Abstract
Bacteriophage MS2 is used in place of pathogenic viruses in a wide variety of studies that range from testing of compounds for disinfecting surfaces to studying environmental transport and fate of pathogenic viruses in groundwater. MS2 is also used as a pathogen simulant in the research, development, and testing (including open air tests) of methods, systems, and devices for the detection of pathogens in both the battlefield and homeland defense settings. PCR is often used as either an integral part of such detection systems or as a reference method to assess the sensitivity and specificity of microbial detection. To facilitate the detection of MS2 by PCR, we describe here a set of real-time fluorogenic reverse transcription-PCR assays. The sensitivity of the assays (performed with primer pairs and corresponding dye-labeled probes) ranged from 0.4 to 40 fg of MS2 genomic RNA (200 to 20,000 genome equivalents). We also demonstrate the usefulness of the primer pairs in assays without dye-labeled probe that included the DNA-binding dye SYBR green. None of the assays gave false-positive results when tested against 400 pg of several non-MS2 nucleic acid targets.
Investigation and production of methods, reagents, and devices for the detection of pathogens require a subject for testing at each phase of technology development. For certain kinds of tasks, such as the development of antibodies that are specific for a pathogen of interest or the determination of assay specificity, usually no organism other than the pathogen to be detected can reasonably be substituted. However, there are phases of technology development in which the use of live, virulent pathogens is not ethically permissible, economically feasible, or politically palatable. For example, it is not permissible now to produce and disseminate aerosols of pathogens out of doors for the purposes of testing a complete pathogen detection system or its components. For such open air work, and in preliminary developmental benchwork such as the engineering of particle collectors and fluidics systems, the development of generalized PCR protocols, or the testing of surface or water decontamination methods, nonpathogenic organisms with generally similar physical and biological characteristics are routinely substituted for more dangerous bacteria or viruses. Such organisms or proteins are termed “simulants” or “surrogates” in the biological defense research community. Examples of simulants that are used widely in biological defense research include nonpathogenic Bacillus spp. (B. atrophaeus, B. subtilis, and B. globigii) (7) in place of Bacillus anthracis, Pantoea agglomerans (formerly Erwinia herbicola) in place of pathogenic gram-negative species (such as Yersinia pestis and Francisella tularensis), hen egg ovalbumin in place of protein toxins such as ricin and botulinum toxin, and the enteric bacteriophage MS2 in place of pathogenic viruses.
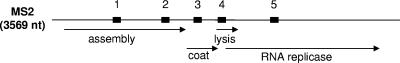
Bacteriophage MS2 (family Leviviridae) is a small, icosahedral, male-specific bacteriophage of Escherichia coli (4). The MS2 genome is comprised of 3,569 bases of single-stranded RNA that encode four proteins (assembly, lysis, coat, and RNA replicase [β chain] proteins) (Fig. 1). The small size of MS2 virions, their simple structure, their RNA genome, and harmlessness to humans, animals, plants, and other higher organisms have made them useful as simulants in place of small RNA viruses (such as Ebola virus, Marburg virus, and the equine encephalitis alphaviruses). Viral simulants in biological defense studies should approximate human pathogens in genome size, nucleic acid composition, and virion size and be nonpathogenic, easy and economical to culture, and easy to detect using common laboratory methods. Bacteriophage MS2 is not a perfect match in these regards to any of the small RNA viruses mentioned above, but has long been used in the biological defense community because it has proved to be a reasonable technical and practical compromise.
FIG. 1.
Linear map of the MS2 genome. The approximate locations and sizes of the amplicon products are indicated by thick bars superimposed on the linear map. Numbers correspond to the assay numbering in Table 1. Approximate locations, sizes, and products of the genes carried by the MS2 genome are indicated by arrows below the map. nt, nucleotides.
MS2 has been used in place of pathogens in a large number of studies: for examining the survival of viruses on produce (1, 11), developing systems and methods to remove viruses from water and surfaces (5, 9, 10, 12, 13, 17-20, 23-25, 29, 30, 32-34, 42), and investigating the fate of pathogens in groundwater (2, 14, 15, 26, 31, 36-39). It has also been used in the testing and development of systems for the detection of biological warfare agents (3, 8, 22, 35, 40, 41).
Depending upon the pathogen detection technology or system under development, reagent sets for the detection of the simulants must be produced in addition to those specific to the pathogens themselves. For systems that rely upon antibody-antigen recognition to confer specificity and sensitivity (35), such reagents include polyclonal, monoclonal, and recombinant antibodies. The use of simulants in the testing of detection systems based on PCR or hybridization to nucleic acid microarrays requires oligonucleotides (primers and dye-labeled probes) whose sequences are complementary to unique sequences found in the genome of the simulant. While Belgrader et al. (3) reported the detection of bacteriophage MS2 in a hand-held real-time PCR thermocycler, they did not report the sequence of the PCR target or the sequences of the primers and probes used. To facilitate the use of bacteriophage MS2 in the development of PCR-based pathogen systems, we describe here five primer and probe sets for real-time fluorogenic reverse transcription (RT)-PCR assays for the detection of bacteriophage MS2.
MATERIALS AND METHODS
Strains, media, and culture conditions.
A seed stock of bacteriophage MS2 and Escherichia coli host strain A/λ were obtained from the Biosensors Team, U.S. Army Edgewood Chemical Biological Center, who obtained the original cultures from the laboratory of D. Peabody, University of New Mexico. To propagate bacteriophage MS2, E. coli strain A/λ was grown in liquid Luria broth at 37°C with shaking, to an optical density of 0.2 to 0.3 (measured spectrophotometrically at 600 nm). MS2 virions were added at a multiplicity of infection of approximately 20, and the culture was incubated a further 16 to 18 h. Cell debris was removed by centrifugation and subsequent filtration through a 0.22-μm filter. Titers of lysates were determined by mixing aliquots of diluted MS2 preparations with suspensions of the host strain in molten Luria top agar (Luria broth, 0.5% Bacto agar), pouring the mixtures onto solid Luria medium (Luria broth plus 1.5% agar), and counting the resulting plaques after overnight incubation at 37°C. Lysates were stored at 4°C. RNA was isolated from virions with the Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
PCR assay design and conditions.
Real-time fluorogenic PCR assays were designed with PrimerExpress software (version 1.0; Applied Biosystems, Foster City, CA), using as a template GenBank accession no. NC_001417, the complete genome sequence of bacteriophage MS2. Primer sets were designed with the following constraints: a maximum amplicon length of 150 bp, a melting temperature (Tm) of approximately 58 to 60°C, and no more than two G or C bases among the last five bases on the 3′ end (to minimize the likelihood of nonspecific priming at non-target sequences). Probe sequences were chosen with a Tm of approximately 68 to 70°C (10°C higher than the primer Tm), more C's than G's (and no more than 3 consecutive G's) in the overall probe sequence, and no 5′-terminal G. Primer and probe sequences in each assay are shown in Table 1. Primers were purchased from Integrated DNA Technologies (Coralville, IA). Probes (with 5′ 6-carboxy-fluorescein [FAM] and 3′-carboxymethylrhodamine [TAMRA]) were purchased from Applied Biosystems. Following the computer-aided design of assays, candidate primer pairs were purchased and the success of PCRs was assessed by examining PCRs for the presence of amplified DNA by agarose gel electrophoresis (29a). Dye-labeled probes were ordered and tested only for those assays observed to yield abundant DNA.
TABLE 1.
Primer and probe sequences for real-time fluorogenic RT-PCR assays for detection of bacteriophage MS2
| Assay | Starta | Probe and primer sequencesb | Product of target genec |
|---|---|---|---|
| 1 | 632 | Forward, GTCGCGGTAATTGGCGC | Assembly protein |
| 708 | Reverse, GGCCACGTGTTTTGATCGA | ||
| 650 | Probe, AGGCGCTCCGCTACCTTGCCCT | ||
| 2 | 1155 | Forward, TGTGGAGAGACAGGGCACTG | Assembly protein |
| 1231 | Reverse, CAGTTGTTGGCCATACGGATT | ||
| 1176 | Probe, TAAGGCCCAAATCTCAGCCATGCATC | ||
| 3 | 1449 | Forward, CGTTCACAGGCTTACAAAGTAACCT | Coat protein |
| 1555 | Reverse, CCAACAGTCTGGGTTGCCAC | ||
| 1498 | Probe, AGAATCGCAAATACACCATCAAAGTCGAGGT | ||
| 4 | 1693 | Forward, CCTCAGCAATCGCAGCAAA | Lysis protein |
| 1807 | Reverse, GGAAGATCAATACATAAAGAGTTGAACTTC | ||
| 1743 | Probe, CAAACATGAGGATTACCCATGTCGAAGACA | ||
| 5 | 2232 | Forward, GCTCTGAGAGCGGCTCTATTG | RNA replicase β chain |
| 2301 | Reverse, CGTTATAGCGGACCGCGT | ||
| 2255 | Probe, CCGAGACCAATGTGCGCCGTG |
Position of first base in each oligonucleotide sequence relative to the whole genome sequence in GenBank accession no. NC_001417.
All sequences are listed in the 5′-to-3′ direction. In the assays as performed, the 5′ base of each probe was labeled with FAM and the 3′ base of each probe was labeled with TAMRA.
Protein names as given in the GenBank genome sequence accession NC_001417. Assembly protein is also known as maturase, or “A-protein.”
To perform a PCR assay, TaqMan Universal PCR Master Mix, MultiScribe reverse transcriptase, RNase inhibitor mix, and nuclease-free water (Applied Biosystems, Foster City, CA) were added to a reaction mixture (50-μl final volume) according to the manufacturer's instructions. Primers were added to a final concentration of 400 nM, and probe was added to a final concentration of 200 nM. To avoid contamination of the reagent stocks, reaction mixtures were prepared in a hood separate from the hood in which template samples were added to each reaction mixture. RNA was added in a volume of 1.0 μl per 50-μl reaction mixture. For negative (no template) control reactions, nuclease-free water was substituted for virions or RNA. Assays were performed on an ABI 7900HT sequence detection system, with a thermocycler profile of 48°C for 30 min and 95°C for 10 min and then 45 cycles of 95°C for 15 s and 60°C for 60 s. In most experiments, assays were prepared in duplicate 50-μl volumes, from which 20 μl was pipetted into two wells of a 384-well assay plate. Therefore, if 1 μl of a 1-ng/μl stock solution of RNA was added to a 50-μl reaction mixture, the amount of target generating a signal in a single well of a 384-well plate was 400 pg. Assays were also tested without probes using SYBR green PCR master mix, Multiscribe reverse transcriptase, and RNase inhibitor according to the manufacturer's instructions (Applied Biosystems). SYBR green assays were run using a thermocycler profile of 48°C for 30 min, 95°C for 10 min, 40 cycles of 95°C for 15 s, 62°C for 60 s, followed by incubations of 95°C for 15 s, 60°C for 15 s, and 95°C for 15 s.
RESULTS
Sensitivity of the RT-PCR assays for bacteriophage MS2.
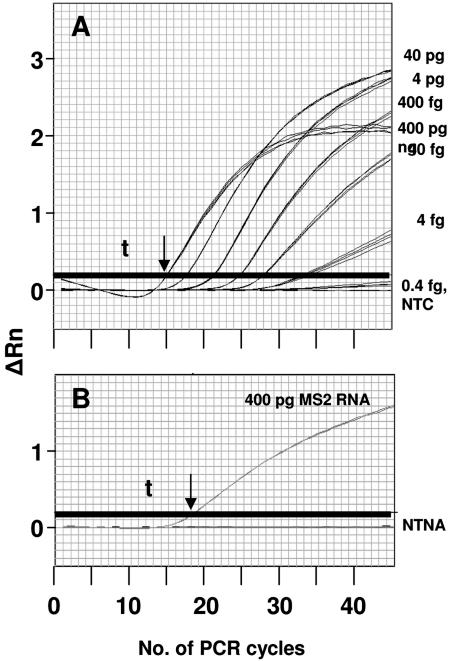
We developed five separate assays for MS2, which among them detect sequences in each of the four genes encoded by the phage genome (assembly protein, coat protein, lysis protein, and RNA replicase) (Table 1). The limits of detection for MS2 genomic RNA for each assay (the lowest amount of template tested that gave positive results in all experiments) were between 40 and 0.4 fg (20,000 to 200 genome equivalents) per 20-μl reaction volume in assays that included the respective probes (Table 2). Representative traces of fluorescence development for assay 1 are shown in Fig. 2. The limit of detection for assay 1 was less than 4 fg of purified genomic RNA (cycle threshold [Ct] of ∼33). (For the sake of brevity, data for only one of the five MS2 assays are shown.)
TABLE 2.
Performance of real-time fluorogenic RT-PCR assays for detection of bacteriophage MS2
| Assaya | Avg CT at sample concnb:
|
||||
|---|---|---|---|---|---|
| 4 pg | 0.4 pg | 40 fg | 4 fg | 0.4 fg | |
| 1 | 20.9 ± 0.4 | 24.5 ± 0.4 | 27.9 ± 0.2 | 33.3 ± 0.5 | ND |
| 2 | 23.8 ± 0.6 | 27.3 ± 0.6 | 32.4 ± 1.3 | ND | ND |
| 3 | 27.4 ± 0.4 | 31.0 ± 0.4 | 36.3 ± 1.3 | ND | ND |
| 4 | 24.6 ± 0.3 | 28.2 ± 0.2 | 31.8 ± 0.1 | 35.5 ± 0.2 | 44.5 ± 1.0 |
| 5 | 26.3 ± 0.5 | 29.9 ± 0.5 | 33.4 ± 0.2 | 37.0 ± 0.8 | 41.0 ± 2.6 |
Assays are numbered as in Table 1.
CT represents the cycle number at which fluorescence increases above the threshold dividing positive from negative results. The values shown are the average result from eight wells (four wells per experiment, two experiments) ± standard deviation. The concentrations reflect the amount of MS2 RNA per 20-μl reaction well. ND, no detection above threshold before cycle 45.
FIG. 2.
Detection of bacteriophage MS2 genomic RNA by real-time fluorogenic RT-PCR. Thick horizontal lines represent the threshold (t) for significant signal detection. Larger amounts of RNA permitted positive detection (signal above threshold) with fewer PCR cycles. The greatest amount of RNA (400 pg) produced a signal with the fewest cycles (arrows). (A) Limit of detection experiments demonstrating the sensitivity of assay MS2-1, performed using 10-fold dilutions of a preparation of bacteriophage MS2 RNA (concentrations at right). Using the probe and primer set in assay 1 (numbered as in Table 1), 4 fg of RNA was repeatably and reproducibly detected. Assay MS-1 did not detect 0.4 fg of MS2 RNA: the assay gave results indistinguishable from the negative (no-template) control (NTC). (B) Cross-reactivity experiments using primers and probe from assay MS2-1. None of the non-target nucleic acid (NTNA) samples were amplified above threshold by the MS2-1 primer set, while 400 pg of MS2 RNA gave a positive reaction. ΔRn, change in fluorescence.
We have also detected MS2 RNA in assays run with each of the primer sets but without the dye-labeled probe oligonucleotides. The assays were performed with reaction mixtures that included SYBR green, a dye that fluoresces more strongly when bound to double-stranded DNA (Table 3). Increases in the amount of double-stranded DNA in a PCR in progress result in more bound dye; therefore, the bulk fluorescence of the reaction mixture increases over the time of the reaction in proportion to the amount of target present. This assay format relieves the requirement for a specific probe molecule and is sometimes more sensitive. Interestingly, we found that the performance of a given primer pair with its corresponding probe did not predict performance without the probe in assays with SYBR green. The primer pairs for assays 4 and 5 gave similar limits of detection when used with either dye-labeled probe or SYBR green. However, the primer pair for assay 3 gave the most sensitive detection of MS2 RNA when used with its corresponding dye-labeled probe, but the least sensitive detection when used with SYBR green. It is possible that differences in local secondary structure in the MS2 RNA template are responsible for this phenomenon, but we have not performed experiments to test this hypothesis.
TABLE 3.
Performance of real-time fluorogenic RT-PCR assays for detection of bacteriophage MS2, performed without probe and including the DNA-binding dye SYBR green
| Assaya | Avg CT at sample concnb:
|
||||
|---|---|---|---|---|---|
| 4 ng | 0.4 ng | 40 pg | 4 pg | 0.4 pg | |
| 1 | 25.0 ± 0.47 | 30.1 ± 0.52 | 35.9 ± 0.64 | ND | ND |
| 2 | 27.4 ± 0.74 | 32.6 ± 0.98 | 38.7 ± 0.98 | ND | ND |
| 3 | 19.5 ± 0.25 | 23.8 ± 0.13 | 28.0 ± 0.27 | 33.0 ± 0.37 | 38.3 ± 0.54 |
| 4 | 22.5 ± 0.67 | 27.3 ± 0.61 | 32.7 ± 0.90 | 38.1 ± 1.00 | ND |
| 5 | 24.2 ± 0.23 | 29.7 ± 0.17 | 34.7 ± 0.29 | 39.9 ± 0.15 | ND |
Assays are numbered as in Table 1.
CT represents the cycle number at which fluorescence increases above the threshold dividing positive from negative results. The values shown are the average result from eight wells (four wells per experiment, two experiments)± standard deviation. The concentrations reflect the amount of MS2 RNA per 20-μl reaction well. ND, no detection above threshold before cycle 40.
Specificity of the RT-PCR assays.
All five assays for MS2 were tested against a panel of DNA or RNA from non-target organisms to determine the specificity of the assays. Assay specificity was determined using assays that incorporated the probe oligonucleotides. The panel included plasmids pXO1 and pXO2 isolated from Bacillus anthracis, and whole genomic DNA isolated from E. coli ATCC 43895, B. cereus ATCC 14579, B. subtilis ATCC 27370, Francisella tularensis strain Schu 4, Clostridium perfringens ATCC 13124, Clostridium tetani ATCC 19406, Yersinia pestis 15-91, Salmonella enterica serovar Typhimurium LT2, Staphylococcus aureus strains ATCC 13566 and ATCC 14458, Bacteroides fragilis ATCC 25285, Bordetella pertussis ATCC 9797, Pseudomonas aeruginosa PAO-1, Streptococcus pyogenes ATCC 12384, Burkholderia cepacia LSPQ-2217, Corynebacterium diphtheriae LSPQ-3083, and Homo sapiens (against the possibility that human DNA, introduced from an operator, might contaminate a sample). None of the non-target nucleic acids produced a signal in any of the assays we developed when added to the assays at a relatively large amount (400 pg DNA or RNA per assay; representative results shown in Fig. 2B).
DISCUSSION
Studies that examine the environmental fate of organisms, the effectiveness of antimicrobial compounds, or water treatment protocols usually require tests for the viability of viruses. However, in some settings, the rapid detection of the presence of a pathogen is of greater importance than determinations of viability. For that reason, many pathogen detection systems currently under development employ either affinity (antibody)-based methods or sequence recognition-based detection methods such as PCR, backed up by sample archiving for confirmatory analysis in a laboratory. PCR is a highly sensitive method for the detection of specific nucleic acid sequences; it has been reported that the use of PCR to detect viral simulants is at least as sensitive as conventional plate assays for bacteriophage (6). While culture-based assays are sensitive and quantifiable, they are also time-consuming and have limited use in industrial-scale settings that require the rapid testing of large numbers of samples. PCR is faster (from 1 to 3 h in the laboratory, but faster instruments are now emerging in the marketplace), can be automated, and is more sensitive per unit of sample. Plaque assays for the detection of bacteriophage require the preparation and/or storage of media, a laboratory for preparation of microbial cultures, incubation periods of several hours, and a trained microbiologist to perform the work, while new instrumentation is beginning to bring PCR out of the laboratory and into the field.
Bacteriophages other than MS2 have also been used as simulants. Phage φX174, for example, has been used in studies examining the passage of virus through surgical and medical exam gloves made from latex and other elastomeric materials to assess their performance as microbial barriers (6, 21, 27). MS2 has been used as a simulant in the biological defense community, however, because, like many of the pathogens of greatest concern, it has a genome made of RNA.
The assays shown here were designed primarily to assist in the development of protocols for the laboratory testing of biological defense equipment (pathogen detection systems, air samplers, etc.). However, they are obviously applicable to studies in which the worker desires to detect bacteriophage MS2 from samples of essentially any origin, provided the samples do not contain substances that inhibit PCR, and contain MS2 above the limits of detection. Experiments in this study were performed only with purified RNA, rather than with titered phage lysates or environmental samples, to eliminate the potentially confounding factors of environmental contaminants or the presence of free MS2 RNA or intact but noninfective phage particles present in phage preparations. The data presented in this study are therefore “best-case” demonstrations of the assays' sensitivity. We acknowledge that the performance of the assays in environmental studies will necessarily vary with the contents and quality of a sample from a given environment, the efficiency of RNA or DNA extraction (or whether extractions take place at all), the possible presence of natural substances that may act as PCR inhibitors (humic acids from soils, for instance), or the presence of proteases and nucleases that may degrade virions in a sample. This study was not intended to demonstrate the optimization of detection of MS2 from a given set of environmental samples because of the limited usefulness of such demonstrations; optimizing the extraction of RNA or of methods for the removal of PCR inhibitors from environmental samples was beyond the scope of this work, and the choice of methods employed will necessarily vary with the needs of each worker.
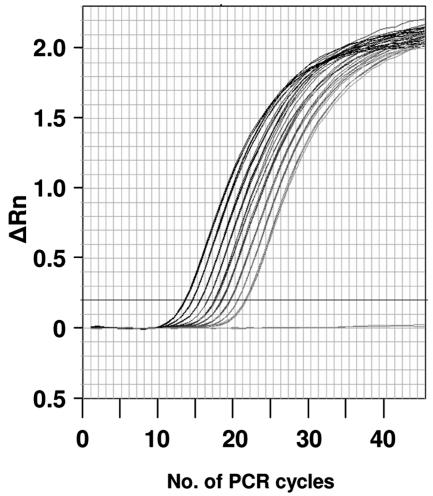
The use of purified RNA in this study allowed the assays to be demonstrated at their limits of performance and precision. The assays were sufficiently repeatable and precise to allow straightforward discrimination of two-fold differences in target concentration (Fig. 3, Table 4). Furthermore, even phage titers in most laboratory preparations are known to decline with the passage of time, and free RNA and noninfective phages in lysates are difficult to quantify for obvious reasons. The lability of RNA and the presence of nucleases in lysates make it likely that the amount of free RNA in a phage lysate will decrease over time, requiring frequent recalibration of the ratio of viable PFU and detectable RNA targets in a given lysate. Such a calibration would be necessary in any given set of experiments in which quantitative PCR would be used to estimate the amount of viable MS2 in a given sample.
FIG. 3.
Resolution of real-time fluorogenic RT-PCR for the quantification of bacteriophage MS2 genomic RNA. MS2 RNA was serially diluted twofold from 400 pg/assay to 1.5635 pg/assay. Each set of curves (nine samples and one negative control) is comprised of four superimposed curves. The CT values for this experiment are given in Table 4. The probe and primer set used in this experiment are for assay 1, as shown in Table 1. ΔRn, change in fluorescence.
TABLE 4.
Resolution of real-time fluorogenic RT-PCR assay 1 for quantification of bacteriophage MS2 RNAa
| Sample concn (pg) | Avg CTb |
|---|---|
| 400 | 13.3 ± 0.16 |
| 200 | 14.2 ± 0.14 |
| 100 | 15.2 ± 0.17 |
| 50 | 16.2 ± 0.15 |
| 25 | 17.2 ± 0.13 |
| 12.5 | 18.1 ± 0.13 |
| 6.25 | 19.2 ± 0.12 |
| 3.125 | 20.1 ± 0.09 |
| 1.5625 | 21.3 ± 0.16 |
The probe and primer set used in this experiment were those of assay 1 (Table 1).
CT represents the cycle number at which fluorescence increases above the threshold dividing positive from negative results. The values shown are the average result from eight wells (four wells per experiment, two experiments) ± standard deviation. The concentrations reflect the amount of MS2 RNA per 20-μl reaction volume.
While the assays described here did not cross-react with DNA from a wide variety of other species, we did not test directly for cross-reactivity with other members of the Leviviridae. However, BLAST (blastn) searches using the five amplicons as queries did reveal significant (and unsurprising) similarities between the MS2 target sequences and sequences in the genomes of other leviviruses (JP501, M12, R17, and fr) (data not shown). It is therefore possible that under some conditions, the genomes of these viruses (or other undescribed, related bacteriophages), when present in environmental samples contaminated by human or animal feces, could give positive signals in these assays. However, methods for phage detection that rely on culture and plaque counting are also susceptible to error introduced by naturally occurring phages, because plaque morphology alone is not a robust tool for phage identification. The RT-PCR assays still provide an advantage over culture methods for phage detection and enumeration, by restricting detection and identification to MS2 and (potentially) its close relatives. The specificity of the assays was also apparent in the lack of amplification of non-target sequences from a large number of other species including H. sapiens and Ricinus communis; we routinely perform PCR assays of 35 to 45 cycles and do not see amplification of non-target sequences from 1-μg quantities of DNA (Fig. 2B).
All of the assays described here are based on the 5′-to-3′ nuclease activity of Taq polymerase, which cleaves a nonextendable, dual-labeled fluorogenic probe (16, 28). The chemistry of these assays is amenable to lyophilization and storage in a stable, dry form with a long shelf life and minimal required infrastructure. However, like all PCR assays, the determination of a positive result can also be obtained by direct observation of the PCR products in an agarose gel or through the use of dyes (such as SYBR green) that do not require the 5′-to-3′ exonuclease activity of a thermostable polymerase. There are currently on the market, or under development, several platforms, including laboratory instruments such as the ABI 7900HT, ABI 7700, and ABI 7300, the LightCycler (Cepheid), so-called “field-ruggedized” instruments (such as the R.A.P.I.D. from Idaho Technologies), and hand-held equipment (for example, the BioSeeq from Smiths-Environmental Technologies Group, Inc.) that are designed to run assays using real-time fluorogenic chemistries, giving workers in public health and the military and first responders a growing selection of rapid pathogen detection and identification capabilities that can be used in a variety of settings. Assays for nonpathogenic microorganisms, such as those described in this study, provide a safer, cost-effective means of testing such equipment in advance of validation with live pathogens.
REFERENCES
- 1.Allwood, P. B., Y. S. Malik, C. W. Hedberg, and S. M. Goyal. 2004. Effect of temperature and sanitizers on the survival of feline calicivirus, Escherichia coli, and F-specific coliphage MS2 on leafy salad vegetables. J. Food Prot. 67:1451-1456. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, M. E., M. Aguilar, A. Fountain, N. Gonzalez, O. Rascon, and D. Saenz. 2000. Inactivation of MS-2 phage and poliovirus in groundwater. Can. J. Microbiol. 46:159-165. [DOI] [PubMed] [Google Scholar]
- 3.Belgrader, P., W. Benett, D. Hadley, G. Long, R. Mariella, Jr., F. Milanovich, S. Nasarabadi, W. Nelson, J. Richards, and P. Stratton. 1998. Rapid pathogen detection using a microchip PCR array instrument. Clin. Chem. 44:2191-2194. [PubMed] [Google Scholar]
- 4.Bollback, J. P., and J. P. Huelsenbeck. 2001. Phylogeny, genome evolution, and host specificity of single-stranded RNA bacteriophage (family Leviviridae). J. Mol. Evol. 52:117-128. [DOI] [PubMed] [Google Scholar]
- 5.Brion, G. M., and J. Silverstein. 2001. Selecting a sensitive bacteriophage assay for evaluation of a prototype water recycling system. Life Support Biosph. Sci. 8:9-14. [PubMed] [Google Scholar]
- 6.Broyles, J. M., K. P. O'Connell, and D. M. Korniewicz. 2002. A PCR-based method for detecting viral penetration of medical exam gloves. J. Clin. Microbiol. 40:2725-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke, S. A., J. D. Wright, M. K. Robinson, B. V. Bronk, and R. L. Warren. 2004. Detection of molecular diversity in Bacillus atrophaeus by amplified fragment length polymorphism analysis. Appl. Environ Microbiol. 70:2786-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cargile, B. J., S. A. McLuckey, and J. L. Stephenson, Jr. 2001. Identification of bacteriophage MS2 coat protein from E. coli lysates via ion trap collisional activation of intact protein ions. Anal. Chem. 73:1277-1285. [DOI] [PubMed] [Google Scholar]
- 9.Casteel, M. J., K. Jayaraj, A. Gold, L. M. Ball, and M. D. Sobsey. 2004. Photoinactivation of hepatitis A virus by synthetic porphyrins. Photochem. Photobiol. 80:294-300. [DOI] [PubMed] [Google Scholar]
- 10.Chaidez, C., M. Moreno, W. Rubio, M. Angulo, and B. Valdez. 2003. Comparison of the disinfection efficacy of chlorine-based products for inactivation of viral indicators and pathogenic bacteria in produce wash water. Int. J. Environ. Health Res. 13:295-302. [DOI] [PubMed] [Google Scholar]
- 11.Dawson, D. J., A. Paish, L. M. Staffell, I. J. Seymour, and H. Appleton. 2005. Survival of viruses on fresh produce, using MS2 as a surrogate for norovirus. J. Appl. Microbiol. 98:203-209. [DOI] [PubMed] [Google Scholar]
- 12.de Roda Husman, A. M., P. Bijkerk, W. Lodder, H. van Den Berg, W. Pribil, A. Cabaj, P. Gehringer, R. Sommer, and E. Duizer. 2004. Calicivirus inactivation by nonionizing (253.7-nanometer-wavelength [UV]) and ionizing (gamma) radiation. Appl. Environ. Microbiol. 70:5089-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drees, K. P., M. Abbaszadegan, and R. M. Maier. 2003. Comparative electrochemical inactivation of bacteria and bacteriophage. Water Res. 37:2291-2300. [DOI] [PubMed] [Google Scholar]
- 14.Gordon, C., and S. Toze. 2003. Influence of groundwater characteristics on the survival of enteric viruses. J. Appl. Microbiol. 95:536-544. [DOI] [PubMed] [Google Scholar]
- 15.Hodgson, C. J., J. Perkins, and J. C. Labadz. 2003. Evaluation of biotracers to monitor effluent retention time in constructed wetlands. Lett. Appl. Microbiol. 36:362-371. [DOI] [PubMed] [Google Scholar]
- 16.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′→3′ exonuclease activity of Thermus aquaticus. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, J. Y., S. L. Ong, L. F. Song, Y. Y. Feng, W. T. Liu, T. W. Tan, L. Y. Lee, and W. J. Ng. 2003. Removal of MS2 bacteriophage using membrane technologies. Water Sci. Technol. 47:163-168. [PubMed] [Google Scholar]
- 18.Huertas, A., B. Barbeau, C. Desjardins, A. Galarza, M. A. Figueroa, and G. A. Toranzos. 2003. Evaluation of Bacillus subtilis and coliphage MS2 as indicators of advanced water treatment efficiency. Water Sci. Technol. 47:255-259. [PubMed] [Google Scholar]
- 19.Jolis, D. 2002. The effect of storage and lag time on MS2 bacteriophage susceptibility to ultraviolet radiation. Water Environ. Res. 74:516-520. [DOI] [PubMed] [Google Scholar]
- 20.Koper, O. B., J. S. Klabunde, G. L. Marchin, K. J. Klabunde, P. Stoimenov, and L. Bohra. 2002. Nanoscale powders and formulations with biocidal activity toward spores and vegetative cells of bacillus species, viruses, and toxins. Curr. Microbiol. 44:49-55. [DOI] [PubMed] [Google Scholar]
- 21.Korniewicz, D. M., B. E. Laughon, W. H. Cyr, C. D. Lytle, and E. Larson. 1990. Leakage of virus through used vinyl and latex examination gloves. J. Clin. Microbiol. 28:787-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuzmanovic, D. A., I. Elashvili, C. Wick, C. O'Connell, and S. Krueger. 2003. Bacteriophage MS2: molecular weight and spatial distribution of the protein and RNA components by small-angle neutron scattering and virus counting. Structure (Cambridge) 11:1339-1348. [DOI] [PubMed] [Google Scholar]
- 23.Lazarova, V., and P. Savoys. 2004. Technical and sanitary aspects of wastewater disinfection by UV irradiation for landscape irrigation. Water Sci. Technol. 50:203-209. [PubMed] [Google Scholar]
- 24.Marchin, G. L., J. Silverstein, and G. M. Brion. 1997. Effect of microgravity on Escherichia coli and MS-2 bacteriophage disinfection by iodinated resins. Acta Astronaut. 40:65-68. [DOI] [PubMed] [Google Scholar]
- 25.Mi, B., C. L. Eaton, J. H. Kim, C. K. Colvin, J. C. Lozier, and B. J. Marinas. 2004. Removal of biological and non-biological viral surrogates by spiral-wound reverse osmosis membrane elements with intact and compromised integrity. Water Res. 38:3821-3832. [DOI] [PubMed] [Google Scholar]
- 26.Nasser, A. M., R. Glozman, and Y. Nitzan. 2002. Contribution of microbial activity to virus reduction in saturated soil. Water Res. 36:2589-2595. [DOI] [PubMed] [Google Scholar]
- 27.O'Connell, K. P., M. Elmasri, J. Broyles, and D. M. Korniewicz. 2004. Testing for viral penetration of non-latex surgical and exam gloves: a comparison of three methods. Clin. Microbiol. Infect. 10:322-326. [DOI] [PubMed] [Google Scholar]
- 28.Olive, D. M., and P. Bean. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryan, J. N., R. W. Harvey, D. Metge, M. Elimelech, T. Navigato, and A. P. Pieper. 2002. Field and laboratory investigations of inactivation of viruses (PRD1 and MS2) attached to iron oxide-coated quartz sand. Environ. Sci. Technol. 36:2403-2413. [DOI] [PubMed] [Google Scholar]
- 29a.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Schijven, J. F., H. A. de Bruin, S. M. Hassanizadeh, and A. M. de Roda Husman. 2003. Bacteriophages and Clostridium spores as indicator organisms for removal of pathogens by passage through saturated dune sand. Water Res. 37:2186-2194. [DOI] [PubMed] [Google Scholar]
- 31.Selas, B., A. Lakel, Y. Andres, and P. Le Cloirec. 2003. Wastewater reuse in on-site wastewater treatment: bacteria and virus movement in unsaturated flow through sand filter. Water Sci. Technol. 47:59-64. [PubMed] [Google Scholar]
- 32.Shin, G.-A., and M. D. Sobsey. 2003. Reduction of Norwalk virus, poliovirus 1, and bacteriophage MS2 by ozone disinfection of water. Appl. Environ. Microbiol. 69:3975-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sommer, R., W. Pribil, S. Pfleger, T. Haider, M. Werderitsch, and P. Gehringer. 2004. Microbicidal efficacy of an advanced oxidation process using ozone/hydrogen peroxide in water treatment. Water Sci. Technol. 50:159-164. [PubMed] [Google Scholar]
- 34.Tanner, B. D., S. Kuwahara, C. P. Gerba, and K. A. Reynolds. 2004. Evaluation of electrochemically generated ozone for the disinfection of water and wastewater. Water Sci. Technol. 50:19-25. [PubMed] [Google Scholar]
- 35.Thomas, J. H., S. K. Kim, P. J. Hesketh, H. B. Halsall, and W. R. Heineman. 2004. Bead-based electrochemical immunoassay for bacteriophage MS2. Anal. Chem. 76:2700-2707. [DOI] [PubMed] [Google Scholar]
- 36.Thurston-Enriquez, J. A., C. N. Haas, J. Jacangelo, K. Riley, and C. P. Gerba. 2003. Inactivation of feline calicivirus and adenovirus type 40 by UV radiation. Appl. Environ. Microbiol. 69:577-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tree, J. A., M. R. Adams, and D. N. Lees. 2003. Chlorination of indicator bacteria and viruses in primary sewage effluent. Appl. Environ. Microbiol. 69:2038-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker, D. C., S.-V. Len, and B. Sheehan. 2004. Development and evaluation of a reflective solar disinfection pouch for treatment of drinking water. Appl. Environ Microbiol. 70:2545-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woolwine, J. D., and J. L. Gerberding. 1995. Effect of testing method on apparent activities of antiviral disinfectants and antiseptics. Antimicrob. Agents Chemother. 39:921-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiang, F., G. A. Anderson, T. D. Veenstra, M. S. Lipton, and R. D. Smith. 2000. Characterization of microorganisms and biomarker development from global ESI-MS/MS analyses of cell lysates. Anal. Chem. 72:2475-2481. [DOI] [PubMed] [Google Scholar]
- 41.Yao, Z. P., C. Afonso, and C. Fenselau. 2002. Rapid microorganism identification with on-slide proteolytic digestion followed by matrix-assisted laser desorption/ionization tandem mass spectrometry and database searching. Rapid Commun. Mass Spectrom. 16:1953-1956. [DOI] [PubMed] [Google Scholar]
- 42.You, Y., G. F. Vance, D. L. Sparks, J. Zhuang, and Y. Jin. 2003. Sorption of MS2 bacteriophage to layered double hydroxides: effects of reaction time, pH, and competing anions. J. Environ. Qual. 32:2046-2053. [DOI] [PubMed] [Google Scholar]