Abstract
Some stimulatory receptors of the innate immune system, such as the NKG2D receptor expressed by NK cells and activated CD8+ T cells, recognize self-molecules that are upregulated in diseased cells by poorly understood mechanisms1. Here we show that mouse and human NKG2D ligands are upregulated in non-tumor cell lines by genotoxic stress and stalled DNA replication, conditions known to activate a major DNA damage checkpoint pathway initiated by ATM (Ataxia telangiectasia, mutated) or ATR (ATM- and Rad3-related) protein kinases2. Ligand upregulation was prevented by pharmacological or genetic inhibition of ATR, ATM or Chk1, the latter a downstream transducer kinase in the pathway. Furthermore, constitutive ligand expression by a tumor cell line was inhibited by ATM siRNA, suggesting that ligand expression in established tumor cells, which often harbor genomic irregularities, may be due to chronic activation of the DNA damage response pathway. Thus, the DNA damage response, previously shown to arrest the cell cycle and enhance DNA repair functions or to trigger apoptosis, may also participate in alerting the immune system to the presence potentially dangerous cells.
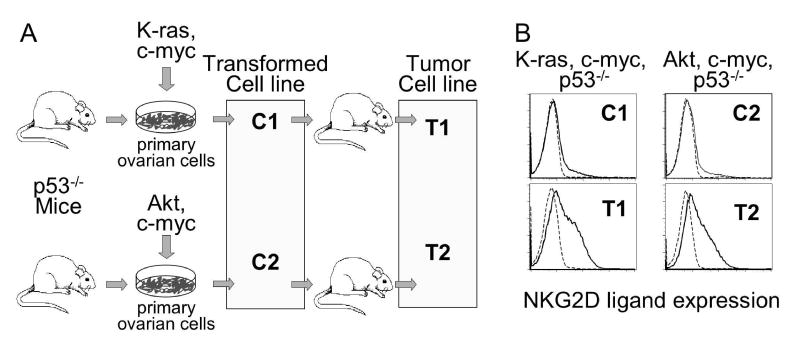
To investigate mechanisms leading to NKG2D ligand upregulation, we examined two transformed ovarian epithelial cell lines from p53−/− mice3. The C1 cell line had been transduced with K-ras and c-myc, whereas the C2 cell line had been transduced with AKT and c-myc (Fig. 1A). Both transformed cell lines grew well in cell culture but did not express appreciable levels of mouse NKG2D ligands as detected by staining with a tetrameric NKG2D reagent that binds to all mouse NKG2D ligands (Rae1, MULT1 and H601) (Fig. 1B). Ligand upregulation failed to occur when C1 and C2 cells were transfected or super-transduced with numerous other oncogenes, including E6, E7, E1A, or Ras V12 (data not shown), some of which interfere with expression of the retinoblastoma tumor suppressor gene. When injected into the ovaries of nude mice, both cell lines generated ovarian epithelial tumors, which were established as cell lines T1 and T23. Both T1 and T2 exhibited significant upregulation of NKG2D ligands (Fig. 1B) including Rae1 (see below). These findings suggested that ligand upregulation was not associated with transformation per se.
Figure 1. NKG2D ligand upregulation is associated with tumorigenesis, not with transformation per se.
A) Cell lines from transgenic mice expressing the avian retrovirus receptor. Ovarian epithelial cells from p53−/− mice were transduced with the indicated genes and established as cell lines C1 and C2. Implantation of C1 and C2 cell into ovaries of nude mice led to formation of tumors, which were established as cell lines T1 and T2. B) Transformed ovarian epithelial cell lines C1 and C2 and T1 and T2 tumor cell lines were stained with NKG2D tetramers (thick trace), or with control tetramers (dashed trace).
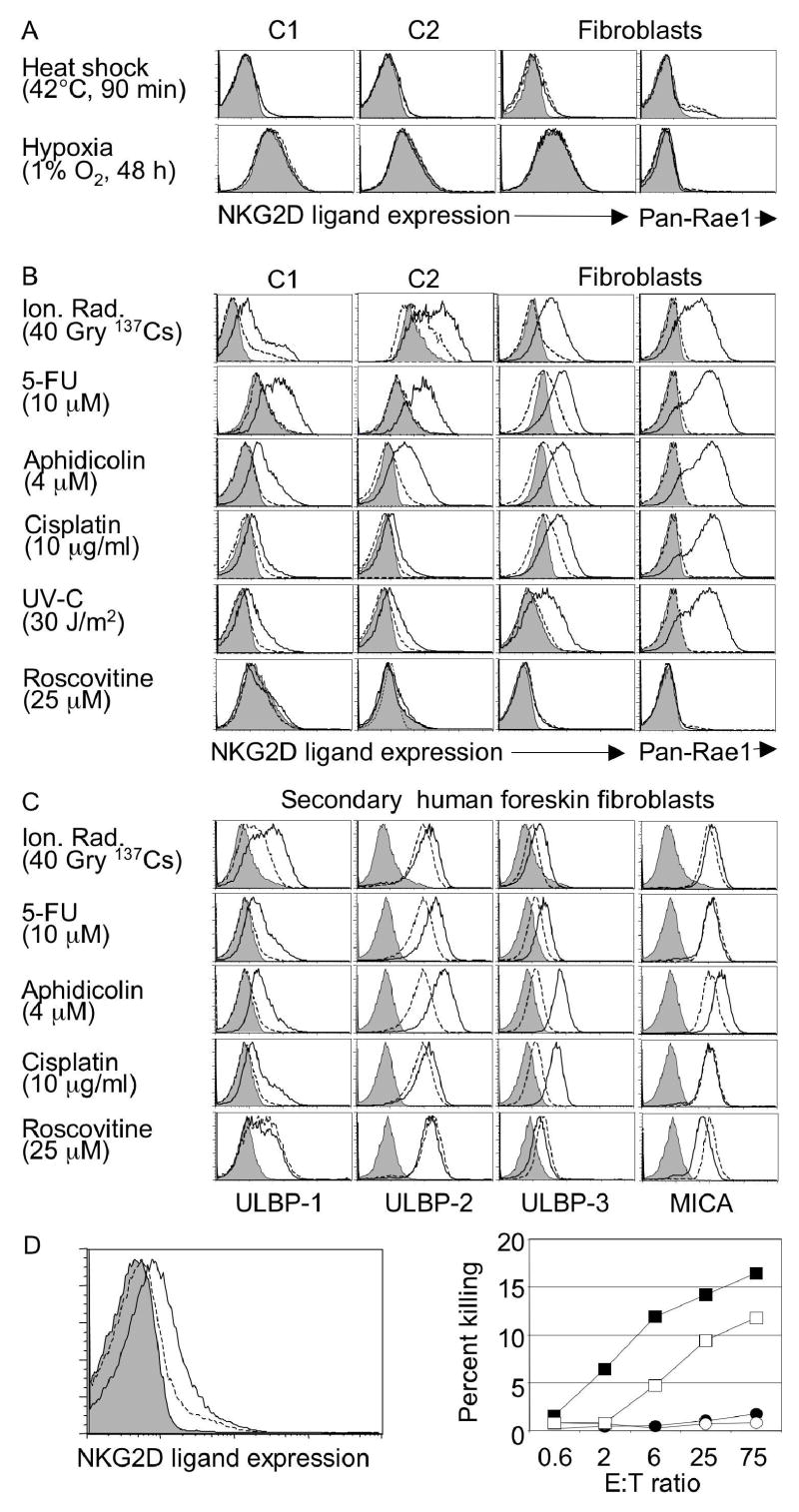
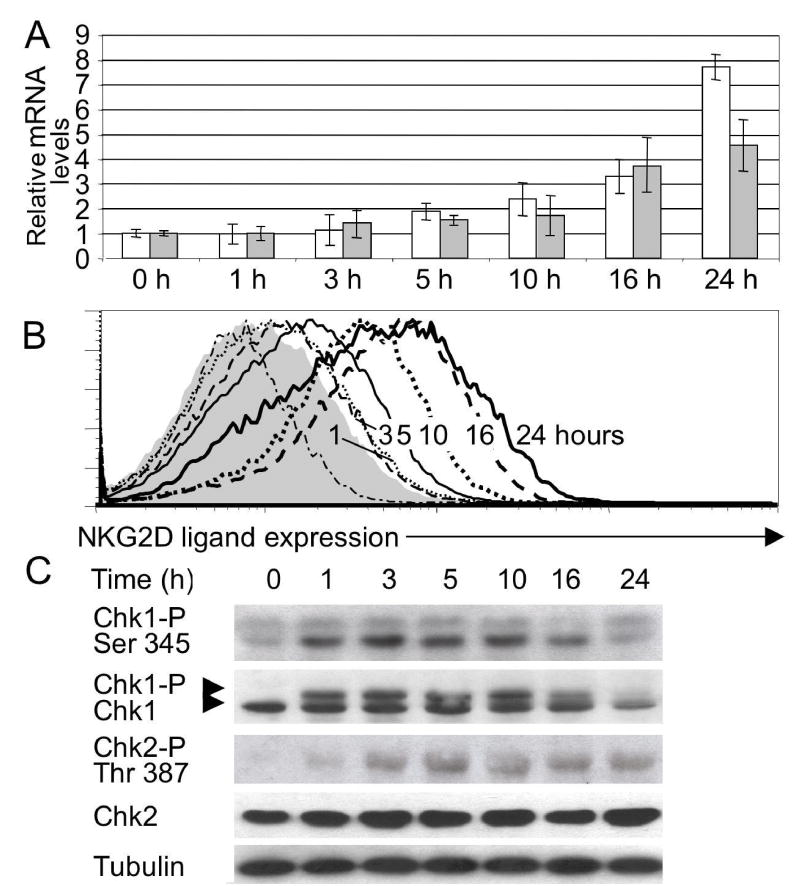
Ligand expression by C1 or C2 cells was not upregulated by numerous cell stress conditions, including heat shock, hyperoxia, hypoxia, inhibition of the cell cycle (by roscovitine), exposure to inflammatory cytokines such as TNF, interferon or IL-6, or incubation in medium of pH6 or pH8.5 (Fig. 2A, S1, data not shown). In contrast, NKG2D ligands were upregulated in C1 or C2 cells exposed to high doses of ionizing radiation, inhibitors of DNA replication such as mitomycin C, hydroxyurea (HU), 5-fluorouracil (5-FU) and the DNA polymerase inhibitor aphidicolin, or chromatin modifying treatments such as trichostatin A, chloroquine and hypotonic conditions4 (Fig. 2B, S1). These same treatments induced ligand upregulation in cultures of adult fibroblasts, showing that neither transformation nor p53 deficiency are essential for ligand upregulation (Fig. 2B). In the case of fibroblasts, but less so with the C1 and C2 cell lines, ligand upregulation was also induced by other DNA damaging conditions such as UV light and the chemotherapy agents cisplatin and ara-C (Fig. 2B and Fig. S1). Cell surface ligand upregulation detected with NKG2D tetramers or Rae1 antibodies (Fig. 2B) was accompanied by increases in both Rae1 and MULT-1 mRNAs (Fig. 3A, S2). Both the mRNAs and protein levels were initially detected at 3–5 h, and reached a plateau after 16–24 h (Fig. 3A, B). Ligand levels reverted to background levels 72 hours after aphidicolin was washed out of the medium (data not shown).
Figure 2. NKG2D ligands are induced by DNA damaging agents and DNA synthesis inhibitors.
A) No induction of ligands in C1 or C2 cells by heat shock, and hypoxia. Cells were stained with NKG2D tetramers or Rae1 antibody. B, C) Upregulation of ligands on mouse cells (B) or human fibroblasts (passage 3-6) (C) by DNA damaging agents and DNA synthesis inhibitors (16 hr treatment). Treated cells (solid lines) were compared to untreated cells (dashed lines) or to treated cells stained with control tetramers or isotype control antibodies (filled histograms). D) Aphidicolin treatment upregulates NKG2D ligands on T cell blasts (left panel, key as defined above) and elevates their sensitivity to IL-2 activated NK cells (right). NK cells were incubated with treated (squares) or untreated (circles) target cells in the absence (filled) or presence (open) of NKG2D antibody. In repetitions of the experiment, untreated cells were killed but always substantially less well than treated cells.
Figure 3. Kinetics of upregulation of NKG2D ligands and phosphorylation of the Chk1 kinase.
A) Levels of Rae1 (open bar) and MULT1 (filled bar) transcripts determined by real-time RT-PCR in cultured fibroblasts treated with aphidicolin for the indicated times. The Rae1 primers amplify all Rae1 isoforms. Data represent means ± SD, n = 3 independent experiments. B) Cell surface expression of NKG2D ligands on cells from panel A. C) Kinetics of phosphorylation of Chk1 in aphidicolin-treated fibroblasts. Phosphorylation was detected by Western blotting as a shift in the mobility of Chk1, or with the use of a phospho-Ser345 Chk1 or phospho-Thr387 Chk2 specific antibody.
Similarly, the homologous human NKG2D ligands, ULBP1, 2 and 3, were often upregulated in secondary human foreskin fibroblasts treated with high doses of ionizing radiation and inhibitors of DNA replication including aphidicolin, but not by roscovitine (Fig. 2C). Mitomycin C and aphidicolin also modestly upregulated the expression of MICA, another human NKG2D ligand.
Mouse T cell blasts treated with aphidicolin also upregulated NKG2D ligands modestly, and exhibited greater sensitivity to lysis by IL-2 activated NK cells (Fig. 2D). Lysis was inhibited by NKG2D antibody, indicating that upregulated ligands induce elevated lysis, but the partial effect suggests that aphidicolin may also induce other NK target ligands.
The treatments that induced ligand upregulation all activate a major DNA damage response pathway initiated by ATR and/or ATM (depending on the treatment), mediated by downstream mediators including the Chk1 and Chk2 kinases and p532,5,6, and resulting in cell cycle arrest, activation of DNA repair pathways, new transcription, and, if damage is extensive, apoptosis. Consistent with a role for this pathway in NKG2D ligand upregulation, phosphorylation of Chk1 on serine 345, which is required for activation of this kinase, occurred before ligand upregulation was detectable within 1 hour of treatment of fibroblasts with aphidicolin (Fig. 3C). Phosphorylation of Chk2 on threonine 387 also occurred but was somewhat delayed.
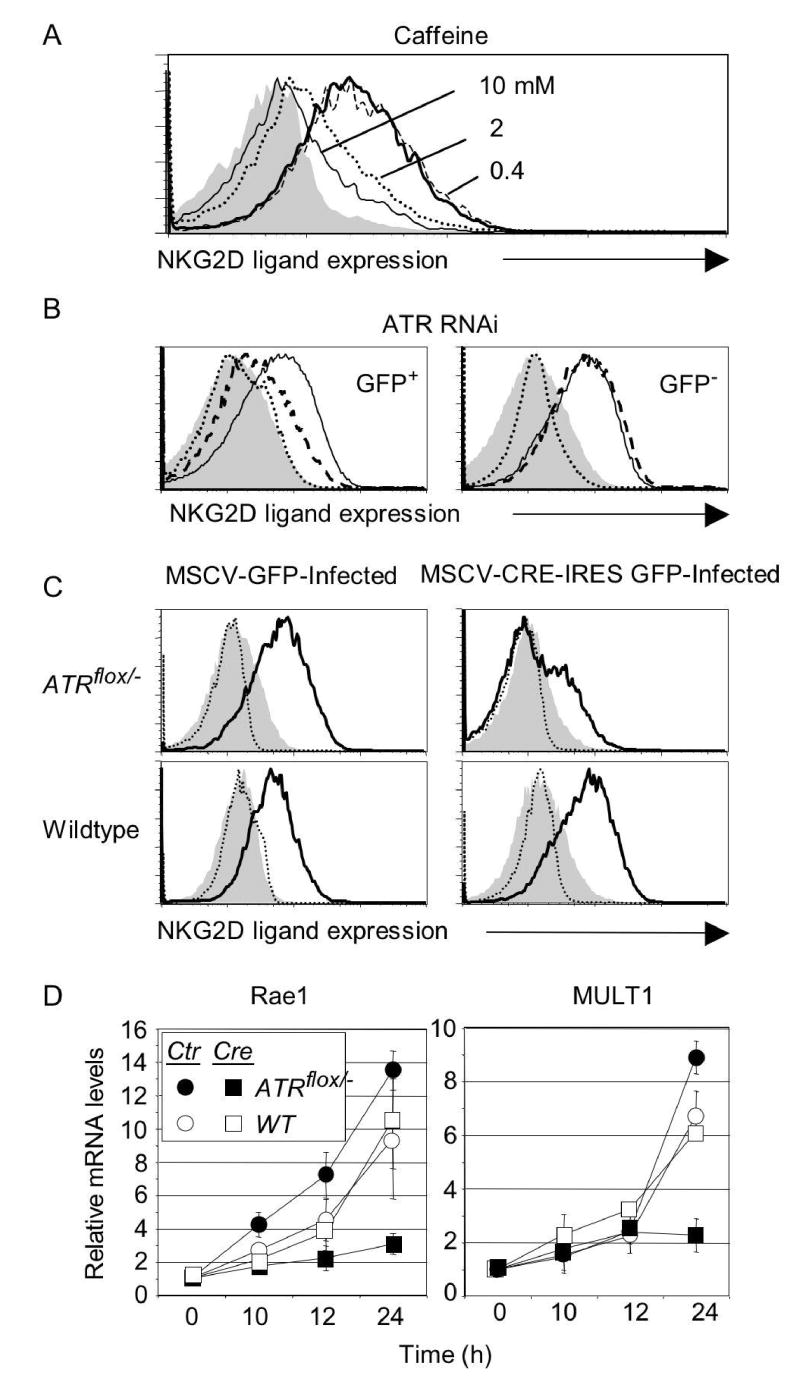
The role of ATR in aphidicolin-induced NKG2D ligand expression was investigated with three independent approaches. Ligand upregulation in response to aphidicolin was blocked by caffeine, an inhibitor of ATR and ATM7, at doses close to the IC50 (inhibitory concentration, 50%) for ATR (1.1 mM, Fig. 4A). In a second approach, ATR gene expression was specifically impaired using short interfering (si) RNAs in adult fibroblast cultures, introduced by transduction with GFP-expressing retroviruses. Upregulation of NKG2D ligands induced by aphidicolin was strongly inhibited in cells transduced with ATR siRNA compared to untransduced (GFP−) cells in the same cultures or to cells transduced with control siRNA (Fig. 4B). Cells infected with the control or ATR siRNA but not treated with aphidicolin did not upregulate ligands (Fig. 4B). As a third approach, the ATR gene was deleted in cultured cells. ATR function is required for cell viability, necessitating the use of a conditional ATR mutation. A fibroblast cell line was derived from adult mice with a gene-targeted ATR allele in which two critical exons were flanked by loxP sites8. Fibroblasts transduced with a Cre recombinase-expressing retrovirus to inactivate the ATR gene and treated with aphidicolin showed minimal upregulation of NKG2D ligand cell surface proteins (Fig. 4C), or mRNAs (Fig. 4D), compared to cells transduced with a control MSCV retrovirus. Control experiments established that both the untreated Cre-transduced ATRflox cells and the untreated MSCV-transduced ATRflox cells proliferated actively (Fig. S3), a necessary condition for activation of the replication checkpoint. Proliferation of both cell populations was blocked with aphidicolin (Fig. S3). Cre-transduction had no effect in aphidicolin-treated wildtype fibroblasts (Fig. 4C, D). Thus, upregulation of NKG2D ligands as a result of replication arrest is ATR-dependent.
Figure 4. Ligand upregulation in aphidicolin-treated fibroblasts is dependent on ATR function.
A) Ligand upregulation inhibited by ATR/ATM inhibitor caffeine added 1 h before aphidicolin. B) Inhibition of ligand upregulation by ATR siRNA. Fibroblasts were transduced with retroviral vectors encoding GFP and ATR siRNA (dashed lines in histograms) or GFP and control siRNA (solid lines), cultured for 5 days and treated with 4 μM aphidicolin for 16 hours. NKG2D ligand expression was determined by gating on transduced (GFP+) and untransduced (GFP−) cells from the same cultures. Filled histogram: untreated cells stained with NKG2D tetramers. Dotted line: aphidicolin-treated cells stained with control tetramers. C) Conditional deletion of the ATR gene prevents ligand upregulation. ATRflox/− or wildtype fibroblasts were transduced with MSCV-IRES-GFP or MSCV-Cre-IRES-GFP, cultured and analyzed as in Figure 4B, gating on transduced (GFP+) cells. D) Reduced induction of Rae1 and MULT1 transcripts in cells deficient for ATR expression. Real-time PCR of cDNA samples from transduced fibroblast cultures described in Figure 4C. Means ± SDs of 3 independent experiments are shown.
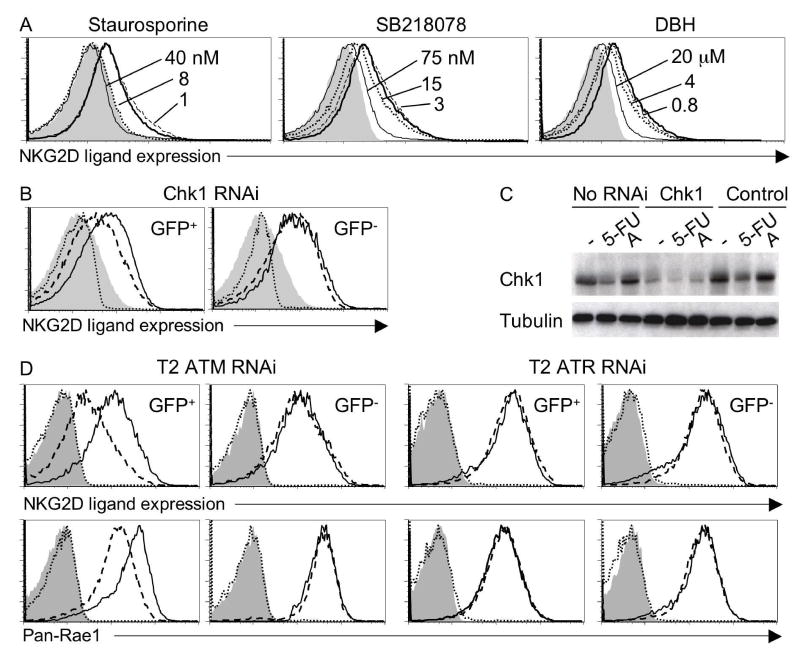
A role for the Chk1 kinase was suggested by the findings that aphidicolin-induced ligand upregulation was inhibited by three independent Chk1 inhibitors (Fig. 5A). The effective doses were close to the IC50s of all three inhibitors for Chk1 (8 nM staurosporine, 15 nM SB-218078 and 3 μM debromohymenialdesine (DBH)) (Fig. 5A). Each of these inhibitors also acts on targets other than Chk1, but the IC50s differ and the other targets are not all shared by the three inhibitors. As a more specific test, transduction of fibroblasts with Chk1 siRNA reproducibly inhibited aphidicolin-induced NKG2D ligand upregulation as compared to untransduced (GFP−) cells in the same culture or cells transduced with control siRNA (Fig. 5B). The Chk1 siRNA did not completely abrogate Chk1 protein expression (Fig. 5C), possibly explaining why pharmacological inhibitors were more effective at blocking ligand upregulation. The inhibitor and siRNA results indicate that Chk1 is necessary for optimal NKG2D ligand upregulation in response to aphidicolin treatment.
Figure 5. Role of Chk1 in expression of NKG2D ligands in aphidicolin treated fibroblasts and tumor cell lines.
A) Chk1 inhibitors prevent upregulation of NKG2D ligands. Fibroblasts were preincubated with staurosporine, SB-218078, debromohymenialdesine, or DMSO before aphidicolin treatment. B) Inhibition of ligand upregulation by Chk1 siRNA. Fibroblasts were transduced with retroviral vectors encoding GFP and Chk1 siRNA (dashed lines in histograms) or GFP and control siRNA (solid lines) before treatment as in Figure 4B. C) Chk1 protein levels are reduced in Chk1 siRNA-tranduced cells. Untransduced or sorted GFP+ transduced fibroblasts were cultured for 2 days and treated for 16 h with DMSO (labeled -), 4 μM aphidicolin (labeled A) or 10 μM 5-FU. Lysates were prepared and Western blotted with Chk1 antibodies. D) ATM siRNA, but not ATR siRNA decreases levels of NKG2D levels in T2 cells. T2 cells were transduced with GFP retroviral vectors encoding control siRNA (solid lines) or either ATM or ATR siRNA (dashed lines in histograms). After culturing for 7 days, NKG2D ligand expression was determined by gating on transduced (GFP+) and untransduced (GFP−) cells from the same cultures. Filled histogram: Control siRNA transduced T2 cells stained with control tetramer. Dotted line: ATR or ATM siRNA transduced T2 cells stained with control tetramers.
Ionizing radiation, hypotonic conditions, and other treatments that activate ATM, in some cases preferentially to ATR4,9, also induced upregulation of NKG2D ligands (Fig. 2, Fig. S1). An ATM siRNA/GFP retrovirus partially inhibited NKG2D ligand upregulation in cells exposed to ionizing radiation or hypotonic conditions, but had no effect on ligand upregulation in cells exposed to aphidicolin or UV-C, which preferentially activate ATR9 (Fig. S4). Ligand upregulation in response to ATM-inducers was also inhibited by caffeine, which blocks both ATR and ATM7 (data not shown). Thus, induction of NKG2D ligands requires ATM or ATR, depending on the nature of the treatment.
It is unlikely that NKG2D ligand upregulation is an indirect consequence of ATR/ATM-induced cell cycle arrest, because cell cycle arrest induced by the cyclin-dependent kinase inhibitor roscovitine or serum deprivation did not trigger ligand upregulation (Fig. 2B,C and S1). It is also unlikely that ligand upregulation is a property of cells entering the cell death pathway, because such cells, defined by staining with Annexin-V, did not upregulate NKG2D ligands significantly (data not shown). Hence, the DNA damage response likely plays a more direct role in the process of ligand upregulation.
Activation of the DNA damage pathway may represent a more distinctive feature of diseased cells than other correlates of tumorigenesis or infection. The pathway is activated in cells infected with several viruses10, raising the possibility that is plays a role in ligand upregulation after virus-infection11. The pathway is also frequently activated in tumor cell lines12–14, possibly due to the greater genomic instability of these cells as compared to transformed cells13,14, and recent reports have demonstrated activation of ATM and other components of the DNA damage response pathway in precancerous lesions, at a stage before genomic instability is apparent15,16. We addressed whether chronic activation of the DNA damage pathway contributes to constitutive expression of NKG2D ligands observed in the T2 ovarian tumor cell line (Fig. 1). T2 cells transduced with ATM siRNA exhibited reduced NKG2D ligand expression, whereas ATR siRNA had no effect (Fig. 5D). Thus, constitutive ligand expression in tumor cell lines may be dependent on ATM activity, at least in some instances. Accelerated loss of genomic stability or other cellular changes that activate the DNA damage response in tumor cells may lead to constitutive ATM activation. Further studies will be necessary to establish the generality of the link between the DNA damage response and NKG2D ligand expression in tumor cells.
Transcriptional regulators including the p53 tumor suppressor, p7317 and p63 are activated by Chk1. p53 is not required for ligand upregulation, which occurred in the p53−/− C1 and C2 cell lines, but p73 or p63 may play redundant or unique or roles in ligand upregulation. The p53-independent component of NKG2D ligand upregulation means that loss of p53 during tumorigenesis should not by itself fully disable the immune-enhancing potential of the cells.
These findings suggest a novel link between the immune response and processes that regulate genome integrity, and may have clinical significance. It is possible that part of the efficacy of some chemotherapies and radiotherapies, most of which activate the DNA damage response18,19, is due to the induction of NKG2D ligands and consequent enhanced sensitivity of the cell to the immune system. The pathway leading to upregulation of NKG2D ligands may be a productive target for design of therapeutic agents to enhance the immunogenicity of tumor cells while reducing overall toxicity.
Materials and Methods
Mice, cells and cell treatments
C57Bl/6 mice from Jackson Laboratory were bred and housed as described20. Ear and tail-derived fibroblasts from adult C57Bl/6 mice or 129SvEv/C57BL/6-Atrflox/- mice8,21 were established as described22. Unless otherwise specified the ear fibroblasts were employed for the experiments. C1, C2, T1 and T2 mouse ovarian cancer cell lines were generated as described3 and treated for 16 h unless stated otherwise. Secondary neonatal human dermal foreskin fibroblasts were purchased (Cascade Biologics, Portland, Or) and cultured in 106 medium.
Hypotonic swelling was carried out for 16 h in PBS containing 0.45% glucose (w/v) and 2% FBS, with the NaCl concentration adjusted to 50 mM or, as a control, 140 mM. For inhibitor experiments, inhibitors were added to subconfluent cultures beginning 1 hour before addition of 4 μM aphidicolin and ligand expression was determined 16 hours later.
Reagents
5-bromo-2'-deoxyuridine (BrdU), 5-fluorouracil (5-FU), aphidicolin, caffeine, cis-Diammineplatinum(II) dichloride, cytosine β-D-arabinofuranoside hydrochloride, mitomycin-C, hydroxyurea (HU), staurosporine and t-butylhydroquinone (TBHQ) were purchased from Sigma. Roscovitine, SB-218078 and debromohymenialdesine were purchased from Calbiochem. NKG2D tetramers and control tetramers were produced as previously described23. For staining, we used Pan-Rae1, MICA, ULBP1, 2 and 3 specific monoclonal antibodies (R&D Systems), mouse IgG and F(ab')2 fragment goat anti-mouse or rat IgG + IgM coupled to phycoerythrin (Jackson ImmunoResearch Laboratories).
MSCV-Cre constructs and transduction of fibroblasts
Cre was subcloned adjacent to the cytomegalovirus promoter in the pMSCV2.2-IRES-GFP proviral vector (gift of W. Sha, University of California, Berkeley). Retroviral supernatants were generated as described24.
siRNA retroviral constructs
Chk1 siRNA (5'-CAACTTGCTGTGAATAGAAT-3') corresponded to the mouse counterpart of a published human Chk1 siRNA25. ATR siRNA (5'-AGGAAGCAATTCCACATTAAGC-3') and ATM siRNA (5’-GAGGTGGCTCTTATTCTAC-3’) were selected based on Dharmacon’s siRNA Design Center algorithm (Dharmacon). The control siRNA (5’-AACAAGTGAAGCAGTCGCAGT-3’) was described in26. The siRNAs were subcloned in the pMND-Banshee vector, a gift of Jose Alberola-Ila (California Institute of Technology), as described previously27. The control RNAi-pMND-Banshee, ATR-pMND-Banshee and Chk1-pMND-Banshee plasmids were transiently transfected into 293 T cells using lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). 48 h after transfection, 50% confluent cultures of fibroblasts or T2 were transduced as described24.
As determined by real time RT-PCR in sorted populations, the level of ATR transcripts in ATR siRNA-transduced cells in Fig. 4B was 37% ± 3% of the level in untransduced (GFP−) cells in the same culture (mean ± SD, n=3). In Fig. 5D, ATM mRNA in ATM siRNA-transduced T2 cells was 53% ± 2% of the level in untransduced (GFP−) cells in the same culture, while ATR mRNA in ATR siRNA transduced cells was 49% ± 5% of the level in untransduced (GFP-) cells in the same culture. Because of impurities in the sorted populations, the knockdowns may be more efficient than indicated.
Western blotting
Whole-cell extracts were prepared from untreated or treated populations, electrophoresed in 6 or 8% SDS-PAGE gels, and blotted onto nitrocellulose membranes. Antibodies against Chk1 (sc-8408, Santa Cruz Biotechnology), Phospho-Chk1-Ser345, Chk2, Phospho-Chk2-Thr387 (Cell Signaling Technology), Tubulin (CP06, Calbiochem) and horseradish peroxidase-coupled second stage reagents were used to develop the blots (SuperSignal West Pico Stable Solution). Blots were exposed on X-ray film.
Quantitative real-time RT-PCR
Total RNA was isolated using the RNeasy kit (Qiagen). Real-time PCR assays were performed using an Applied Biosystems 5700 sequence detector (Applied Biosystems). Two μg of total RNA was reverse transcribed with random hexamers using a Transcriptor First Strand cDNA Synthesis Kit (Roche). Each amplification mixture (25 μl) contained 25 ng of reverse transcribed RNA, 8 μM forward primer, 8 μM reverse primer and 12.5 μl of iTaq SYBR Green Supermix with ROX (Bio-Rad). PCR thermocycling parameters were 50oC for 2 min, 95oC for 10 min, and 40 cycles of 95oC for 15 sec and 60oC for 15 sec and 72oC for 1 min. All samples were normalized to the signal generated from the housekeeping genes, β-actin or GAPDH (for ATR and ATM levels). SYBR Green PCR was performed in triplicate using the ABI PRISM® 7700 Sequence Detection System. The following primers were used: β-actin-5’: tgtttgagaccttcaacacc; β-actin-3’: taggagccagagcagtaatc; GAPDH-5’: gaaggtcggtgtgaacgga: GAPDH-3’: gttagtggggtctcgctcct: MULT1-5’: caatgtctctgtcctcggaa; MULT1-3’: ctgaacacgtctcaggcact; Pan-Rae1-5’: tggacactcacaagaccaatg; Pan-Rae1-3’: cccaggtggcactaggagt; ATR-5’: tgcgctctgctagagcacggt; ATR-3’: agtgctggctggctgtgctg; ATM-5’ atccaggccctgcagaatttggg; ATM-3’: ctccacgccgcctggtaacg. Samples prepared without reverse-transcription served as negative control templates.
Cytolysis assay
BALB/c spleen cells were activated for three days with 2.5 μg/ml Concanavalin A, washed with 50 mM α-methyl mannoside and recultured with 10 u/ml IL-2 plus either aphidicolin or DMSO for 18 hrs before labeling with 51Cr for use as target cells. NK cells were prepared as described20. Thirty min before adding target cells to initiate the 3 h cytolysis assay20, antibody MI-6 (anti-NKG2D20) was added to a concentration of 50 μg/ml in some groups. Spontaneous release was 5% for DMSO-treated target cells and 15% for aphidicolin-treated target cells.
Supplementary Material
Acknowledgments
We thank Jeremy Thorner and Stuart Linn for helpful discussions, Yana Natanzon and Lily Zhang for excellent technical assistance, Hector Nolla for assistance with flow cytometry and Lillian Fritz-Laylin for help with transductions. This work was supported by a National Institutes of Health grant (RO1 CA093678) to DHR and an award from the Prostate Cancer Foundation.
References
- 1.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3:781–90. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 2.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–96. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 3.Xing D, Orsulic S. A genetically defined mouse ovarian carcinoma model for the molecular characterization of pathway-targeted therapy and tumor resistance. Proc Natl Acad Sci U S A. 2005;102:6936–41. doi: 10.1073/pnas.0502256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 5.Osborn AJ, Elledge SJ, Zou L. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 2002;12:509–16. doi: 10.1016/s0962-8924(02)02380-2. [DOI] [PubMed] [Google Scholar]
- 6.Sancar A, Lindsey-Boltz LA, Unsal-Kaccmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 7.Sarkaria JN, et al. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–82. [PubMed] [Google Scholar]
- 8.Brown EJ, Baltimore D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 2003;17:615–28. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Toward maintaining the genome: DNA damage and replication checkpoints. Annu Rev Genet. 2002;36:617–56. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 10.Weitzman MD, Carson CT, Schwartz RA, Lilley CE. Interactions of viruses with the cellular DNA repair machinery. DNA Repair (Amst) 2004;3:1165–73. doi: 10.1016/j.dnarep.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Lodoen M, et al. NKG2D-mediated natural killer cell protection against cytomegalovirus is impaired by viral gp40 modulation of retinoic acid early inducible 1 gene molecules. J Exp Med. 2003;197:1245–53. doi: 10.1084/jem.20021973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–9. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 13.Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunol Rev. 2003;194:77–95. doi: 10.1034/j.1600-065x.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 14.Sieber OM, Heinimann K, Tomlinson IP. Genomic instability--the engine of tumorigenesis? Nat Rev Cancer. 2003;3:701–8. doi: 10.1038/nrc1170. [DOI] [PubMed] [Google Scholar]
- 15.Gorgoulis VG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 16.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez S, Prives C, Cordon-Cardo C. p73alpha regulation by Chk1 in response to DNA damage. Mol Cell Biol. 2003;23:8161–71. doi: 10.1128/MCB.23.22.8161-8171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Bignami M, Casorelli I, Karran P. Mismatch repair and response to DNA-damaging antitumour therapies. Eur J Cancer. 2003;39:2142–9. doi: 10.1016/s0959-8049(03)00569-0. [DOI] [PubMed] [Google Scholar]
- 19.Zhou BB, Bartek J. Targeting the checkpoint kinases: chemosensitization versus chemoprotection. Nat Rev Cancer. 2004;4:216–25. doi: 10.1038/nrc1296. [DOI] [PubMed] [Google Scholar]
- 20.Jamieson AM, et al. The role of the NKG2D immunoreceptor in immune cell activation and natural killing. Immunity. 2002;17:19–29. doi: 10.1016/s1074-7613(02)00333-3. [DOI] [PubMed] [Google Scholar]
- 21.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 22.Lander MR, Moll B, Rowe WP. A procedure for culture of cells from mouse tail biopsies: brief communication. J Natl Cancer Inst. 1978;60:477–8. [PubMed] [Google Scholar]
- 23.Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nature Immunology. 2000;1:119–126. doi: 10.1038/77793. [DOI] [PubMed] [Google Scholar]
- 24.Diefenbach A, Hsia JK, Hsiung MY, Raulet DH. A novel ligand for the NKG2D receptor activates NK cells and macrophages and induces tumor immunity. Eur J Immunol. 2003;33:381–91. doi: 10.1002/immu.200310012. [DOI] [PubMed] [Google Scholar]
- 25.Krause DR, et al. Suppression of Tousled-like kinase activity after DNA damage or replication block requires ATM, NBS1 and Chk1. Oncogene. 2003;22:5927–37. doi: 10.1038/sj.onc.1206691. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, et al. Artemis Is a Phosphorylation Target of ATM and ATR and Is Involved in the G2/M DNA Damage Checkpoint Response. Mol Cell Biol. 2004;24:9207–20. doi: 10.1128/MCB.24.20.9207-9220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/s1074-7613(03)00176-6. [DOI] [PubMed] [Google Scholar]
- 28.Tough DF, Sprent J. Turnover of naive- and memory-phenotype T cells. J Exp Med. 1994;179:1127–1135. doi: 10.1084/jem.179.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.