholate resistance requires the yegMNOB gene cluster.
A group of four genes—yegM, yegN, yegO, and yegB—is located immediately upstream of baeS and may be cotranscribed with baeS and baeR (see Discussion). These genes encode a putative membrane fusion protein (yegM), two RND-type transporters (yegN and yegO), and an MFS-type transporter (yegB). The locus has been shown to encode a multidrug efflux system (26): when the yegMNOB genes were introduced into a ΔacrAB mutant on a multicopy vector, the resistance of the cells to novobiocin increased 16 times and the resistance to deoxycholate increased at least 32 times. To test whether these genes were responsible for the drug resistance induced by BaeR, we created an AG100A-derived gene knockout strain, AG100A ΔyegMNOB::cat, in which cat is substituted for the yegM, yegN, yegO, and yegB genes by the λ Red-based system described in Materials and Methods. In this strain, the deletion starts 40 bp downstream of the yegM start codon and ends 170 bp upstream of the yegB stop codon, and the orientation of cat coincides with the direction of transcription of the yegMNOB locus (Fig. 1), as revealed by a series of PCR amplifications of areas around this locus. This disruption of the yegMNOB locus completely obliterated the effect of BaeR on novobiocin and deoxycholate resistance (Table 2).
To examine the role of each of the three putative transporters within this locus—yegN, yegO, and yegB—in BaeR-induced drug resistance, we created knockout strains AG100A ΔyegB::cat and AG100A ΔyegOB::cat by using the same approach based on the λ Red system as outlined above. In these strains, DNA segments starting at nucleotides +47 of yegB and +43 of yegO, respectively, and extending to the same position as the deletion in the AG100A ΔyegMNOB::cat strain were deleted and replaced by the cat gene (Fig. 1). The knockout strains were transformed with pTrcBaeR or an empty vector for control, and the MICs of novobiocin and deoxycholate were determined in the presence of 100 μM IPTG (Table 3). Disruption of the yegB gene alone did not affect the resistance of the cells to these two agents. In contrast, disruption of yegO completely abolished the increase in both novobiocin and deoxycholate MICs caused by BaeR (Table 3).
While the integrity of yegO was clearly crucial for BaeR-induced drug resistance, it was not clear whether yegN played any part in it. To examine whether yegN was necessary for the effect of BaeR, we created the AG100A ΔyegN ΔyegB::cat strain, in which all of the yegN coding sequence except the start codon was deleted (see Materials and Methods) and the coding sequence of yegO, starting with codon 2, was fused in frame with the initiation ATG codon of yegN. The strain also contained a deletion of yegB (Fig. 2). The pTrcBaeR plasmid introduced into AG100A ΔyegN ΔyegB::cat cells had no effect on its drug resistance (Table 3). This result suggests that both the yegO and yegN transporters are needed for BaeR-induced drug resistance, although we have no experimental proof for the successful expression of YegO in this strain. On the basis of these results, we propose to rename yegMNOB as mdtABCD (mdt for multidrug transporter).
The overexpression of mdtABCD genes in a multicopy vector was reported by Nishino and Yamaguchi (26) to increase resistance not only to norfloxacin and deoxycholate but also to nalidixic acid and norfloxacin (twofold) and to SDS (fourfold). In our experiments, in which only the activator BaeR and not its putative target, MdtABCD, was overexpressed, there was no detectable change in resistance to these latter compounds. Also, although the increase in novobiocin resistance induced by BaeR in our experiments was similar to the increase caused by MdtABCD overexpression observed by Nishino and Yamaguchi (26), resistance to deoxycholate in cells overexpressing only BaeR went up only eightfold.
BaeR activates transcription from the mdtA (yegM) promoter.
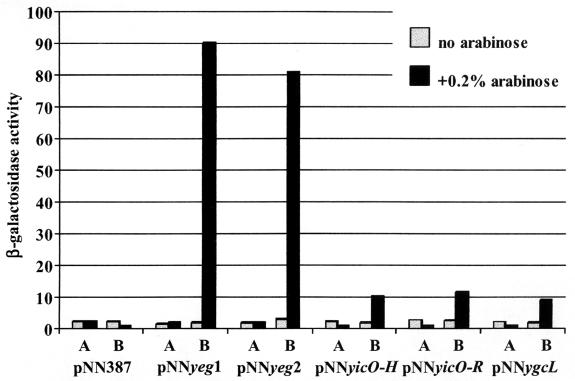
Our data suggested that BaeR activated transcription of the mdtABCD genes. To demonstrate this, we cloned a genomic fragment spanning nucleotides −632 to +163 relative to the mdtA start codon in front of the lacZ reporter gene in a single-copy pNN387 vector. The resulting pNNyeg1 plasmid was introduced into LMG194 cells containing either pBADBaeR or, as a control, pBAD/Myc-His A vector. When cells carrying pBADBaeR and pNNyeg1 were incubated with 0.2% l-arabinose for 4 h to induce production of BaeR, the specific β-galactosidase activity in the culture increased approximately 45 times compared to that of the cells grown without arabinose (Fig. 4). No increase in activity was observed in control cells. We were able to further narrow down the location of BaeR-responsive transcriptional elements to the DNA region including the 275 bp immediately upstream of the mdtA start codon. Plasmid pNNyeg2, which contained this DNA fragment, was assayed as described above and produced a similar increase in β-galactosidase activity in response to the induction of BaeR synthesis (Fig. 4). It was therefore likely that all of the elements necessary for BaeR-inducible transcription of mdtA were located within this 275-bp fragment. This fragment was further used to investigate the interaction between BaeR and the mdtA promoter in vitro in the experiments described below.
FIG. 4.
Effect of BaeR on the activity of different reporter constructs. LMG194 cells combining the indicated reporter plasmids with either pBAD/Myc-His A (bars labeled A) or pBADBaeR (bars labeled B) were grown in LB medium at 37°C to the mid-exponential phase. l-Arabinose (0.2%) was added to one-half of each culture, and β-galactosidase activity was measured 3 h later.
BaeR binds the mdtA (yegM) promoter region in vitro.
To facilitate the purification of BaeR for use in the in vitro experiments, we constructed pBADBaeR-His plasmid and expressed the protein with a hexahistidine affinity tag (His6) at its carboxy terminus. We chose to introduce the His tag at the protein's carboxy terminus because of the observation that alteration of the BaeR sequence at this position in pTrcBaeR vector did not affect its biological activity. To test the activity of BaeR-His6 in vivo, we introduced pBADBaeR-His into WZM120. Growing these cells with 0.2% arabinose to induce BaeR-His6 expression increased the MIC of novobiocin against them eightfold, from 2 to 16 μg/ml. Thus, the His6 tag does not appear to interfere with the biological function of BaeR.
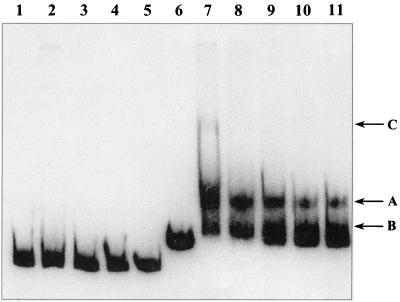
BaeR-His6 was purified by nickel chelate affinity chromatography to essential homogeneity, as revealed by SDS-polyacrylamide gel electrophoresis (see Materials and Methods). Electrophoretic mobility shift assays were performed with purified BaeR-His6 and a 275-bp DNA fragment upstream of mdtA mentioned above. As a control, a 242-bp fragment including the acrA promoter was used. Assays were performed in the presence of a 250-fold excess of a nonspecific competitor, poly(dI-dC). At all protein concentrations tested, we observed the formation of a discrete complex between BaeR-His6 and the mdtA promoter that remained stable during electrophoresis (Fig. 5, arrow A, lanes 7 to 11). In the presence of increasing concentrations of BaeR-His6, more probe apparently became associated with the protein in a complex that was unstable during electrophoresis, as indicated by the smearing and the slight upward shift of the lowest migrating band (Fig. 5, arrow B, lanes 7 to 11). At the highest tested concentration of BaeR-His6 (1.32 μM), the formation of a high-molecular-weight complex could be observed (Fig. 5, arrow C, lane 7). No interaction between the control fragment and BaeR could be detected (Fig. 5). This confirms that complexes formed by BaeR-His6 and the mdtA promoter in this experiment are specific.
FIG. 5.
Electrophoretic mobility shift assay of the interaction of BaeR-His6 with the acrA (lanes 1 to 5) and yegM (mdtA) (lanes 6 to 11) promoter regions. 32P-labeled DNA fragments were present at 1 nM in reaction mixtures that contained no BaeR-His6 (lanes 1 and 6), 16 nM BaeR-His6 (lane 11), 49 nM BaeR-His6 (lanes 5 and 10), 0.15 μM BaeR-His6 (lanes 4 and 9), 0.44 μM BaeR-His6 (lanes 3 and 8), and 1.32 μM BaeR-His6 (lanes 2 and 7). A 250-fold excess (50 μg/ml) of unlabeled competitor [poly(dI-dC)] was included in all reaction mixtures. Arrows indicate the positions of migration of discrete complexes formed between BaeR-His6 and the probe containing the yegM (mdtA) promoter region.
BaeR activates promoters of the yicO and ygcL genes.
Many of the two-component regulatory systems in E. coli control the expression of multiple genes and operons. To test the possibility that BaeR may be involved in the regulation of other promoters in the E. coli genome, we cloned a library of short genomic fragments into the pNN387 transcription reporter vector and examined whether promoters in these fragments responded to the overproduction of BaeR (see Materials and Methods). From this screening, we isolated two reporter constructs with inserts including the upstream region of the gene yicO. One of these constructs (pNNyicO-H) contained an HaeIII fragment spanning nucleotides −324 to +982 relative to the yicO translation start site, and the other (pNNyicO-R) had an RsaI fragment including nucleotides −611 to +132 of yicO. β-Galactosidase activity, expressed from the lacZ gene in pNN387, increased four to five times in response to the induction of BaeR with 0.2% arabinose in the cells doubly transformed with pBADBaeR and either pNNyicO-H or pNNyicO-R (Fig. 4). Another reporter construct isolated in these screening experiments contained a 785-bp HaeIII fragment including a 649-bp upstream sequence of the gene ygcL and the first 136 bp of its coding sequence. This construct was named pNNygcL. The activity of β-galactosidase in LMG194 cells containing both pBADBaeR and pNNygcL increased 4.5 times following incubation with 0.2% arabinose for 3 h (Fig. 4).
DISCUSSION
In this study, we made a genome-wide search for a regulator of multidrug resistance in E. coli by random shotgun cloning and discovered BaeR, which upregulated the locus mdtABCD and thereby increased resistance to novobiocin and deoxycholate. BaeR exhibits sequence similarity to the response regulators and is likely to constitute a two-component regulatory system with a sensor kinase, BaeS. Genes baeS and baeR were first identified in a random screening for the two-component signal transduction genes in E. coli based on their ability to phenotypically suppress mutational lesions of the sensor kinase genes envZ and phoR/creC (24). BaeR and BaeS were shown to exhibit the in vitro phosphotransfer reaction in the presence of ATP (24). Genes baeS and baeR are found immediately downstream of the mdtABCD (yegMNOB) genes (Fig. 1), and in this cluster of six genes, the start codons of every subsequent gene are immediately adjacent to or overlap the stop codon of the preceding gene. Thus, these genes probably form an operon. We found that an overexpression of BaeR strongly stimulated promoter activity of the 275-bp fragment immediately upstream of mdtA. This fragment was also shown to specifically interact with purified BaeR in vitro. BaeR thus directly binds to and stimulates the transcriptional activity of the mdtA promoter.
Typically in two-component systems, a specific signal binds to the sensor kinase, which undergoes autophosphorylation at a specific histidine residue. This phosphoryl group is then transferred to an aspartate residue of the cognate response regulator (16), resulting in its activation. At this time, we do not know what stimuli activate the BaeS-BaeR system. We have tested subinhibitory concentrations of novobiocin and deoxycholate, substrates of the MdtABC-mediated transport, for the ability to stimulate transcription from the mdtA promoter in wild-type cells, with negative results (data not shown). Interestingly, we found that the presence of the sensor kinase BaeS was not required for the full activity of the overexpressed BaeR in intact cells. Possibly, the overexpressed BaeR is phosphorylated by other sensor kinases present in E. coli; such cross talks occur, especially when one of the noncognate partners is present in excess (32, 40). Also, some sensor kinases may be involved in physiological cross-communication. For example, ArcB can efficiently phosphorylate the chemotaxis protein CheY and the osmoregulator OmpR in addition to its cognate effector ArcA (32). Alternatively, even low levels of activity of nonphosphorylated BaeR might be sufficient when the protein is present in large excess.
The observed interaction between the mdtA promoter and BaeR in vitro (Fig. 5) most likely involved unphosphorylated protein, as response regulators usually possess autophosphatase activity that limits the lifetime of the phosphorylated states (40). Interaction of other unphosphorylated response regulators with their cognate promoters has been observed in vitro, with phosphorylation increasing the affinity of binding (8, 11, 15).
The mdtABCD multidrug resistance cluster is unusual in that it encodes three different transporters. Genes mdtB and mdtC code for RND-type transporters, and mdtD encodes a transporter of the MFS type. We found that mdtD was not involved in novobiocin and deoxycholate resistance, since the resistance of the cells overexpressing BaeR was not compromised by the deletion of this gene. This is in agreement with the finding of Nishino and Yamaguchi (26) that expression of MdtD (YegB) alone does not confer drug resistance on the cells. Deletion of both mdtC and mdtD completely abolished the elevation of drug resistance by BaeR, demonstrating that mdtC plays a crucial role in this resistance. This cluster, however, contains another RND-type pump gene, mdtB. Therefore, we constructed strain AG100A ΔyegN (mdtB) ΔyegB (mdtD)::cat, in which genes mdtB and mdtD were both disrupted while the mdtA gene was left intact and the complete coding sequence of the gene mdtC, starting with the second codon, was fused to the remaining start codon of mdtB, putting mdtC under the control of mdtB translational signals (Fig. 2). Overexpression of BaeR in this strain failed to induce either deoxycholate or novobiocin resistance. Although we do not yet have a proof of MdtC expression in this strain, these results suggest that both MdtB and MdtC transporters are required to produce a fully functional multidrug transporter. In this connection, we note that Nishino and Yamaguchi (26) found that overexpression of MdtC alone conferred 4-fold greater resistance to deoxycholate (compared to a 32-fold increase in MIC produced by the mdtABCD overexpression plasmid) but no increase in novobiocin resistance. Our AG100A ΔyegN ΔyegB::cat strain was expected similarly to produce MdtC but not MdtB, yet BaeR overexpression in this strain did not result in increased deoxycholate resistance. This difference may be due to the different levels of MdtC present in the cells, the protein overexpressed from a multicopy vector in the Nishino and Yamaguchi study. Since MdtB and MdtC are RND-type transporters, they likely function in a complex with a membrane fusion protein, probably MdtA; the effect of nonpolar mdtA disruption remains to be tested.
Another two-component system, EvgSA, is known to affect the multidrug efflux phenotype of E. coli through the regulation of the YhiUV transporter (27, 28). The present result thus represents the second example of such control of multidrug efflux processes by two-component systems.
Many of the two-component signal transduction systems in E. coli control the expression of multiple target genes (11, 18, 19). We have found evidence that the BaeS-BaeR system is also involved in the regulation of more than one operon. In addition to the mdtA promoter, we found two other genes—yicO and ygcL—whose transcription is stimulated by the overexpression of BaeR. Neither the yicO nor the ygcL gene product has been characterized. The gene ygcL is likely to be the first gene of an operon of unknown function which also includes ygcK, ygcJ, ygcI, ygcH, ygbT, and ygbF. YicO exhibits sequence similarity to the members of Nucleobase:cation symporter family 2 (TC: 2.A.40 of Saier et al. [34]), which includes bacterial xanthine and uracil permeases. A divergently transcribed gene, yicP, found upstream of yicO is predicted by homology to be a probable adenine deaminase involved in the synthesis of xanthine.
The requirement for two RND-type transporters was unexpected, because chemical cross-linking gave no evidence for a possible oligomeric nature of the RND transporter AcrB (43); this requirement warrants further biochemical studies. Further investigation of both the function of the BaeR-induced genes yicO and ygcL and the nature of the signal that activates BaeS is needed in order to understand the biological significance of the BaeS-BaeR signal transduction system and may provide further insights into the role of multidrug transporters in the physiology of the cell.
ADDENDUM
After the completion of this study, we learned that a similar set of results was obtained by Nagakubo and coworkers (23).
Acknowledgments
This study was supported in part by a research grant from the U.S. Public Health Service (AI-09644).
We thank Xian-Zhi Li for a useful suggestion and A. Yamaguchi for communicating the results of his group before publication.
REFERENCES
- 1.Ahmed, M., C. M. Borsch, S. S. Taylor, N. Vazquez-Laslop, and A. A. Neyfakh. 1994. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J. Biol. Chem. 269:28506-28513. [PubMed] [Google Scholar]
- 2.Alekshun, M. A., and S. B. Levy. 1999. Alteration of the repression activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J. Bacteriol. 181:4669-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbosa, T. M., and S. B. Levy. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182:3467-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooun, A., J. J. Tomashek, and K. Lewis. 1999. Purification and ligand binding of EmrR, a regulator of a multidrug transporter. J. Bacteriol. 181:5131-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, M. H., I. T. Paulsen, and R. A. Skurray. 1999. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol. Microbiol. 31:394-395. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, S. P., H. Hachler, and S. B. Levy. 1993. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 175:1484-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, S. P., S. B. Levy, J. Foulds, and J. L. Rosner. 1993. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J. Bacteriol. 175:7856-7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahl, J. L., B.-Y. Wei, and R. J. Kadner. 1997. Protein phosphorylation affects binding of the Escherichia coli transcription activator UhpA to the uhpT promoter. J. Biol. Chem. 272:1910-1919. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demple, B. 1996. Redox signalling and gene control in the Escherichia coli soxRS oxidative stress regulon—a review. Gene 179:53-57. [DOI] [PubMed] [Google Scholar]
- 11.Drapal, N., and G. Sawers. 1995. Purification of ArcA and analysis of its interaction with the pfl promoter-regulatory region. Mol. Microbiol. 16:597-607. [DOI] [PubMed] [Google Scholar]
- 12.Elledge, S. J., and R. W. Davis. 1989. Position and density effects on repression by stationary and mobile DNA-binding proteins. Genes Dev. 3:185-197. [DOI] [PubMed] [Google Scholar]
- 13.George, A. M., and S. B. Levy. 1983. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J. Bacteriol. 155:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grkovic, S., M. H. Brown, N. J. Roberts, I. T. Paulsen, and R. A. Skurray. 1998. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J. Biol. Chem. 273:18665-18673. [DOI] [PubMed] [Google Scholar]
- 15.Head, C. G., A. Tardy, and L. J. Kenny. 1998. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J. Mol. Biol. 281:857-870. [DOI] [PubMed] [Google Scholar]
- 16.Hoch, J. A. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3:165-170. [DOI] [PubMed] [Google Scholar]
- 17.Lomovskaya, O., K. Lewis, and A. Matin. 1995. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J. Bacteriol. 177:2328-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynch, A. S., and E. C. C. Lin. 1996. Responses to molecular oxygen, p. 1526-1538. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, Jr., B. Magasanik, W. S. Reznikoff, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 19.Lynch, A. S., and E. C. C. Lin. 1996. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding at target promoters. J. Bacteriol. 178:6238-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma, D., M. Alberti, C. Lynch, H. Nikaido, and J. E. Hearst. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19:101-112. [DOI] [PubMed] [Google Scholar]
- 21.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16:45-55. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1992. A short course in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Nagakubo, S., K. Nishino, T. Hirata, and A. Yamaguchi. 2002. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J. Bacteriol. 184:4161-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagasawa, S., K. Ishige, and T. Mizuno. 1993. Novel members of the two-component signal transduction genes in Escherichia coli. J. Biochem. 114:350-357. [DOI] [PubMed] [Google Scholar]
- 25.Nikaido, H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishino, K., and A. Yamaguchi. 2001. Overexpression of the response regulator EvgA of the two-component signal transduction system modulates multidrug resistance transporters. J. Bacteriol. 183:1455-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishino, K., and A. Yamaguchi. 2002. EvgA of the two-component signal transduction system modulates production of the YhiUV multidrug transporter in Escherichia coli. J. Bacteriol. 184:2319-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance phenotype (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paulsen, I. T., J. Chen, K. E. Nelson, and M. H. Saier, Jr. 2001. Comparative genomics of microbial drug efflux systems. J. Mol. Microbiol. Biotechnol. 3:145-150. [PubMed] [Google Scholar]
- 31.Paulsen, I. T., M. K. Sliwinski, and M. H. Saier, Jr. 1998. Microbial genome analyses: global comparisons of transport capabilities based on phylogenies, bioenergetics and substrate specificities. J. Mol. Biol. 277:573-592. [DOI] [PubMed] [Google Scholar]
- 32.Perraud, A.-L., V. Weiss, and R. Gross. 1999. Signalling pathways in two-component phosphorelay systems. Trends Microbiol. 7:115-120. [DOI] [PubMed] [Google Scholar]
- 33.Reeves, V. B. 1995. Liquid chromatographic procedure for the determination of novobiocin residues in bovine milk: interlaboratory study. J. AOAC Int. 78:55-58. [PubMed] [Google Scholar]
- 34.Saier, M. H., Jr., B. H. Eng, S. Fard, J. Garg, D. A. Haggerty, W. J. Hutchinson, D. L. Jack, E. C. Lai, H. J. Liu, D. P. Nusinew, A. M. Omar, S. S. Pao, I. T. Paulsen, J. A. Quan, M. Sliwinski, T.-T. Tseng, S. Wachi, and G. B. Young. 1999. Phylogenetic characterization of novel transport protein families revealed by genome analyses. Biochim. Biophys. Acta 1422:1-56. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Sitnikov, D. M., J. B. Schineller, and T. O. Baldwin. 1996. Control of cell division in Escherichia coli: regulation of transcription of ftsQA involves both rpoS and SdiA-mediated autoinduction. Proc. Natl. Acad. Sci. USA 93:336-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka, T., T. Horii, K. Shibayama, K. Sato, S. Ohsuka, Y. Arakawa, K. Yamaki, K. Takagi, and M. Ohta. 1997. RobA-induced multiple antibiotic resistance largely depends on the activation of the AcrAB efflux. Microbiol. Immunol. 41:697-702. [DOI] [PubMed] [Google Scholar]
- 38.Wang, X. D., P. A. de Boer, and L. I. Rothfield. 1991. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 10:3363-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei, Y., J.-M. Lee, D. R. Smulski, and R. A. LaRossa. 2001. Global impact of sdiA amplification revealed by comprehensive gene expression profiling of Escherichia coli. J. Bacteriol. 183:2265-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signalling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]
- 41.White, D. G., J. D. Goldman, B. Demple, and S. Levy. 1997. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 179:6122-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiong, A., A. Gottman, C. Park, M. Baetens, S. Pandza, and A. Matin. 2000. The EmrR protein represses the Escherichia coli emrRAB multidrug resistance operon by directly binding to its promoter region. Antimicrob. Agents Chemother. 44:2905-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zgurskaya, H. I., and H. Nikaido. 2000. Cross-linked complex between oligomeric periplasmic lipoprotein AcrA and the inner-membrane-associated multidrug efflux pump AcrB from Escherichia coli. J. Bacteriol. 182:4264-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]