Abstract
Representatives of the genus Legionella were detected by use of a real-time PCR method in all water samples collected directly after treatment from 16 surface water (SW) supplies prior to postdisinfection and from 81 groundwater (GW) supplies. Legionella concentrations ranged from 1.1 × 103 to 7.8 × 105 cells liter−1 and were significantly higher in SW treated with multiple barriers at 4°C than in GW treated at 9 to 12°C with aeration and filtration but without chemical disinfection. No Legionellae (<50 CFU liter−1) were detected in treated water by the culture method. Legionella was also observed in untreated SW and in untreated aerobic and anaerobic GW. Filtration processes in SW and GW treatment had little effect or increased the Legionella concentration, but ozonation in SW treatment caused about 1-log-unit reduction. A phylogenetic analysis of 16S rRNA gene sequences of 202 clones, obtained from a selection of samples, showed a high similarity (>91%) with Legionella sequences in the GenBank database. A total of 40 (33%) of the 16S rRNA gene sequences obtained from treated water were identified as described Legionella species and types, including L. bozemanii, L. worsleiensis, Legionella-like amoebal pathogen types, L. quateirensis, L. waltersii, and L. pneumophila. 16S rRNA gene sequences with a similarity of below 97% from described species were positioned all over the phylogenetic tree of Legionella. Hence, a large diversity of yet-uncultured Legionellae are common members of the microbial communities in SW and GW treated at water temperatures of below 15°C.
Reported annual incidences of Legionnaires’ disease, a life-threatening pneumonia, typically range from about 5 per million (e.g., in the United States) to nearly 20 per million (in several European countries), but many cases are either not diagnosed or not reported (34, 61). Sporadic community-acquired cases with unidentified origin of the etiologic agent prevail, but large outbreaks continue to occur as well (19). The outbreak of legionellosis among visitors and participants at a flower show in The Netherlands in 1999 caused 31 deaths and more than 200 cases of disease. This demonstrated once again the potential public health risk of the presence of Legionella pneumophila in water, especially when usage leads to aerosolization. A whirlpool on display at the entrance of the flower show was identified as the source of the outbreak (15). L. pneumophila is responsible for more than 90% of the reported cases of legionellosis (17, 61). Currently, about 50 Legionella species have been defined, nearly half of which have been associated with cases of disease (e.g., L. pneumophila, L. micdadei, L. bozemanii, L. longbeachae, and L. dumoffii) (17, 35, 40).
L. pneumophila is ubiquitous in natural freshwater environments, including hot springs, and is also a common inhabitant of engineered water systems, such as treated sewage, cooling towers, and hot water systems (17, 18, 42, 44, 50, 58). The organism has been isolated from these environments at temperatures below 10°C (surface water) to 60°C (engineered water systems), but growth of L. pneumophila is restricted to temperatures between 20 and 43°C (49, 59). L. pneumophila has been detected only sporadically by use of the cultivation method in untreated groundwater (GW) and in treated water at temperatures below 20°C (4, 14, 20, 24, 56). However, cultivation-independent techniques, including immunological methods, fluorescent in situ hybridization (FISH), and PCR-based methods, clearly revealed the common presence of Legionella species in aquatic environments, even at temperatures below 20°C (11, 12, 14, 44, 50). In addition, FISH methods showed that undefined Legionella species may represent up to 7% of biofilms grown in treated water (52). 16S rRNA gene sequences related to L. parisiensis, L. maceachernii and Legionella-like amoebal pathogen (LLAP) species were observed in slow sand filters operating at temperatures below 20°C and were used for fungal plant pathogen suppression (13).
GW, both aerobic and anaerobic, and surface water (SW) are used as sources for the production of drinking water in The Netherlands at a ratio of approximately two to one. GW is treated and distributed without chemical disinfection. SW is treated with multiple barriers against chemical and microbiological contaminants and distributed without or with a very low disinfectant residual. Contamination of drinking water in the distribution system is prevented by a series of protective measures, including maintenance of high pressure and cross-connection control. Regrowth is limited by a far-reaching removal of growth-promoting compounds from the water to achieve biological stability (57). A number of reports showed that L. pneumophila multiplying in hot-water systems in buildings originated from the water supply (27, 27, 37). The aim of our study was to elucidate the role of drinking water in the distribution of Legionella. For this purpose, an investigation on the presence and identity of Legionella in treated water was conducted using a quantitative real-time PCR method as well as a standardized culture method to ensure that all types of Legionella were included in the analysis. Furthermore, the genetic diversity of Legionella was studied to determine (i) the presence of pathogenic Legionella species and (ii) the diversity of the indigenous Legionella types.
MATERIALS AND METHODS
Water sampling sites.
Samples of treated water were collected from a total of 82 GW treatment plants and 16 SW plants, covering 67% of the total drinking water production in The Netherlands. The GW plants included 72 plants using anaerobic GW, which in most cases is treated with aeration to introduce oxygen and to remove methane. Subsequently, water is treated by one or two stages of rapid sand filtration to remove ammonia, iron, and manganese. Aerobic GW is either distributed without treatment or aerated to add oxygen and to remove CO2, followed by limestone filtration. No chemical disinfectant is used in GW treatment and distribution. The concentration of dissolved natural organic matter, measured as nonparticulate organic carbon, in treated GW ranges from below 0.5 mg liter−1 in aerobic GW to about 7 mg liter−1 in an anaerobic GW supply. The temperature of treated GW ranges from 9 to 12°C throughout the year, attaining a maximum of 15°C in a few plants, whereas the temperature of treated SW ranges from about 3°C in winter to 22°C in summer. SW treatment includes either storage in open reservoirs or soil passage, followed by a combination of the following processes: coagulation and sedimentation, chlorination, ozonation, dual medium filtration, granular activated carbon (GAC) filtration, and slow sand filtration. In one treatment plant, reverse osmosis is used as the final treatment process. Postdisinfection with a low residual concentration (<0.1 mg liter−1) of chlorine dioxide or chlorine is applied when GAC filtration is the final treatment step. Each water sample was taken directly after the final treatment step but before postdisinfection when applicable. In addition, SW samples were collected from different rivers and from open basins for storage or collection of pretreated SW. Untreated GW was sampled from anaerobic and aerobic sources.
Water filtration and DNA extraction.
At each sampling location a volume of 2 liters was collected in a glass container, which had been heat treated (4 h at 150°C) to ensure the absence of DNA contamination. The samples were stored at 4°C and processed within 24 h. DNA-free water was analyzed in each experiment to check for possible DNA contamination during filtration, DNA extraction, and PCR amplification. Samples were filtered through a 25-mm polycarbonate filter (0.22-μm pore size, type GTTP; Millipore, Amsterdam, The Netherlands) in volumes of approximately 1 liter for treated water and GW and 10 ml for samples of SW and open storage reservoirs. Subsequently, DNA was isolated and purified using a MagNA Pure LC DNA isolation kit III following the instructions of the manufacturer (Roche Diagnostics, Almere, The Netherlands). In brief, filters were transferred to a six-well multidish (Nuclon; Nalge Nunc International, Neerijse, Belgium). To each well 120 μl lysis buffer and 30 μl proteinase K were added, and the dishes were incubated at 65°C for 10 min. Following incubation, the suspension of lysed cells was transferred from each well to a new microtube. The filters in the wells of the multidish were washed with 300 μl binding/lysis buffer, and this buffer was transferred to the same microtubes. Washing of the filter with the binding/lysis buffer was performed to maximize DNA recovery. Magnetic glass beads (150 μl) were added to bind DNA. After incubation for 10 min at room temperature, the magnetic glass particles with bound DNA were concentrated using a magnet (Dynal Biotech S.A., Compiègne, France). Subsequently, beads were washed twice with washing buffer. Finally, the DNA was eluted from the beads in 100 μl elution buffer and analyzed by PCR.
Detection of Legionella in SW and reservoir water.
A semiquantitative (dilution) PCR was applied for assessing the concentrations of DNAs of Legionella and L. pneumophila in SW samples and samples from open basins. In this test, undiluted and decimal dilutions (100, 10−1, 10−2, and 10−3) of the isolated DNA were analyzed in duplicate in the PCR. A second dilution series (10−4 and 10−5) was analyzed when all dilutions were positive in the first series. Legionella was detected using LEG-225 (5′ AAGATTAGCCTGCGTCCGAT) and LEG-858 (5′ GTCAACTTATCGCGTTTGCT) Legionella-specific primers (38) targeting the 16S rRNA gene. The amplification resulted in a DNA fragment of approximately 654 bp, enabling genetic analysis to determine the diversity of the detected Legionella. The primers LmipL920 (5′ GCTACAGACAAGGATAAGTTG) and LmipR1548 (5′ GTTTTGTATGACTTTAATTCA), targeting the mip gene, were used for the specific detection of L. pneumophila (32). Identical amplification mixtures were used for both specific reactions. A 25-μl reaction mixture contained 10 μl of each template DNA solution, 3 U of Platinum Taq DNA polymerase (Invitrogen, Breda, The Netherlands) with the supplied PCR buffer, 0.2 μM of primers LEG-225 and LEG-858 or LmipL920 and LmipR1548, 0.2 mM of each deoxynucleoside triphosphate, 3 mM of MgCl2, and 0.4 mg ml−1 bovine serum albumin (PCR grade; Roche Diagnostics, Almere, The Netherlands). Amplification was performed in an ABI 9700 thermocycler (Applied Biosystems, Foster City, Calif.) with a PCR thermal profile consisting of an initial incubation for 2 min at 94°C; 40 cycles of 20 s at 94°C, 30 s at 60°C, and 40 s at 72°C; and finally a postamplification step of 2 min at 72°C. Subsequently, PCR products were analyzed by agarose gel electrophoresis. The initial DNA concentration was calculated from the highest DNA dilution that yielded a PCR product.
The semiselective buffered charcoal yeast extract (BCYE) agar medium supplemented with antibiotics was used to detect culturable Legionella (16, 41). Untreated SW and water from open storage reservoirs contain nonspecific bacteria which hamper the growth of Legionella on this medium. Therefore, no concentration step was included in the procedure, and sample volumes of 100 μl were spread directly over 10 plates of BCYE agar medium, resulting in a detection limit of 1,000 CFU/liter. The plates were examined for the presence of typical Legionella colonies after 5 and 7 days of incubation at 37°C.
Detection of Legionella in GW and in treated water.
Untreated GW and all samples of treated GW and SW were analyzed for the presence of Legionella species (DNA) by using real-time PCR with SYBR green. The above-mentioned dilution PCR method was used to detect the presence of L. pneumophila, and the culture method was also used with these samples. In the Legionella species real-time PCR assay, the primers LEG-225 and LEG-858 were also used. Amplification, detection, and data analysis were performed with the iCycler IQ real-time detection system (Bio-Rad Laboratories B.V., Veenendaal, The Netherlands). The amplification mixture of 50 μl contained 25 μl of 2× Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen, Breda, The Netherlands), 10 μl of DNA template, 0.2 μM of primers LEG-225 and LEG-858, and 0.4 mg ml−1 bovine serum albumin (PCR grade; Roche Diagnostics, Almere, The Netherlands). The number of PCR cycles after which the amplified DNA is detected in the real-time PCR system (threshold cycle) is used to quantify the concentration of Legionella. This threshold cycle depends on the initial concentration of the target DNA. The undiluted sample and the 10−1 dilution of each DNA sample were analyzed in duplicate, resulting in four different PCRs. The PCR thermal profile was identical to the profile used for the dilution PCR method.
The average number of threshold cycles for each water sample was transformed into numbers of cells by using a calibration curve obtained with known concentrations of L. pneumophila cells. For this purpose a freshly grown colony was collected from the BCYE medium and suspended in distilled water free of DNase and RNase (Invitrogen, Breda, The Netherlands). The cells were enumerated by the acridine orange direct count method using epifluorescence microscopy (22). Volumes of 1.5 ml of the suspension were stored at −80°C. For each sample series, 1 ml of the reference cell suspension containing 3.5 × 104 ± 9.1 × 103 Legionella cells was filtered, and DNA was isolated by the procedure described above. Subsequently, DNA was eluted in 50 μl DNA-free water, and 10 μl was added to each PCR. A dilution series containing approximately 104, 103, 101, 102, and 1 cells was prepared to generate a calibration curve, and each DNA dilution was analyzed in duplicate. The calibration curve was calculated automatically with the software supplied by the iCycler IQ real-time detection system (Bio-Rad Laboratories B.V., Veenendaal, The Netherlands), according to the average threshold cycle and the highest possible r2 value. The concentration of the initial Legionella DNA in each sample was calculated using the threshold cycle values of the duplicates of the undiluted and diluted DNA solutions and the calibration curve.
The culture method with BCYE medium was also used to detect Legionella bacteria in treated water (41). For this purpose, 500 ml of the sample was filtered through a 47-mm polycarbonate filter with pore size of 0.2 μm (Sartorius Technologies BV, Nieuwegein, The Netherlands). The filters were sonicated upside down in 5 ml of sample water for 2 min using an ultrasonic water bath (42 KHz, 135 W) (Branson 5510; FMH Medical, Veenendaal, The Netherlands). Volumes of 100 μl of the obtained suspension were spread over BCYE medium with and without antibiotics and cysteine, as described above (detection limit, 50 CFU liter−1).
Cloning and sequencing of PCR products.
The genetic diversity of the Legionella community in SW, storage basins, and treated water was determined by cloning and sequence analysis of the PCR products of the genus-specific primer pair LEG-225 and LEG-858. The PCR products obtained from the undiluted target DNA sample were cloned into pGEM-T Easy Vector System II (Promega, Leiden, The Netherlands) according to the instructions of the manufacturer. Briefly, the PCR products were ligated overnight into the pGEM-T plasmid at 4°C and were subsequently transferred to competent Escherichia coli JM109 cells by heat shock treatment. The transformed cells were incubated overnight at 37°C on standard LB medium with ampicillin, IPTG (isopropyl-β-d-thiogalactopyranoside), and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). White colonies were screened for the correct insert size by PCR using the standard primers T7 and SP6 located outside the cloning site of the pGEM-T plasmid, followed by agarose gel electrophoreses. The DNA insert was sequenced in both the forward and reverse directions using primers T7 and SP6. If possible, the sequences of 5 clones of each selected sample of treated water and 10 clones of each selected SW sample were analyzed.
Nucleotide sequence data analysis.
The two sequences of a cloned 16S rRNA gene sequence were assembled and edited by using the software package DNAStar Seqman II (Lasergene Inc., Madison, Wis.). The edited sequences were aligned and manually checked using ClustalW (http://www.ebi.ac.uk/clustalw/). A BlastN search (4a) was performed to analyze the similarity of the clone sequences to 16S rRNA gene sequences available in the GenBank database. The primer sequences at both sides of the sequence were removed, and the obtained sequences of approximately 614 bp were imported and aligned into the 16S rRNA ARB database of January 2004 (31). The aligned sequences were added to the main tree by using the parsimony tool with local optimization included in the ARB program. Sequences of species of Piscirickettsia, Wolbachia, and Coxiella which were most closely related to the Legionella cluster were used as an outgroup in this tree. 16S rRNA gene sequences with 99% and more similarity were grouped.
Nucleotide sequence accession numbers.
All partial 16S rRNA gene sequences from the 16S rRNA gene sequences determined in this study have been deposited in GenBank under accession no. AY923985 to AY924186.
RESULTS
Legionella in SW and in open storage basins.
Legionella species were detected by the dilution PCR method in all 10 SW samples and in all 4 samples from open basins (Table 1). These samples were all collected in the winter period (January and February) at a water temperature of 3 to 4°C. The concentration of Legionella in the river water samples was in most cases greater than 106 16S rRNA gene copies liter−1. In the open storage basins, containing pretreated SW, clearly lower concentrations were observed, ranging from 2.5 × 103 to 1.4 × 104 16S rRNA gene copies liter−1. L. pneumophila was detected at concentrations of between 1.0 × 103 and 2.0 × 104 mip gene copies liter−1 in four SW samples, comprising 0.1 to 1% of the total Legionella concentration. In the other samples the L. pneumophila concentration was below the detection limit of 1 × 103 16S rRNA gene copies liter−1. Culturable Legionella bacteria were not detected in any of these samples (detection limit, 1,000 CFU liter−1).
TABLE 1.
Legionella concentrations in untreated and treated water as determined with the quantitative real-time PCR method
| Water type (no. of samples) |
Legionella concn (cells liter−1)
|
||
|---|---|---|---|
| Minimum | Median | Maximum | |
| Surface water (10)a | 2.0 × 104 | 1.0 × 106 | 2.5 × 106 |
| Storage basin (4)a | 2.5 × 103 | 1.9 × 104 | 1.4 × 105 |
| Anaerobic groundwater (3 out of 7) | <2.0 × 102 | 5.5 × 102 | 2.4 × 103 |
| Aerobic groundwater (8 out of 9) | <2.0 × 102 | 8.3 × 103 | 2.5 × 104 |
| Treated surface water (16) | 2.1 × 104 | 4.1 × 104 | 7.8 × 105 |
| Treated anaerobic groundwater (70) | 1.1 × 103 | 1.4 × 104 | 1.7 × 105 |
| Treated aerobic groundwater (11) | 2.9 × 103 | 2.3 × 104 | 9.8 × 104 |
Water samples analyzed with the dilution PCR method (concentrations in 16S rRNA gene copies liter−1).
Legionella in GW and in treated water.
A total of 16 samples of untreated GW, 82 samples of treated GW, and 16 samples of treated SW taken directly after treatment were quantitatively analyzed for the presence of Legionella by using a real-time PCR method. The amplification efficiency as determined with dilutions of the suspension of L. pneumophila was above 85% in all sample series, and the correlation coefficient (r2value) of the standard curve was high (0.99). Each DNA sample was analyzed undiluted and as a 10−1 dilution, both in duplicate. The relative standard deviations (RSDs) of the concentrations obtained for the PCR duplicates and for the two DNA dilutions were calculated to assess the quality of the quantitative measurements. The average RSDs of the obtained concentrations were 13% (for the undiluted duplicates; n = 97), 26% (for the 10−1 duplicates; n = 79) and 6% (for the two DNA concentrations; n = 79). These values demonstrate that the quantification of the DNA concentration is highly reproducible.
Legionella was detected in three out of seven samples of anaerobic GW at a maximum concentration of 2.4 × 103 cells liter−1 (Table 1). In one sample the Legionella concentration was below the detection limit of 2.0 × 102 cells liter−1, and in three water samples the PCR was inhibited, as was demonstrated by spiking with the L. pneumophila suspension (results not shown). The Legionella concentrations in eight of nine samples of aerobic GW were above the detection limit and ranged from 2.7 × 103 to 2.5 × 104 cells liter−1 (Table 1). Legionella was detected in 33 of 34 samples of water collected from different treatment stages in 25 treatment plants (8 SW and 17 GW). Legionella was below the detection limit in water sampled after reverse osmosis. Increased Legionella concentrations (>1 log unit) were observed directly after aeration and after sand filtration of anaerobic GW. Effects of granular activated carbon filtration, applied in SW treatment, ranged from a decrease to no effect or an increase (1 log unit) of the Legionella concentration. Ozonation caused a clear (>1-log-unit) reduction (results not shown).
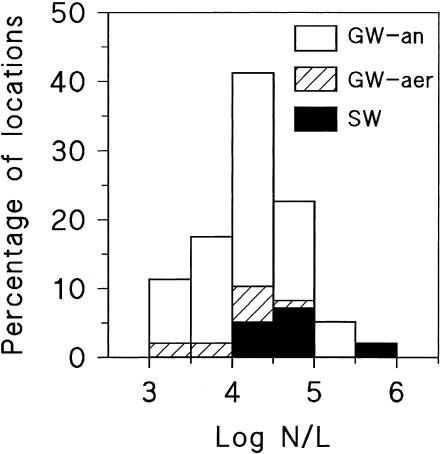
All samples of treated water taken from 97 different treatment plants contained Legionella species at concentrations ranging from 1.1 × 103 to 7.8 × 105 cells liter−1 (Fig. 1). The highest Legionella concentration was observed in treated SW, with GAC filtration as the final treatment step (before postdisinfection). L. pneumophila was not detected in any of these water samples by using the dilution PCR method (detection limit, 100 mip gene copies liter−1). Furthermore, no culturable Legionella organisms were detected (detection limit, 50 CFU liter−1) in treated water. The concentrations in treated SW differed significantly (P < 0.05) from those in treated aerobic and anaerobic GW, which did not significantly differ from each other.
FIG. 1.
Frequency distributions of Legionella concentrations (N, cells/16S rRNA gene copies per liter) in treated water from 97 treatment plants analyzed by a real-time PCR method using the 16S rRNA gene-targeting primers LEG-225 and LEG-858. GW-an, groundwater supply with anoxic source water; GW-aer, groundwater supply with aerobic source water; SW, surface water supply.
Sequence diversity of detected Legionella 16S rRNA genes.
Cloning and sequencing of the PCR products of the Legionella genus-specific PCR primers LEG-225 and LEG-858 (654 bp) resulted in 49 16S rRNA gene sequences from seven SW samples and 29 sequences from three storage basins. Furthermore, a total of 123 16S rRNA gene sequences were obtained from treated water samples collected at eight SW treatment plants (50 sequences), 10 anaerobic GW treatment plants (49 sequences), and 6 aerobic GW treatment plants (25 sequences). All 202 16S rRNA gene sequences exhibited the greatest similarity to 16S rRNA-encoding genes from defined Legionella spp., confirming the specificity of the PCR primers used. Similarities between the 16S rRNA gene sequences and Legionella sequences deposited in the GenBank database ranged from 91% to a maximum of 99%. Even 16S rRNA gene sequences with a similarity value of 91% had Legionella species as the nearest relative. Sequences with a similarity equal to or above 97% are considered to represent the same species (55). A total of 30 (38%) 16S rRNA gene sequences obtained from SW and open collection basins and 40 (32%) of the 16S rRNA gene sequences from treated water samples showed similarity values of ≥97% with 18 Legionella species and a number of LLAP types (Table 2). Relatedness to L. pneumophila was observed in four 16S rRNA gene sequences originating from three samples of untreated SW (97 to 99% similarity) and in one from a sample of treated GW (98% similarity). Most (18 out of 20) 16S rRNA gene sequences related to LLAP types had SW-related origins, viz., untreated SW (9 sequences), open storage basins (4 sequences), or treated SW (5 sequences). Other Legionella species with high similarities to 16S rRNA gene sequences from SW(-related) samples were L. worsleiensis (5 sequences), L. bozemanii (5 sequences), and L. donaldsonii (3 sequences). Legionella species most frequently related to 16S rRNA gene sequences from treated GW were L. worsleiensis (6 sequences), L. bozemanii (5 sequences), L. quateirensis (4 sequences), and L. adelaidensis (2 sequences). The 16S rRNA gene sequences related to L. bozemanni predominated in two samples, viz., the GAC filtrate of an SW supply and the rapid sand filtrate of an anaerobic GW supply. Furthermore, 16S rRNA gene sequences related to L. lytica and L waltersii predominated in GAC filtrates of SW supplies, and L. quateirensis predominated in the limestone filtrate of an aerobic GW supply.
TABLE 2.
Legionella species or types related to 16S rRNA gene sequences (similarities of >97%) obtained from untreated surface water, pretreated surface water from open storage basins, treated surface water, and treated groundwater, in decreasing frequency of occurrence
| Species or type | Human pathogena | No. of sequences obtained from:
|
Total no. of sequences (%) | ||||
|---|---|---|---|---|---|---|---|
| SW | Open storage basin | Treated SW | Treated anaerobic GW | Treated aerobic GW | |||
| L. worsleiensis | − | 2 | 2 | 1 | 5 | 1 | 11 (5.5) |
| L. bozemanii | + | 1 | 1 | 3b | 5 (3b) | 10 (5.0) | |
| L. lytica (LLAP 9) | + | 1 | 1 | 3b | 5 (2.5) | ||
| L. pneumophila | + | 3 | 1 | 1 | 5 (2.5) | ||
| L. waltersii | − | 1 | 3b | 1 | 5 (2.5) | ||
| L. quateirensis | − | 1 | 1 | 3b | 5 (2.5) | ||
| L. donaldsonii | − | 1 | 1 | 1 | 3 (1.5) | ||
| LLAP 2c | − | 1 | 1 | 1 | 3 (1.5) | ||
| LLAP 7c | − | 2 | 1 | 3 (1.5) | |||
| L. adelaidensis | − | 1 | 1 | 2 (1.0) | |||
| L. dumoffii | + | 1 | 1 | 2 (1.0) | |||
| L. londiniensis | − | 1 | 1 | 2 (1.0) | |||
| LLAP 4d | + | 1 | 1 | 2 (1.0) | |||
| L. rowbothamii (LLAP 6) | − | 2 | 2 (1.0) | ||||
| L. anisa | + | 1 | 1 (0.5) | ||||
| L. fairfieldensis | − | 1 | 1 (0.5) | ||||
| L. drozanski (LLAP 1) | + | 1 | 1 (0.5) | ||||
| LLAP 3c | + | 1 | 1 (0.5) | ||||
| LLAP 8 | − | 1 | 1 (0.5) | ||||
| L. fallonii (LLAP 10) | + | 1 | 1 (0.5) | ||||
| LLAP 11d | − | 1 | 1 (0.5) | ||||
| L. micdadei | + | 1 | 1 (0.5) | ||||
| L. steigerwaltii | − | 1 | 1 (0.5) | ||||
| L. wadsworthii | − | 1 | 1 (0.5) | ||||
| Total identified (%) | 17 (34.7) | 13 (44.8) | 15 (30) | 17 (34.7) | 8 (32) | 70 (34.7) | |
| Total isolated | 49 | 29 | 50 | 49 | 25 | 202 (100) | |
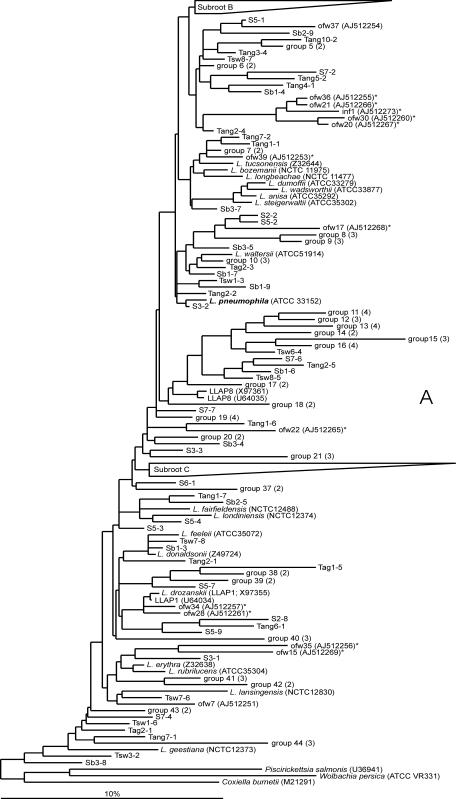
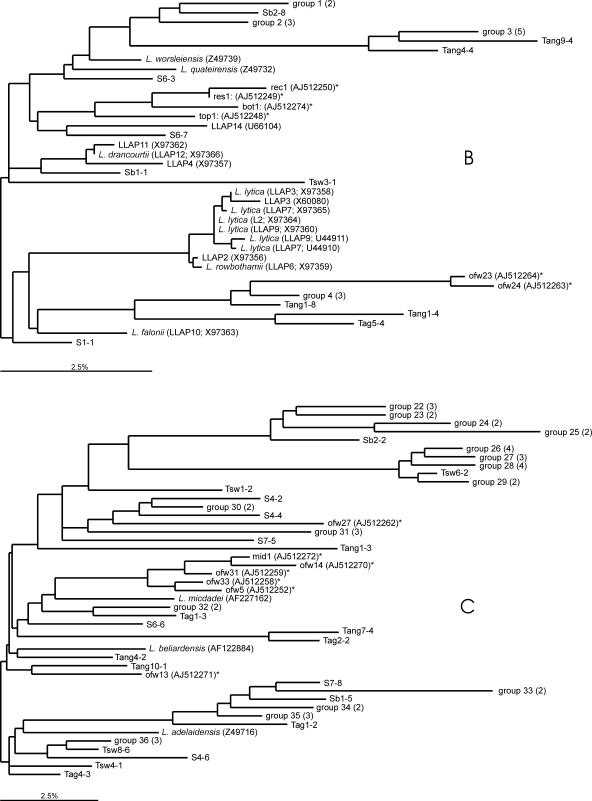
A total of 65% of the 202 16S rRNA gene sequences had similarities of below 97% with defined Legionella species and may belong to not-yet-described Legionella types. A phylogenetic tree was calculated using the ARB program (31) (Fig. 2). Parts of the tree, e.g., LLAP-related sequences, are presented separately (Fig. 2B and C). The various 16S rRNA gene sequences created a highly diverse tree without major clusters. A total of 44 groups, containing 16S rRNA gene sequences with similarities of ≥99%, are present in the tree. Sixteen of these groups contain a total of 38 16S rRNA gene sequences originating from identical water samples, and 28 groups contain sequences from different water samples. Hence, certain groups predominated in specific samples, e.g., groups 3 and 4 in treated aerobic GW (limestone filtrate); groups 8 and 19 in treated anaerobic GW supplies (filtrates of rapid sand filters); and groups 13, 27, 31, 40, and 41 in treated SW (GAC filtrates). Groups 22 to 25, with a total of 14 16S rRNA gene sequences, mostly originating from SW samples and samples of treated SW, formed a distinct branch in the tree (Fig. 2C). This branch, with a minimal internal sequence similarity of 96% and a minimal 91 to 93% similarity with Legionella sequences deposited in the GenBank database, possibly represents a surface water phylotype. Another distinct branch is formed by groups 26, 27, 28, and 29. The 16S rRNA gene sequences in these groups all originated from different types of SW or treated SW, with no 16S rRNA gene sequences from untreated SW and treated SW at specific plants. In most water samples, a variety of different 16S rRNA gene sequences were detected. The observations show that certain defined Legionella species as well as distinct, yet-unknown Legionella species are common members of the microbial communities in SW and GW, both untreated and treated at temperatures below 15°C.
FIG. 2.
Phylogenetic trees showing the positions of the 202 16S rRNA gene sequences of the partial 16S RNA gene amplified using Legionella-specific primers LEG-225 and LEG-858. The subtree in panel B includes mostly LLAP-related 16S rRNA gene sequences, and that in panel C contains sequences mostly originating from SW and samples from treated SW. The 16S rRNA gene sequences shown in the trees are derived from seven SW samples (S1 to S7), three storage basins (Sb1 to Sb3), eight SW treatment plants (Tsw1 to Tsw8), ten anaerobic GW treatment plants (Tang1 to Tang10), and eight aerobic GW treatment plants (Tag1 to Tag8). The trees were constructed by using the parsimony tool with local optimization. The bar represents percentage sequence difference. Asterisks indicate 16S rRNA gene sequences from slow sand filters (13). Origin of 16S rRNA gene sequences included in groups (≥99% similarity): 1, S2- and S7-1; 2, Tang10-3, Tag3-3, and Sb3-10; 3, Tang8-5, Tag4-1, Tag4-2, and Tag4-5; 4, Tag3-1, Tag3-2, and Tag3-5; 5, Tsw5-3 and Tang6-2; 6, Tang10-4 and Tang7-3; 7, Tsw8-1 and Tsw8-2; 8, Tang8-1, Tang8-3, and Tang8-4; 9, Tang8-2, Tsw1-8, and Tang4-5; 10, Tsw4-2, Tsw4-4, and Tsw4-5; 11, Tag2-4, Tag2-5, Tag6-1, and Tang6-3; 12, Tsw7-4, Tag3-4, and Sb1-2; 13, Tsw5-2, Tsw5-4, Tsw5-5, and Tsw4-3; 14, Sb3-3 and Sb3-6; 15, Tang9-3, Tag5-3, and Tang2-3; 16, Tang3-1, Tang5-1, Tang5-3, and Tang5-5; 17, Tang10-5 and Sb3-1; 18, S2-1 and S2-6; 19, Tang9-2, Tang3-2, Tang3-3, and Tang3-5; 20, Sb3-2 and Sb3-9; 21, Tang5-4, Tang1-2, and Tang1-5; 22, S4-1, Tang4-3, and S4-8; 23, S7-3 and Tsw1-5; 24, Tsw3-4 and Tag1-4; 25, Sb2-2 and Sb2-6; 26, S5-5, S6-2, S4-9, and S2-9; 27, Tsw6-1, Tsw6-6, and Tsw6-9; 28, Tsw6-7, S1-2, S1-3, and Tsw6-8; 29, Tsw6-3 and Tsw6-5; 30, S5-6 and S5-8; 31, Tsw1-1, Tsw1-4, and Tsw1-7; 32, S2-4 and S2-5; 33, Tsw8-3 and Tsw8-4; 34, S2-3 and S7-9; 35, S4-5, S4-3, and S4-7; 36, Tsw5-1, Tang9-1, and Tsw3-3; 37, Sb1-8 and Sb1-10; 38, Tag5-1 and Tag5-2; 39, S6-4 and S6-5; 40, Tsw7-1, Tsw7-2, and Tsw7-3; 41, Tsw7-5, Tsw7-7, and Tsw7-9; 42, Tsw2-1 and Tsw2-2; 43, Tsw2-3 and Tag1-1; and 44, Sb2-1, Sb2-4, and Sb2-7.
DISCUSSION
Detection of Legionella in aquatic environments.
A steadily increasing number of Legionella species, presently a total of about 50, have been defined since the discovery of L. pneumophila in 1976. About half of these species are associated with disease, but L. pneumophila still is by far the most prominent reported pathogen (17, 61). A variety of methods, including guinea pig inoculation, fluorescent-antibody (FA) techniques, culture techniques, FISH, and PCR-based assays, have been applied to detect L. pneumophila in environmental samples. Culture methods, which enable the quantitative detection of culturable bacteria and isolation of strains, are commonly used but have a number of limitations, viz., a long incubation period, growth of competing background bacteria, and recovery reduction by antibiotics and sample treatment (10, 16, 28, 48, 60). Reduced recoveries or inability to grow on solid media strongly limit the use of these methods for detection of non-L. pneumophila species (28, 39). FA-based methods are available for rapid and specific detection of L. pneumophila, resulting in concentrations exceeding the culturable counts, but do not differentiate between dead and viable cells (3, 5, 9). A number of defined culturable, non-L. pneumophila species can also be detected with FA-based methods (42). Uncultured yet undefined Legionella types, however, remain undetected with this method. Several so-called LLAPs can be isolated in amoebal coculture, but this method is not attractive for quantitative detection (26, 51). FISH methods have a potential for the detection of individual Legionella species and differentiate between active and inactive (dead) cells, but their application is laborious and the detection level is relatively high (12, 62). PCR-based methods, which enable quantitative detection even at low concentrations, therefore were used to determine the presence of L. pneumophila, non-L. pneumophila species, and yet-undefined Legionella types in treated water, despite the inability to differentiate between dead and viable cells.
Quantitative real-time PCR analysis.
The primers LEG-225 and LEG-858 were used to detect Legionella by a semiquantitative dilution PCR method and by a quantitative real-time PCR method. The suitability of the primer set in detecting Legionella in environmental samples has unequivocally been demonstrated (13, 38, 45). The results of the present investigation shows that real-time PCR is a suitable method for selective and quantitative detection of representatives of the genus Legionella, both defined species and yet-undefined types. In three of seven anaerobic GW samples, however, major PCR inhibition was observed, which might have resulted in false-negative PCR results. The high concentrations of iron (up to 9 mg liter−1) and manganese (0.5 mg liter−1) in these water samples could be the cause of the observed inhibition. Amplification and detection of the 16S rRNA genes of Legionella were highly effective and reliable for the other samples, as was demonstrated by the small RSDs of the concentrations obtained from the collected DNA solutions and the decimal dilutions. A suspension of L. pneumophila was used for calibration and calculation of the concentration of Legionella cells from the threshold cycle values. This approach incorporates the efficiency of the DNA extraction in the method. As indicated above, the composition of the water may have an impact on the efficiency of the DNA extraction; tests with samples spiked with L. pneumophila showed that the efficiency of isolation of DNA from drinking water did not differ much from that of the control (data not shown), but random variations were observed. In this respect, the method resembles the culture method (10, 48), and improvement requires further research.
Legionella in raw and in treated water.
The present study confirmed earlier reports that L. pneumophila is a common member of the microbial community of SW, even at low temperatures. The ubiquitous presence of L. pneumophila in rivers and lakes in the United States at a temperature range of 10 to 29°C, with concentrations ranging from 104 to 107 cells liter−1, has been demonstrated for the first time with an FA-based method (18). Concentrations of up to 108 cells liter−1 have been observed with this method in subtropical environments at temperatures between 23 and 30°C (42). L. pneumophila does not multiply at temperatures below 20°C but can survive for very long periods in water at low temperature (46). Consequently, L. pneumophila organisms detected in SW at low temperatures may be survivors from the summer period when water temperatures above 20°C are reached in The Netherlands, and/or they may originate from discharges in surface water of treated and untreated sewage with relatively high Legionella concentrations (36, 43, 50). In the present study, L. pneumophila was not detected in samples of treated SW and was detected in only one sample of treated GW (Table 2). Multiple-barrier SW treatment in The Netherlands causes a 3- to 6-log-unit reduction of vegetative cells (E. coli) and spores (sulfite-reducing clostridia) (21). Hence, in the absence of growth, L. pneumophila is not likely to be detectable in treated SW. L. pneumophila has been detected by use of the culture method in water from GW wells in the United States (11, 47) but was only rarely observed in GW samples (1%) in Germany (53). The results of this study using PCR-based methods show that L. pneumophila is not a common member of the microbial community in raw or treated GW in The Netherlands at temperatures of <12°C. In contrast, non-L. pneumophila species and types were observed in all samples (Table 1). Aerobic GW originates from sand soils and is highly oligotrophic, with low concentrations of dissolved organic carbon (DOC) (<0.5 mg liter−1) and inorganic nutrients and very low heterotrophic plate count values, and it is free from fecal contamination. The presence of Legionella spp. in untreated aerobic GW, with a median concentration of 8.3 × 103 cells liter−1 and with even higher concentrations after limestone filtration, demonstrates the adaptation of these organisms to oligotrophic conditions. Examples of such organisms include 16S rRNA gene sequences related to L. quateirensis (Table 2) and groups 3, 4, 11, and 38 (Fig. 2A and B). Legionella was also detected at relatively low concentrations (median value, 5.5 × 102 cells liter−1) in raw anaerobic GW at three different sites. Untreated anaerobic GW contains methane, ammonia, iron, and manganese and has a DOC concentration of up to 7 mg liter−1. All cultured Legionella species need molecular oxygen for multiplication, and therefore the most likely explanation for its detection is the introduction of (trace amounts of) oxygen at the well head, the transportation pipe, or the sampling location (tap), enabling some local growth.
Representatives of uncultured Legionella spp. were detected in treated SW and in treated GW at all locations. Given the above-mentioned effects of multiple-barrier SW treatment, it must be concluded that these organisms most likely multiply during water treatment. This conclusion is supported by the differences between the 16S rRNA gene sequences isolated from SW and treated SW at the involved treatment plants. The presence of Legionella in untreated GW in the absence of fecal contamination and its presence in treated GW also demonstrate its ability to multiply in the involved aquatic environments. Filtration processes, including dual-medium filtration, rapid sand filtration, GAC filtration, or slow sand filtration, which are operated without chemical disinfectant, are the environments for growth. Such growth occurs at a wide range of concentrations of dissolved organic carbon (nonparticulate organic carbon concentrations of <0.5 to 7 mg liter−1). No significant correlation was observed between the DOC concentration and the Legionella concentration in treated GW (results not shown). Growth of the Legionella species detected in treated water occurs at relatively low temperatures (<5°C for SW and <12°C for GW). These observations correspond with results of other studies where PCR was used to quantify Legionella. In these studies, it was demonstrated that Legionella species contributed to 7% of the total biomass in biofilms grown on pieces of polyvinyl chloride and polyethylene exposed to treated surface water and multiplied at temperatures of <20°C in slow sand filters used for horticulture as well as in experimental sand columns (13, 29, 52). The ability of Legionella spp. to multiply in biofilms in low-pH environments has recently been demonstrated, and those authors also concluded that water temperature affected species composition (54).
Sequence diversity of the detected Legionella.
The genus Legionella presently includes about 50 defined species. This number increases continuously with newly described species, including LLAPs (1, 2, 6, 17, 23, 26, 30). LLAPs, which initially were isolated in coculture with protozoa (51), have been obtained from various environmental sources. Several of these organisms create a monophyletic subgroup within the phylogenetic tree of the genus Legionella (7). The LLAP species are presented in Table 2 and in Fig. 2 in connection with 16S rRNA gene sequences obtained in this study. In the present study all 202 16S rRNA gene sequences of a 654-bp fragment showed the greatest similarity to 16S rRNA-encoding genes of Legionella sequences deposited in the GenBank database. None of the 16S rRNA gene sequences were more than 99% similar with cultured Legionella species, and 65% of the 16S rRNA gene sequences had similarity values of below 97% (Table 2). The predominance of 16S rRNA gene sequences related to defined species, e.g., L. bozemanii, L. lytica, L. quateirensis, and groups of unidentified types in specific situations, may provide information about the conditions favoring the growth of the organisms harboring these 16S rRNA gene sequences. L. quateirensis was also observed in GW from wells in the United States and Canada (11), but most Legionella species observed in these wells differed from those described in the present study. Furthermore, Legionella species and sequences identified in slow sand filters (13) differed from those obtained in this study (Fig. 2). The high genetic diversity of Legionella phylotypes multiplying in aquatic environments at low temperature is further demonstrated by (i) positioning of the obtained 16S rRNA gene sequences all over the phylogenetic tree and (ii) different sequence types being obtained from individual samples. The groups of 16S rRNA gene sequences identified in the present study may represent yet-undefined Legionella spp. Hence, continuing studies using PCR-based methods most likely will cause a large increase of the number of species within the genus Legionella.
Public health significance.
L. pneumophila is not commonly detected in treated water and represented only a small fraction (<1%) of the total number of Legionellae detected in this study. Still, the organism has been observed in many hot water installations, and the large majority of reported cases of legionellosis are caused by L. pneumophila (17, 61). A number of non-L. pneumophila species, including LLAP types, have been observed in association with disease or antigen titer increase (1, 17, 33, 35). Sequences with a high similarity to the disease-associated L. bozemanii predominated in several samples of treated water (Table 2). However, it is not clear if these or other organisms represent potential pathogens, because representatives of L. pneumophila serogroups and genotypes also show distinct differences in infectivity (8, 17, 25, 61). Furthermore, none of these organisms were detected with the BCYE medium. Some of the organisms may have been nonviable, but other explanations include (i) absence of certain specific growth requirements, (ii) inhibition of growth by certain medium components, or (iii) inability to multiply at 37°C. In a recent study, more isolates were obtained from GW samples incubated at 30°C than from those incubated at 35°C (11). Bacteria unable to multiply at body temperature are not likely to be human pathogens. The Legionella types observed in this study may include psychrophilic species that are unable to multiply at elevated temperatures. Elucidation of the properties of these organisms is needed to assess their potential public health significance and explain the conditions favoring their growth in the aquatic environments including water treatment.
Acknowledgments
This study was financed by the water supply companies in The Netherlands as part of the Joint Research Program (BTO).
We are much indebted to Remko Voogt for skillful technical assistance and to Leo Heijnen and Paul van der Wielen for critical reading of the manuscript.
REFERENCES
- 1.Adeleke, A., J. Pruckler, R. Benson, T. Rowbotham, M. Halablab, and B. Fields. 1996. Legionella-like amebal pathogens—phylogenetic status and possible role in respiratory disease. Emerg. Infect. Dis. 2:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adeleke, A. A., B. S. Fields, R. F. Benson, M. I. Daneshvar, J. M. Pruckler, R. M. Ratcliff, T. G. Harrison, R. S. Weyant, R. J. Birtles, D. Raoult, and M. A. Halablab. 2001. Legionella drozanskii sp. nov., Legionella rowbothamii sp. nov. and Legionella fallonii sp. nov.: three unusual new Legionella species. Int. J. Syst. Evol. Microbiol. 51:1151-1160. [DOI] [PubMed] [Google Scholar]
- 3.Alary, M., and J. R. Joly. 1992. Comparison of culture methods and an immunofluorescence assay for the detection of Legionella pneumophila in domestic hot water devices. Curr. Microbiol. 25:19-23. [DOI] [PubMed] [Google Scholar]
- 4.Althaus, H. 1987. Legionellas in drinking, bathing and warm water. Offentl. Gesundheitswes 49(Suppl. 1):8-13. [PubMed] [Google Scholar]
- 4a.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aurell, H., P. Catala, P. Farge, F. Wallet, M. Le Brun, J. H. Helbig, S. Jarraud, and P. Lebaron. 2004. Rapid detection and enumeration of Legionella pneumophila in hot water systems by solid-phase cytometry. Appl. Environ. Microbiol. 70:1651-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson, R. F., and B. S. Fields. 1998. Classification of the genus Legionella. Semin. Respir. Infect. 13:90-99. [PubMed] [Google Scholar]
- 7.Birtles, R. J., T. J. Rowbotham, D. Raoult, and T. G. Harrison. 1996. Phylogenetic diversity of intra-amoebal Legionellae as revealed by 16S rRNA gene sequence comparison. Microbiology 142:3525-3530. [DOI] [PubMed] [Google Scholar]
- 8.Bollin, G. E., J. F. Plouffe, M. F. Para, and R. B. Prior. 1985. Difference in virulence of environmental isolates of Legionella pneumophila. J. Clin. Microbiol. 21:674-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bopp, C. A., J. W. Sumner, G. K. Morris, and J. G. Wells. 1981. Isolation of Legionella sp. from environmental water samples by low-pH treatment and use of a selective medium. J. Clin. Microbiol. 13:714-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boulanger, C. A., and P. H. Edelstein. 1995. Precision and accuracy of recovery of Legionella pneumophila from seeded tap water by filtration and centrifugation. Appl. Environ. Microbiol. 61:1805-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brooks, T., R. A. Osicki, V. S. Springthorpe, S. A. Sattar, L. Fillion, D. Abrail, and S. Riffard. 2004. Detection and identification of Legionella species from groundwaters. J. Toxicol. Environ. Microbiol. Health 67(Part A):1845-1849. [DOI] [PubMed] [Google Scholar]
- 12.Buchbinder, S., K. Trebesius, and J. Heesemann. 2002. Evaluation of detection of Legionella sp. in water samples by fluorescence in situ hybridization, PCR amplification and bacterial culture. Int. J. Med. Microbiol. 292:241-245. [DOI] [PubMed] [Google Scholar]
- 13.Calvo-Bado, L. A., J. A. Morgan, M. Sergeant, T. R. Pettitt, and J. M. Whipps. 2003. Molecular characterization of Legionella populations present within slow sand filters used for fungal plant pathogen suppression in horticultural crops. Appl. Environ. Microbiol. 69:533-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colbourne, J. S., and R. M. Trew. 1986. Presence of Legionella in London’s water supplies. Isr. J. Med. Sci. 22:633-639. [PubMed] [Google Scholar]
- 15.Den Boer, J. W., E. P. Yzerman, J. Schellekens, K. D. Lettinga, H. C. Boshuizen, J. E. Van Steenbergen, A. Bosman, S. Van den Hof, H. A. Van Vliet, M. F. Peeters, R. J. Van Ketel, P. Speelman, J. L. Kool, and M. A. Conyn-Van Spaendonck. 2002. A large outbreak of Legionnaires’ disease at a flower show, the Netherlands, 1999. Emerg. Infect. Dis. 8:37-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edelstein, P. H. 1981. Improved selective medium for isolation of Legionella pneumophila from contaminated clinical and environmental samples. J. Clin. Microbiol. 14:293-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires’ disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fliermans, C. B., W. B. Cherry, L. H. Orrison, S. J. Smith, D. L. Tison, and D. H. Pope. 1981. Ecological distribution of Legionella pneumophila. Appl. Environ. Microbiol. 41:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Fulgueiras, A., C. Navarro, D. Fenoll, J. Garcia, P. Gonzales-Diego, T. Jimenez-Bunuelas, M. Rodriguez, R. Lopez, F. Pacheco, J. Ruiz, M. Segovia, B. Balandron, and C. Pelaz. 2003. Legionnaires’ disease outbreak in Murcia, Spain. Emerg. Infect. Dis. 9:915-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henke, M., and K. M. Seidel. 1986. Association between Legionella pneumophila and amoebae in water. Isr. J. Med. Sci. 22:690-695. [PubMed] [Google Scholar]
- 21.Hijnen, W. A. M., G. J. Medema, and D. vander Kooij. 2004. Qualitative assessment of the removal of indicator bacteria in full-scale treatment plants. Water Sci. Technol. 4:47-54. [Google Scholar]
- 22.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hookey, J. V., N. A. Saunders, N. K. Fry, R. J. Birtles, and T. G. Harrison. 1996. Phylogeny of Legionellaceae based on small-subunit ribosomal DNA sequences and proposal of Legionella lytica comb. nov. for Legionella-like amoebal pathogens. Int. J. Syst. Bacteriol. 46:526-531. [Google Scholar]
- 24.Hsu, S. C., R. Martin, and B. B. Wentworth. 1984. Isolation of Legionella species from drinking water. Appl. Environ. Microbiol. 48:830-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, B., B. A. Heron, B. R. Gray, S. Eglezos, J. R. Bates, and J. Savill. 2004. A predominant and virulent Legionella pneumophila serogroup 1 strain detected in isolates from patients and water in Queensland, Australia, by an amplified fragment length polymorphism protocol and virulence gene-based PCR assays. J. Clin. Microbiol. 42:4164-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.La Scola, B., R. J. Birtles, G. Greub, T. J. Harrison, R. M. Ratcliff, and D. Raoult. 2004. Legionella drancourtii sp. nov., a strictly intracellular amoebal pathogen. Int. J. Syst. Evol. Microbiol. 54:699-703. [DOI] [PubMed] [Google Scholar]
- 27.Lawrence, C., M. Reyrolle, S. Dubrou, F. Forey, B. Decludt, C. Goulvestre, P. Matsiota-Bernard, J. Etienne, and C. Nauciel. 1999. Single clonal origin of a high proportion of Legionella pneumophila serogroup 1 isolates from patients and the environment in the area of Paris, France, over a 10-year period. J. Clin. Microbiol. 37:2652-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, T. C., R. M. Vickers, V. L. Yu, and M. M. Wagener. 1993. Growth of 28 Legionella species on selective culture media: a comparative study. J. Clin. Microbiol. 31:2764-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehmann, U., and H. Kreipe. 2001. Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods 25:409-418. [DOI] [PubMed] [Google Scholar]
- 30.Ludwig, W., and E. Stackebrandt. 1983. A phylogenetic analysis of Legionella. Arch. Microbiol. 135:45-50. [DOI] [PubMed] [Google Scholar]
- 31.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahbubani, M. H., A. K. Bej, R. Miller, L. Haff, J. DiCesare, and R. M. Atlas. 1990. Detection of Legionella with polymerase chain reaction and gene probe methods. Mol. Cell. Probes 4:175-187. [DOI] [PubMed] [Google Scholar]
- 33.Marrie, T. J., D. Raoult, B. La Scola, R. J. Birtles, and E. de Carolis. 2001. Legionella-like and other amoebal pathogens as agents of community-acquired pneumonia. Emerg. Infect. Dis. 7:1026-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marston, B. J., H. B. Lipman, and R. F. Breiman. 1994. Surveillance for Legionnaires’ disease. Risk factors for morbidity and mortality. Arch. Intern. Med. 154:2417-2422. [PubMed] [Google Scholar]
- 35.McNally, C., B. Hackman, B. S. Fields, and J. F. Plouffe. 2000. Potential importance of Legionella species as etiologies in community acquired pneumonia (CAP). Diagn. Microbiol. Infect. Dis. 38:79-82. [DOI] [PubMed] [Google Scholar]
- 36.Medema, G., B. Wullings, P. Roeleveld, and D. van der Kooij. 2004. Risk assessment of Legionella and enteric pathogens in sewage treatment works. Water Sci. Technol. 4:125-132. [Google Scholar]
- 37.Meenhorst, P. L., A. L. Reingold, D. G. Groothuis, G. W. Gorman, H. W. Wilkinson, R. M. McKinney, J. C. Feeley, D. J. Brenner, and R. van Furth. 1985. Water-related nosocomial pneumonia caused by Legionella pneumophila serogroups 1 and 10. J. Infect. Dis. 152:356-364. [DOI] [PubMed] [Google Scholar]
- 38.Miyamoto, H., H. Yamamoto, K. Arima, J. Fujii, K. Maruta, K. Izu, T. Shiomori, and S. Yoshida. 1997. Development of a new seminested PCR method for detection of Legionella species and its application to surveillance of Legionellae in hospital cooling tower water. Appl. Environ. Microbiol. 63:2489-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morrill, W. E., J. M. Barbaree, B. S. Fields, G. N. Sanden, and W. T. Martin. 1990. Increased recovery of Legionella micdadei and Legionella bozemanii on buffered charcoal yeast extract agar supplemented with albumin. J. Clin. Microbiol. 28:616-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muder, R. R., and V. L. Yu. 2002. Infection due to Legionella species other than L. pneumophila. Clin. Infect. Dis. 35:990-998. [DOI] [PubMed] [Google Scholar]
- 41.Nederlands Normalisatie Instituut. 1991. Bacteriological examination of water: examination on the presence and the number of colony forming units (CFU) of Legionella bacteria. NEN 6265. Nederlands Normalisatie Instituut, Delft, The Netherlands.
- 42.Ortiz-Roque, C. M., and T. C. Hazen. 1987. Abundance and distribution of Legionellaceae in Puerto Rican waters. Appl. Environ. Microbiol. 53:2231-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer, C. J., G. F. Bonilla, B. Roll, C. Paszko-Kolva, L. R. Sangermano, and R. S. Fujioka. 1995. Detection of Legionella species in reclaimed water and air with the EnvoroAmp Legionella PCR kit and direct fluorescent-antibody staining. Appl. Environ. Microbiol. 61:407-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmer, C. J., Y. L. Tsai, C. Paszko-Kolva, C. Mayer, and L. R. Sangermano. 1993. Detection of Legionella species in sewage and ocean water by polymerase chain reaction, direct fluorescent-antibody, and plate culture methods. Appl. Environ. Microbiol. 59:3618-3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pascual, L., S. Perez-Luz, A. Amo, C. Moreno, D. Apraiz, and V. Catalan. 2001. Detection of Legionella pneumophila in bioaerosols by polymerase chain reaction. Can. J. Microbiol. 47:341-347. [PubMed] [Google Scholar]
- 46.Paszko-Kolva, C., M. Shahamat, and R. R. Colwell. 1992. Long-term survival of Legionella pneumophila serogroup 1 under low-nutrient conditions and associated morphological changes. FEMS Microbiol. Ecol. 102:45-55. [Google Scholar]
- 47.Riffard, S., S. Douglass, T. Brooks, S. Springthorpe, L. G. Filion, and S. A. Sattar. 2001. Occurrence of Legionella in groundwater: an ecological study. Water Sci. Technol. 43:99-102. [PubMed] [Google Scholar]
- 48.Roberts, K. P., C. M. August, and J. D. Nelson, Jr. 1987. Relative sensitivities of environmental Legionellae to selective isolation procedures. Appl. Environ. Microbiol. 53:2704-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogers, J., A. B. Dowsett, P. J. Dennis, J. V. Lee, and C. W. Keevil. 1994. Influence of temperature and plumbing material selection on biofilm formation and growth of Legionella pneumophila in a model potable water system containing complex microbial flora. Appl. Environ. Microbiol. 60:1585-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roll, B. M., and R. S. Fujioka. 1995. Detection of Legionella bacteria in sewage by polymerase chain reaction and standard culture method. Water Sci. Technol. 31:409-416. [Google Scholar]
- 51.Rowbotham, T. J. 1993. Legionella-like amoebal pathogens, p. 137-140. In J. M. Barbaree, R. F. Breiman, and A. P. Dufour (ed.), Legionella: current status and emerging perspectives. American Society for Microbiology, Washington, D.C.
- 52.Schwartz, T., S. Hoffmann, and U. Obst. 1998. Formation and bacterial composition of young, natural biofilms obtained from public bank-filtered drinking water systems. Water Res. 32:2787-2797. [Google Scholar]
- 53.Seidel, K., W. Bornert, G. Baz, A. Blankenburg, and I. Alexander. 1986. Presence of Legionella pneumophila in ground water, cold and warm water. Vom Wasser 67:39-48. [Google Scholar]
- 54.Sheehan, K. B., J. M. Henson, and M. J. Ferris. 2005. Legionella species diversity in an acidic biofilm community in Yellowstone National Park. Appl. Environ. Microbiol. 71:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stackebrandt, E., and B. M. Goebel. 1994. A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 56.Tison, D. L., and R. J. Seidler. 1983. Legionella incidence and density in potable drinking water supplies. Appl. Environ. Microbiol. 45:337-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van der Kooij, D., J. H. M. Lieverloo, J. Schellart, and P. Hiemstra. 1999. Maintaining quality without a disinfectant residual. J. Am. Water Works Assoc. 91:55-64. [Google Scholar]
- 58.Verissimo, A., G. Marrao, F. G. da Silva, and M. S. da Costa. 1991. Distribution of Legionella sp. in hydrothermal areas in continental Portugal and the island of Sao Miguel, Azores. Appl. Environ. Microbiol. 57:2921-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wadowsky, R. M., R. Wolford, A. M. McNamara, and R. B. Yee. 1985. Effect of temperature, pH, and oxygen level on the multiplication of naturally occurring Legionella pneumophila in potable water. Appl. Environ. Microbiol. 49:1197-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wadowsky, R. M., and R. B. Yee. 1981. Glycine-containing selective medium for isolation of Legionellaceae from environmental specimens. Appl. Environ. Microbiol. 42:768-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.World Health Organization. 2000. ‘Legionnaires’ disease, Europe, 1999. Wkly. Epidemiol. Rec. 2000:347-352. [PubMed] [Google Scholar]
- 62.Wullings, B., R. Voogt, H. Veenendaal, and D. van der Kooij. 2002. The fluorescent in situ hybridization test in comparison with culture for the detection of Legionella pneumophila in water samples, p. 263-266. In R. Marre, Y. A. Kwaik, C. Bartlett, N. P. Cianciotto, B. S. Fields, M. Frosch, J. Hacker, and P. C. Lück (ed.), Legionella. ASM Press, Washington, D.C.