Abstract
Previous molecular analysis of the Octopus Spring cyanobacterial mat revealed numerous genetically distinct 16S rRNA sequences from predominant Synechococcus populations distantly related to the readily cultivated unicellular cyanobacterium Synechococcus lividus. Patterns in genotype distribution relative to temperature and light conditions suggested that the organisms contributing these 16S rRNA sequences may fill distinct ecological niches. To test this hypothesis, Synechococcus isolates were cultivated using a dilution and filtration approach and then shown to be genetically relevant to natural mat populations by comparisons of similarities of 16S rRNA genes and 16S-23S internal transcribed spacer (ITS) regions. Most isolates were identical or nearly identical at both loci to predominant mat genotypes; others showed 1- to 2-nucleotide differences at the 16S rRNA locus and even greater difference in ITS sequences. Isolates with predominant mat genotypes had distinct temperature ranges and optima for growth that were consistent with their distributions in the mat. Isolates with genotypes not previously detected or known to be predominant in the mat exhibited temperature ranges and optima that were not representative of predominant mat populations and also grew more slowly. Temperature effects on photosynthesis did not reflect temperature relations for growth. However, the isolate with the highest temperature optimum and upper limit was capable of performing photosynthesis at a higher temperature than other isolates. Growth rate and photosynthetic responses provided evidence for light acclimation but evidence of, at best, only subtle light adaptation.
Molecular analysis of the Octopus Spring cyanobacterial mat community revealed that the dominant Synechococcus spp. constructing the mat were genetically quite distinct (on the order of an 8 to 10% difference in 16S rRNA sequences) from readily cultivated Synechococcus lividus strains (8, 9, 31-34). Five cyanobacterial genotypes readily observed in molecular analyses (genotypes A, A′, A″, B, and B′), and shown to be predominant in the mat (18), are closely related (on the order of ≤3% difference in their 16S rRNA sequences). The sequences form two distinct phylogenetic clusters containing A-like (A, A′, and A″) and B-like (B and B′) genotypes (29, 31). Oligonucleotide probe analysis demonstrated that A-like sequences were distributed at higher temperatures and B-like sequences at lower temperatures (22). In addition, shifting samples from low to high temperature for 1 week resulted in a disappearance of B-like sequences but a rise in A-like sequence levels, providing evidence that distribution patterns may reflect adaptation. A more detailed pattern was revealed when PCR-amplified 16S rRNA gene segments from different temperature sites were examined with denaturing gradient gel electrophoresis (DGGE) (10). At increasingly higher temperatures from ∼50°C to the upper temperature limit of the mat (∼72°C), a progression of genotypes from B to B′ to A to A′ and finally A″ was noted. These findings were interpreted as suggesting that these closely related cyanobacterial genotypes represent different temperature-adapted populations (ecotypes) (10, 31).
DGGE was also used to study the vertical structure of the top 1 mm of the 60°C mat system in Mushroom Spring (20), which is located ∼0.5 km from Octopus Spring and has strains with similar chemical composition and identical or nearly identical cyanobacterial 16S rRNA genotypes (7). DGGE analysis of 16S rRNA genotypes obtained from 100-μm-thick horizontal sections revealed that the B′ genotype was detected through the entire 1-mm depth interval, but the A sequence was only found at depths between 400 and 800 μm. The subsurface positioning of the A genotype corresponded to a layer of brightly autofluorescent Synechococcus cells extending from 400 to 700 μm (20). The high autofluorescence of these cells and their vertical orientation at midday suggested either adaptation or acclimation to low light. Taken together with the extreme light extinction in the upper 1 to 2 mm of these mats (7), the co-occurrence of the A genotype with the subsurface population suggested the possibility that this genotype might be associated with a low-light-adapted Synechococcus population. A second example of genetically and phenotypically distinct Synechococcus populations occurring at different depths was observed for the 68°C Mushroom Spring mat (7). Here, distinctly pigmented A′-like populations found at different depths were so closely related that it was necessary to use the more rapidly evolving internal transcribed spacer (ITS) region separating the 16S and 23S rRNA genes to discern them.
The discovery that very closely related 16S rRNA or ITS-defined genotypes might correspond to unique ecotypes adapted to different temperature and/or light intensity conditions challenges the notion that a 2 to 3% difference in 16S rRNA sequences should be used to demarcate bacterial species (28). An ecological species concept (i.e., species are ecologically specialized populations that occupy unique niches (26, 27) would seem a sensible alternative (4, 28).
It is impossible to determine from distributions alone whether organisms are adapted to specific environmental conditions; however, adaptation can be studied with isolates in a laboratory setting. Temperature-adapted Synechococcus populations from Hunter’s Hot Springs, Oregon, have been cultivated (14, 19). Light adaptation has been observed in filamentous hot spring cyanobacterial isolates described as Plectonema notatum (23, 24) and in Prochlorococcus populations cultivated from (and predominant in) different depths in the marine water column (15). We tested the hypothesis that 16S rRNA-defined Synechococcus genotypes in the Octopus Spring microbial mat correspond to distinct ecotypes that are adapted to specific temperature and/or light conditions by obtaining genetically relevant Synechococcus cultures from the Octopus Spring mat system and determining temperature and light relationships for their growth and photosynthesis.
MATERIALS AND METHODS
Sample collection.
Samples were obtained from the microbial mat in Octopus Spring, Yellowstone National Park, by use of a no. 4 cork borer (8 mm inside diameter). Six different temperature sites were sampled: 49 to 56°C (sampled on 22 October 2000), 58 to 65°C (on 24 September 2000), 52 to 65°C and 51 to 61°C (on 10 July 2002), 58 to 65°C (on 25 July 2002), and 59 to 70°C (on 30 October 2002 and 13 January 2003). Samples were returned to the laboratory in a thermos containing spring water at ∼65°C, which cooled to ∼48°C during transit. In situ light intensity (irradiance) measurements were taken using a LI-250 light meter with a LI-190SA quantum sensor (LI-COR, Lincoln, Nebr.); summer midafternoon intensities were approximately 1,450 μmol photons m−2 s−1 (no measurements were taken in fall and winter).
Cultivation.
We used a filter cultivation approach (5), which physically isolated cells from one another, thereby preventing competitive exclusion suspected in previous liquid enrichments, even with highly diluted samples (9). Immediately upon arrival at the laboratory, the top green layer (approximately 1 to 2 mm) of each core was removed using a sterile razor blade and homogenized in 10 ml of Octopus Spring water by use of a Dounce tissue grinder until no visible cell clumps remained. Homogenized cells were then serially diluted 10-fold in preautoclaved Octopus Spring water (kept at a temperature matching that of the sampling site) to 10−7 of the cell density in the original suspension and filtered through presterilized 47-mm-diameter, 0.2-μm-pore-size Nuclepore polycarbonate filters by use of a Millipore vacuum system. Since it was impractical to sterilize the filtration unit between samples, filtration was performed in the order of highest (10−7) to lowest (100) dilution to minimize contamination of the samples with the rare individual cells that might be particularly well suited to laboratory culture. Inoculated filters were transferred to 47-mm-diameter petri dishes (Millipore) containing Whatman glass fiber filters presaturated with 1.5 ml of medium DH (Castenholz’s medium D) (3) supplemented with HEPES buffer at 1.2 g/liter and pH 8.2 (the approximate pH of the mat at this site). The petri dishes were placed in plastic bags with wetted paper towels and incubated at 55°C with ∼20 to 50 μmol photons m−2 s−1 of light or 60°C with ∼60 μmol photons m−2 s−1 of light (59 to 70°C site). Microcolonies of Synechococcus (confirmed microscopically) approximately 0.2 mm in diameter developed about 2 weeks after inoculation. Colony growth was maintained by adding 0.5 ml of medium DH every 2 to 3 days.
Isolates cultivated from samples collected in 2000 were revived in 2002 or 2003 from stocks that had been frozen in 0%, 5%, 8%, 12%, or 15% dimethyl sulfoxide in distilled water and stored in a −80°C freezer in a manner similar to that described by Brand (http://www-cyanosite.bio.purdue.edu/protocols/cryo.html; posted May 2002). Samples were quickly warmed to room temperature and centrifuged in a microcentrifuge at a relative centrifugal force of 82 for approximately 5 to 10 s. The pellet was washed once with 1 ml of sterile medium DH, resuspended in 1 ml medium DH, and then incubated at either 50°C or 55°C ∼30 μmol photons m−2 s−1 of light for several weeks until growth was observed. Liquid medium was added every 2 to 3 days to prevent drying of cultures.
All cultures contained small, nonfluorescing cells about as numerous as, but easily distinguished from, the larger autofluorescent Synechococcus cells (Fig. 1). Contaminants with 16S rRNA sequences identical in BLAST analysis (2) to those of Rubrobacter taiwanensis (GenBank accession number AF465803), Meiothermus taiwanensis (AF18001) and Geobacillus uralicus (AY079151) were isolated in pure culture and subcultured on 0.1% (wt/vol) tryptone-yeast extract-dextrose medium (broth or solidified with 1.6% [wt/vol] Difco agar) (17). We tried unsuccessfully to obtain pure Synechococcus cultures by using antibiotic treatment (Primaxin [imipenem and cilastatin]; Merck Pharmaceuticals) (final concentration, 100 μg/ml) similar to that described by Ferris and Hirsch (6) and medium supplements (B vitamins, NaHCO3 as a CO2 source, contaminant, and contaminated culture supernatants) as detailed by Allewalt (1).
FIG. 1.
Phase-contrast photomicrographs of Synechococcus isolates of different genotypes. Arrows identify contaminant organisms.
Genotypic analysis.
DNA was extracted using bead beating, ammonium acetate precipitation of proteins, and isopropanol precipitation of nucleic acids by methods similar to those described by Moré et al. (16; also see reference 10). PCR amplification of a portion of the 16S rRNA gene and the ITS region was performed using primers 1070F (5′-ATGGCTGTCGTCAGCT) (8) and L23cyR (5′-TGCCTAGGTATCCACC) (18). Samples were presumed to be unicyanobacterial when only one band appeared in gel electrophoresis analysis. Such PCR products were purified using a QIAquick PCR purification kit (QIAGEN) and sequenced using primers 1070F to obtain 16S rRNA sequences and 1505F (5′-GTGAAGTCGTAACAAGG) to obtain ITS sequences, Big Dye version 3.1 terminators, and an Applied Biosystems 310 genetic analyzer (Applied Biosystems, Foster City, CA). Sequences were analyzed using Sequencher 3.0 (Gene Codes) software. Culture sequences were compared to sequences for all Synechococcus genotypes directly retrieved from Yellowstone microbial mats.
Temperature and light effects on growth.
The growth of Synechococcus isolates with respect to different temperatures and light intensities was assessed by inoculating triplicate tubes containing 9 ml of medium DH with cell material from stock cultures of each isolate to yield an initial cell density of 1.0 × 106 to 1.6 × 106 cells/ml. Stock cultures were pregrown in 50 ml of medium DH at 55°C with ∼50 μmol photons m−2 s−1 of light. For temperature adaptation experiments tubes were placed at a slight angle in boxes under 48 to 55 μmol photons m−2 s−1 of light in incubators held at different temperatures. Light-intensity-adaptation experiments were conducted in a similar manner at 55°C, except that light intensities of up to ∼385 μmol photons m−2 s−1 were achieved by incubating tubes close to the light source, and light intensity was reduced using layers of plastic screen mesh. Tubes were removed from the incubator at 24-h intervals and the contents were thoroughly mixed, and then 100 μl was transferred into a 0.5 ml tube and 10 μl of 37% formaldehyde was added to preserve the sample. Synechococcus cell counts were performed using a Petroff-Hausser counting chamber (Sperm/Bacteria counter; Hausser Scientific, Horsham, Pa.). Each sample was counted three times, generating nine counts per isolate for each set of temperature and light conditions. Simple linear regression was used to determine the relationship between the logarithm of cell count and time during exponential growth under a particular set of temperature or light conditions. Slope estimates between isolates of different genotypes under specific incubation conditions or within individual isolates of particular genotypes incubated under different conditions of temperature and light intensity were compared using Tukey’s multiple comparison test. Comparisons reported to be significantly different differ at the 95% confidence level. Temperature relations of pure-cultured heterotrophic contaminants were assessed in a similar manner except that growth was approximated as turbidity (at 595 nm, an absorption minimum for all contaminant isolates).
Microsensor analysis of oxygenic photosynthesis.
A small experimental chamber (Fig. 2) was designed to facilitate rapid microsensor measurements of oxygen produced during photosynthesis at various light intensities and temperatures. The chamber consisted of two microscope coverslips kept apart at a distance of 0.7 mm by thin solid glass rods that were glued in place with UV-curing cement, resulting in a thin vessel that minimized self shading and maximized temperature exchange. The glass rods were placed so as to form a small triangular chamber, and a tapered 6-mm (inside diameter) glass tube was inserted into one corner to enable introduction of an oxygen microsensor (21). The other end of the glass tube penetrated the wall of a 2-liter transparent acrylic water bath filled with water so that one flat side of the chamber was facing the water surface. The microsensor could then be inserted into the coverslip chamber through the glass tube by use of a micromanipulator placed next to the acrylic water bath. The temperature of the water bath was controlled using a coiled stainless steel tube connected to a heating bath so that it functioned as a heat exchanger. The chamber was illuminated from above by a 20 W fiber-optic halogen lamp equipped with a collimator lens at the end of the light guide (Schott KL 200). The light intensity was regulated by distance and, for the lowest light intensities, also by reducing the voltage. The largest inaccuracy was probably a function of the repeated positioning of the lamp, which resulted in a reproducibility of only ± 15% in light intensity.
FIG. 2.
View from above of the apparatus used for temperature-controlled microsensor measurement of oxygenic photosynthesis in small volumes of Synechococcus cultures. Inset shows detail of triangular culture chamber with sleeve to accommodate microsensor.
Photosynthesis was determined in two ways: (i) the rate of increase in oxygen concentration after 1 min in the light (net photosynthesis) and (ii) the rate of increase in oxygen concentration after 1 min in the light plus the rate of decrease in oxygen concentration just before the light period (gross photosynthesis). Absolute rates cannot be converted to rates per unit of biomass or chlorophyll, as the cells were not evenly distributed in the chamber (Fig. 2), likely creating an uneven distribution of biomass in the entire chamber relative to oxygen kinetics measured locally near the microsensor tip. The culture was placed in the chamber for at least 15 min before measurements were made to allow for sedimentation and possible aggregation of the cells. We waited at least 1 min after changing conditions before readings were taken, since the average diffusion time for an O2 molecule from top to bottom in the chamber is 49 s, as calculated using the Einstein-Smoluchowski equation (for example, see reference 11) as follows: s =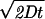 , where D is 5 × 10 − 5 cm−2 s−1 (diffusivity in water at 60°C) and s is 0.07 cm. Reproducibility was checked by conducting triplicate measurements on one culture at a single temperature and a variety of light intensities; relative standard errors averaged 3.9 and 1.8% for mean gross and net photosynthesis rates, respectively.
, where D is 5 × 10 − 5 cm−2 s−1 (diffusivity in water at 60°C) and s is 0.07 cm. Reproducibility was checked by conducting triplicate measurements on one culture at a single temperature and a variety of light intensities; relative standard errors averaged 3.9 and 1.8% for mean gross and net photosynthesis rates, respectively.
Nucleotide sequence accession numbers.
Sequence data have been deposited with GenBank (accession no. AY884052 to AY884060).
RESULTS AND DISCUSSION
Synechococcus cultivation and genotypic analysis.
Synechococcus isolates were obtained from all except the highest-temperature site (59 to 70°C). Colony growth was highest on filters from the first two dilutions (100 and 10−1), typically resulting in a dense collection of microcolonies. Comparisons to direct microscopic counts revealed that only up to 10% of Synechococcus cells inoculated onto the filters produced microcolonies. Well-isolated colonies appeared on filter plates from higher dilutions (10−2 to 10−5), and these were used for subculture and purification. Colony growth was never achieved for samples diluted more than 10−5, and colony frequency was lower than expected on the basis of dilution conditions. We considered that these patterns (i.e., growth limited to low-dilution filters and colony number less than proportional with dilution) might reflect nutrient insufficiency of the medium and/or quorum sensing. However, medium supplementation with vitamins, NaHCO3, and pure-cultured contaminant and contaminated Synechococcus culture extracts did not permit recovery of more Synechococcus cells or purification free from heterotrophic contaminants (1).
Most isolates were identical at the 16S rRNA locus to the Synechococcus genotypes A, B′, and B previously detected in the mat (Table 1). Isolates with 16S rRNA genotypes A and B′ were also identical or nearly identical to predominant mat genotypes at the ITS locus. Two new genotypes not previously detected in the mat, labeled B″ and B‴, had 16S rRNA sequences that were 1 and 2 nucleotides (nt) different from 16S rRNA genotype B′ and ITS sequences that were even more divergent from the predominant mat sequences. Slight differences in cell morphology were noted among isolates of different genotypes (Fig. 1). Cells of the genotype A, B′, and B‴ isolates were typically approximately 8 to 9 μm long and rod shaped, sometimes with slight curvature. Cells of the genotype B isolate appeared to remain connected after division, forming short spiral-shaped chains. Cells of the genotype B″ isolate were usually 8 to 10 μm long and had a propensity to form small clusters. The failure to cultivate Synechococcus strains with genotypes A′ and A″ from high-temperature sites could be related to the need for higher light intensities. Meeks and Castenholz (12) found that a high-temperature Synechococcus strain from Hunter’s Hot Springs would grow only at 72°C at intensities greater than ∼146 μmol photons m−2 s−1; our cultures were provided ∼60 μmol photons m−2 s−1.
TABLE 1.
Genotypes of Synechococcus isolates obtained in this study
| Genotype-isolatea | Results for closest relative at locus
|
Collection date (mo/day/yr) | Site temperature (°C) | Strain(s)c | |||
|---|---|---|---|---|---|---|---|
| 16S rRNA type | Similarity/ no. of nt differences | ITSb | Similarity/ no. of nt differences | ||||
| A-NACy05a | A | 100/0 | NACy05a | 100/0 | 9/24/2000 | 58-65 | TS-64, -65, -66, -67, -71, -72, -75, -77, -83, -85, -88, -89, -94, -96 |
| 10/22/2000 | 49-56 | TS-37, -39, -41, -47, -49 | |||||
| 7/10/2002 | 52-65 | JA-1-3Aa, -1-3Ab, -1-3Ac, -1-3Ad | |||||
| 7/25/2002 | 58-65 | JA-3-3Aa, -3-3Ab, -3-3Ac | |||||
| A-NACy05d | A | 100/0 | NACy05a | 99/1 | 9/24/2000 | 58-65 | TS-62, -70, -76, -78, -81, -87, -93 |
| B′-NACy10b-1d | B′ | 100/0 | NACy10b | 100/0 | 7/10/2002 | 52-65 | JA-1-3B′a, -b, -c, -d, -e, -f, -g, 1-5 B′h, -i, -j |
| B′-NACy10b-2d | B′ | 100/0 | NACy10b | 100/0 | 9/24/2000 | 58-65 | TS-97, -98, -99, -100, -101, -102, -103, -107 |
| B′-NACy10o | B′ | 100/0 | NACy10b | 99/1 | 7/10/2002 | 51-61 | JA-2-3B′ a(2-13), -2-3 B′b, -2-4 B′c, -2-5 B′d |
| B″ | B′ | 99/1 | NACy10j | 99/4 | 7/10/2002 | 51-61 | JA-2-3B″ a(2-F6), -2-3 B′(1 diff)b |
| B″-104e | B′ | 99/1 | NACy10b | 99/3 | 9/24/2000 | 58-65 | TS-104 |
| B‴ | B′ | 99/2 | Unalignable | Unalignable | 9/24/2000 | 58-65 | TS-63, -73, -80, -84, -86, -91, -92 |
| 10/22/2000 | 49-56 | TS-42, -44, -45, -46 | |||||
| 7/10/2002 | 51-61 | JA2-3 B‴a | |||||
| B-P2 | B | 100/0 | P2 | 100/0 | 10/22/2000 | 49-56 | TS-11, -12, -15, -19, -28, -30, -34 |
Genotype designations match those previously used in our laboratory: A and B are used to designate 16S rRNA genotypes associated with the A and B Synechococcus clades, respectively (primes are used to signify variants within these clades).
ITS genotypes are designated with NA for North America and Cy (for cyanobacterial sequence) and a number-letter combination (e.g., 05a, 10b) indicating unique genotype, as in reference 18. Please note that genotypes B″ and B‴ were referred to as genotypes B′-1diff and B′-2diff in reference 1.
Designations in boldface characters represent strains used in experiments examining relationships between temperature and light.
One base difference between these two variants exists outside the region for which there is data for ITS type NACy10b.
One base different in ITS from B″.
Temperature and light effects on growth.
One isolate of each unique 16S rRNA genotype was selected for analysis of adaptation to temperature and light (Table 1). Synechococcus isolates with genotypes typically observed in situ, genotypes A, B′, and B, grew significantly faster at temperatures typical of the collection sites than did B″ and B‴ isolates, whose genotypes have never been detected in direct molecular analysis of the mat and are presumed to represent numerically less relevant populations (Fig. 3a). If strains with genotypes A, B′, and B also grow faster under in situ conditions, it could explain why these genotypes are more prevalent in the Octopus Spring microbial mat. It is interesting that strains with only 1- to 2-nt differences in their 16S rRNA sequences nevertheless have quite different thermal relations that are not representative of the temperature relations of isolates with the predominant mat genotypes (compare, for instance, the B′ isolate results with the B″ and B‴ isolate results in Fig. 3a).
FIG. 3.

Comparison of Synechococcus growth rates with respect to temperature: (a) Octopus Spring isolates B′, B″, and B‴; (b) Octopus Spring isolates with genotypes A, B′, and B predominating in situ; (c) Synechococcus groups II (B-like), III (A-like), and IV (A′-like) from Hunter’s Hot Springs, Oregon (modified from reference 14). Error bars correspond to ±1 standard error.
The A isolate had a higher upper temperature limit and broader temperature optimum (range, 40 to 65°C; optimum range, 50 to 60°C) than the B′ isolate (range, 40 to 60°C; optimum range, 50 to 55°C) (Fig. 3b). Similarly, the B′ isolate had a higher upper-temperature limit and broader optimum temperature range than the B isolate (range, 40 to 55°C; optimum, 50°C). These patterns were also observed in a repeat experiment (1). The differences in upper temperature limits observed for Synechococcus were not due to temperature relations of contaminants, as (i) contaminants cultivated from mixed cultures of each Synechococcus genotype exhibited a broad range of temperature optima (50 to 65°C) and maxima of 65 to 70°C and (ii) contaminants from the type B′ Synechococcus culture grew faster at higher temperatures than did contaminants from the type A Synechococcus culture. At 50°C, B and B′ isolates grew significantly faster than the A isolate, whereas at 55°C the A and B′ isolates grew significantly faster than the B isolate. At 60°C the A isolate grew significantly faster than B′ and B isolates. The A isolate was the only one capable of growth at 65°C. These results support the hypothesis that these predominant Synechococcus genotypes represent different temperature-adapted ecotypes and explain (at least in part) their distribution in the Octopus Spring thermal gradient (10). Competition and niche partitioning appear to play a major role. At temperatures at which all genotypes can exist (i.e., 50 to 60°C), the ability to grow faster ensures the success of one ecotype over another (i.e., the realized niche is narrower than the fundamental niche measured in our experiments).
Our results can be compared to data collected by Miller and Castenholz (14) on Synechococcus isolates from Hunter’s Hot Springs, which are related, but not identical, to the Octopus Spring type A- and B-like Synechococcus isolates (Fig. 3c). Growth was observed for their B-like (group II) isolates between 40°C and 61°C (optimum, 57°C), for their A-like (group III) isolates between 45°C and 63°C (optimum, 57°C), and for their A′-like (group IV) isolates between 55°C and 70°C (optimum, 65°C). Miller and Castenholz noted that as the upper temperature limit for growth was extended, an even larger shift upward in the minimum temperature was observed, leading them to suggest that increases in thermal specialization resulted in a decrease in the overall temperature range for growth. The Octopus Spring isolates obtained in our study had equivalent lower temperature limits, and only the upper temperature limit was extended. Unless this was a consequence of differences in experimental design, it does not appear that increased thermal adaptation must always correlate with a reduction in the ability to grow at lower temperatures (i.e., the evolutionary cost of extending the upper temperature limit may not always require a tradeoff of a narrowing niche breadth). This might be due to differences in detail between Octopus Spring, which undergoes periodic flow surges that cause temperatures to vary ∼5 to 10°C over ∼5-min cycles, and Hunter’s Hot Springs, where short-term temperature changes are not observed (R.W. Castenholz and S.R. Miller, personal communication). In both studies, however, isolates with an extended temperature limit appear to have a broad optimum range.
In general, results for Octopus Spring and Hunter’s Hot Springs correlate with the distribution of genotypes in Octopus Spring mat. The type A′-like Hunter’s Hot Springs isolate (group IV) shows a higher temperature optimum and range than do the type A-like isolates observed in both studies, consistent with DGGE results showing a higher temperature distribution for genotype A′ than for genotype A (10). The A-like isolates from both Octopus Spring and Hunter’s Hot Springs (group III) are able to grow at slightly higher temperature than the B-like isolates (group II Oregon isolates and B′ Octopus Spring isolate). Miller and Castenholz (14) noted the high degree of similarity between their Oregon group II and the Octopus Spring B genotype based on the number of 16S rRNA nucleotide differences. But direct assignment of comparable genotypes between the two systems is complicated by the fact that divergence can be due to geographic isolation as well as adaptation (14, 18). The Octopus Spring B′ isolate is able to grow at slightly higher temperature than the Octopus Spring B isolate, consistent with the in situ genotype distribution.
Isolates of all genotypes grew at all light intensities investigated (10 to 385 μmol photons m−2 s−1), and no obvious differences in ranges and optima were observed, such as the distinct differences reported for low- and high-light-adapted marine Prochlorococcus populations (15). If our isolates are differently adapted to light the adaptations must be subtle (see below). Isolates of all genotypes showed visual evidence of acclimation to light in that cells were greener when incubated at low light intensities, indicating higher chlorophyll content per cell. Type A and B′ isolates also showed elevated CO2 fixation at low light intensities when pregrown at low light intensities (1).
Photosynthesis responses to temperature and light.
Genotype A, B′, and B isolates showed increased gross photosynthesis with increasing light intensity until saturation occurred (Fig. 4, left panels). 14CO2 fixation experiments, which could also be used to estimate photosynthesis, showed similar results except that some evidence for reduced uptake at high light intensity was noted (1). The type A isolate was able to conduct gross photosynthesis at a higher temperature than the type B′ and B isolates, as a small net rate of oxygen production in light was still observed at 75°C for the A culture whereas the upper limit was 70°C for the B culture. The type B′ isolate was similar to the B isolate, but the photosynthetic activity decreased with time at 70°C, so we could not make reproducible determinations. For all isolates the highest photosynthetic rates (both net and gross) at the highest applied light intensity (1,100 mmol photons m−2 s−1) were measured at 65°C. All the cultures photosynthesized very poorly at 40°C. Temperature effects on photosynthetic rates at 440 μmol photons m−2 s−1 are shown in Fig. 5. 14CO2 fixation experiments also showed that the type A isolate had a higher upper temperature limit (70°C) than did the B′ and B isolates (65°C) (1). Miller has been able to use these and other temperature-adapted Synechococcus strains to observe evidence of molecular adaptation in the large subunit of the ribulose biphosphate carboxylase (13; D. Bhaya, M. M. Bateson, F. M. Cohan, A. R. Grossman, J. Johnson, A. Koeppel, N. Hamamura, J. Hostetler, N. Khuri, M. Kühl, M. C. Melendrez, S. R. Miller, S. Y. Rhee, C. Simpson, A.-S. Steunou, R. Stewart, L. J. Tallon, D. M. Ward, and J. Heidelberg, unpublished results) that could explain such differences in CO2 fixation with temperature. High levels of photosynthetic activity at temperatures that are not permissive of growth were found for other thermophilic Synechococcus strains (12, 25). This suggests that photosynthetic activity, a short-term measure of metabolic performance, is not a good determinant of growth rate adaptations; the individual components of the photosynthetic apparatus are apparently not restricted by temperature in the same manner as the organism as a whole.
FIG. 4.
Microsensor measurements of gross (left panels) and net (right panels) oxygenic photosynthesis as a function of light intensity and temperature for A, B′, and B isolates.
FIG. 5.
Gross photosynthetic rate of oxygenic photosynthesis at 440 μmol photons m−2 s−1 as a function of temperature measured for A, B′, and B isolates by use of an oxygen microsensor.
The respiration rate in the cultures increased with increasing temperature, and it was therefore worthwhile to study how net photosynthesis varied as a function of light intensity at different temperatures (Fig. 4, right panels) (Table 2). The compensation point (the light intensity where the rate of photosynthesis balances the rate of respiration) was only 5 to 15 μmol photons m−2 s−1 for all cultures at 50°C and increased only slightly until the upper temperature limit was approached. High intensities of ∼300 μmol photons m−2 s−1 were needed to induce a net oxygen evolution in the A culture at 75°C and in the B culture at 70°C. The photosynthetic activity was declining at 70°C for the B′ culture, so we could not estimate a compensation point. However, just 5°C below the upper temperature limits of the cultures, the compensations points were as low as 20 to 40 μmol photons m−2 s−1. Although quite low, the compensation points at 5°C below the upper temperature limits are still very close to the intensities we used for laboratory culture. Higher light intensities should thus be used for growth near the upper temperature limits. The B′ isolate showed a higher half-saturation light intensity for maximum photosynthesis than the A and B genotypes, but this might have been due to small differences in pregrowth conditions among the isolates.
TABLE 2.
Light adaptation parameters from microsensor studies of net photosynthesis
| Isolate | Compensation point (μmol photons m−2 s−1) at indicated temp (°C)
|
Half-saturation value (μmol photons m−2 s−1) | ||
|---|---|---|---|---|
| 65 | 70 | 75 | ||
| A | ∼15 | 35 | ∼300 | 70 |
| B′ | ∼20 | 220 | ||
| B | ∼40 | ∼300 | 80 | |
Conclusions.
This study yielded clear evidence of temperature-adapted Synechococcus ecotypes that is consistent with predominant genotypes found in situ at corresponding temperatures. Some of these ecotypes exhibit less than a 3% difference in 16S rRNA sequence (e.g., genotypes B and B′), providing another example of the problem with quantitative molecular cutoffs as species concepts (4, 30). Neither our growth nor our activity experiments yielded obvious evidence (e.g., differences in light range and optima) that would support the hypothesis that distinct Synechococcus genotypes might be either low-light or high-light adapted. Photosynthesis measurements are, at best, possibly consistent with subtle adaptation of the A and B isolates to lower light intensity than the B′ isolate. Since the A isolate displayed an acclimation response to light, it is possible that it is simply differently acclimated when found in different light regimens in different positions in the mat vertical profile. It is also possible that high- and low-light-adapted Synechococcus ecotypes of the same 16S rRNA genotype exist (see above and reference 7) and that our cultivation procedure favored the isolation of a particular type A ecotype that is not low-light adapted. The light intensity we used to retrieve Synechococcus cultures was near that at which subsurface Synechococcus populations orient vertically, possibly to evade high light at the brightest part of the day (20).
By isolating only a single variable (either temperature or light), a complete picture of the fitness (i.e., growth response) of Synechococcus populations with different genotypes relative to natural variations in both of these conditions might not have been obtained. Furthermore, environmental parameters other than temperature and light (e.g., nutrient availability, pH, and interactions with other microorganisms) may influence the overall fitness of these organisms and thus affect their distribution in the mat. Forthcoming genome data for the Octopus Spring Synechococcus A and B′ ecotypes characterized in this study (http://landresources.montana.edu/FIBR/; D. Bhaya, M. M. Bateson, F. M. Cohan, A. R. Grossman, J. Johnson, A. Koeppel, N. Hamamura, J. Hostetler, N. Khuri, M. Kühl, M. C. Melendrez, S. R. Miller, S. Y. Rhee, C. Simpson, A.-S. Steunou, R. Stewart, L. J. Tallon, D. M. Ward, and J. Heidelberg, unpublished results) may reveal further evidence of adaptation to temperature (and light?) and could give insight into other environmental factors affecting the niche adaptations of these organisms in situ.
Acknowledgments
This project was completed with funding from the National Science Foundation (Ecology Program award BSR-9708136) and from the Thermal Biology Institute at Montana State University (NASA Exobiology Program award NAG5-8807).
We appreciate the help of Tony Scotti in cultivating Synechococcus isolates from year 2000 samples, Preben Sørensen for making microsensors, Jim Robison-Cox and Melinda Yager for assisting us with the statistical analyses, and Scott Miller and two anonymous reviewers for providing comments on the manuscript.
REFERENCES
- 1.Allewalt, J. A. 2004. Temperature and light adaptations of Synechococcus isolates from a hot spring microbial community. M.S. thesis. Montana State University, Bozeman, MT.
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myersand, and D. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Castenholz, R. W. 1969. Thermophilic blue-green algae and the thermal environment. Bacteriol. Rev. 33:476-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohan, F. M. 2002. What are bacterial species? Annu. Rev. Microbiol. 56:457-487. [DOI] [PubMed] [Google Scholar]
- 5.de Bruyn, J., F. C. Boogerd, P. Bos, and J. G. Kuenen. 1990. Floating filters, a novel technique for isolation and enumeration of fastidious, acidophilic, iron-oxidizing, autotrophic bacteria. Appl. Environ. Microbiol. 56:2891-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferris, M. J., and C. F. Hirsch. 1991. Method for isolation and purification of cyanobacteria. Appl. Environ. Microbiol. 57:1448-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferris, M. J., M. Kühl, A. Wieland, and D. M. Ward. 2003. Cyanobacterial ecotypes in different optical microenvironments of a 68oC hot spring community revealed by 16S-23S rRNA internal transcribed spacer region variation. Appl. Environ. Microbiol. 69:2893-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferris, M. J., A. L. Ruff-Roberts, E. D. Kopczynski, M. M. Bateson, and D. M. Ward. 1996. Enrichment culture and microscopy conceal diverse thermophilic Synechococcus populations in a single hot spring microbial mat habitat. Appl. Environ. Microbiol. 62:1045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferris, M. J., and D. M. Ward. 1997. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 63:1375-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Islam, M. A. 2004. Einstein-Smluchowski diffusion equation: a discussion. Physica Scripta 70:120-125. [Google Scholar]
- 12.Meeks, J. C., and R. W. Castenholz. 1971. Growth and photosynthesis in an extreme thermophile, Synechococcus lividus (Cyanophyta). Arch. Mikrobiol. 78:25-41. [DOI] [PubMed] [Google Scholar]
- 13.Miller, S. R. 2003. Evidence for the adaptive evolution of the carbon fixation gene rbcL during diversification in temperature tolerance of a clade of hot spring cyanobacteria. Mol. Ecol. 12:1237-1246. [DOI] [PubMed] [Google Scholar]
- 14.Miller, S. R., and R. W. Castenholz. 2000. Evolution of thermotolerance in hot spring cyanobacteria of the genus Synechococcus. Appl. Environ. Microbiol. 66:4222-4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore, L. R., G. Rocap, and S. W. Chisholm. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464-467. [DOI] [PubMed] [Google Scholar]
- 16.Moré, M. I., J. B. Herrick, M. C. Silva, W. C. Ghiorse, and E. L. Madsen. 1994. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl. Environ. Microbiol. 60:1572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nold, S. C., and D. M. Ward. 1996. Photosynthate partitioning and fermentation in hot spring microbial mat communities. Appl. Environ. Microbiol. 62:4598-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papke, R. T., N. B. Ramsing, M. M. Bateson, and D. M. Ward. 2003. Geographical isolation in hot spring cyanobacteria. Environ. Microbiol. 5:650-659. [DOI] [PubMed] [Google Scholar]
- 19.Peary, J. A., and R. W. Castenholz. 1964. Temperature strains of a thermophilic blue-green alga. Nature 202:720-721. [Google Scholar]
- 20.Ramsing, N. B., M. J. Ferris, and D. M. Ward. 2000. Highly ordered vertical structure of Synechococcus populations within the one-millimeter-thick photic zone of a hot spring cyanobacterial mat. Appl. Environ. Microbiol. 66:1038-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Revsbech, N. P. 1989. An oxygen microelectrode with a guard cathode. Limnol. Oceanogr. 34:472-476. [Google Scholar]
- 22.Ruff-Roberts, A. L., J. G. Kuenen, and D. M. Ward. 1994. Distribution of cultivated and uncultivated cyanobacteria and Chloroflexus-like bacteria in hot spring microbial mats. Appl. Environ. Microbiol. 60:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheridan, R. P. 1976. Sun-shade ecotypes of a bluegreen alga in a hot spring. J. Phycol. 12:279-285. [Google Scholar]
- 24.Sheridan, R. P. 1979. Seasonal variation in sun-shade ecotypes of Plectonema notatum (Cyanophyta). J. Phycol. 15:223-226. [Google Scholar]
- 25.Sheridan, R. P., and T. Ulik. 1976. Adaptive photosynthesis responses to temperature extremes by the thermophilic cyanophyte Synechococcus lividus. J. Phycol. 12:255-261. [Google Scholar]
- 26.Simpson, G. G. 1961. Principles of animal taxonomy. Columbia University Press, New York, N.Y. [DOI] [PubMed]
- 27.Van Valen, L. 1976. Ecological species, multispecies, and oaks. Taxon 25:233-239. [Google Scholar]
- 28.Ward, D. M. 1998. A natural species concept for prokaryotes. Curr. Opin. Microbiol. 1:271-277. [DOI] [PubMed] [Google Scholar]
- 29.Ward, D. M., and R. W. Castenholz. 2000. Cyanobacteria in geothermal habitats, p. 37-59. In B. A. Whitton and M. Potts (ed.), Ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 30.Ward, D. M., and F. M. Cohan. Microbial diversity in hot spring cyanobacterial mats: pattern and prediction, p. 185-202. In W. P. Inskeep and T. R. McDermott (ed.), Geothermal biology and geochemistry in Yellowstone National Park, in press. Thermal Biology Institute, Montana State University, Bozeman, MT.
- 31.Ward, D. M., M. J. Ferris, S. C. Nold, and M. M. Bateson. 1998. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 62:1353-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward, D. M., R. T. Papke, U. Nübel, and M. C. McKitrick. 2002. Natural history of microorganisms inhabiting hot spring microbial mat communities: clues to the origin of microbial diversity and implications for microbiology and macrobiology, p. 25-48. In J. T. Staley and A.-L. Reysenbach (ed.), Biodiversity of microbial life. Wiley-Liss, Inc., New York, N.Y.
- 33.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 344:63-65. [DOI] [PubMed] [Google Scholar]
- 34.Weller, R., M. M. Bateson, B. K. Heimbuch, E. D. Kopczynski, and D. M. Ward. 1992. Uncultivated cyanobacteria, Chloroflexus-like inhabitants, and spirochete-like inhabitants of a hot spring microbial mat. Appl. Environ. Microbiol. 58:3964-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]






