Abstract
Deep-subsurface samples obtained by deep drilling are likely to be contaminated with mesophilic microorganisms in the drilling fluid, and this could affect determination of the community structure of the geothermal microflora using 16S rRNA gene clone library analysis. To eliminate possible contamination by PCR-amplified 16S rRNA genes from mesophiles, a combined thermal denaturation and enzyme digestion method, based on a strong correlation between the G+C content of the 16S rRNA gene and the optimum growth temperatures of most known prokaryotic cultures, was used prior to clone library construction. To validate this technique, hot spring fluid (76°C) and river water (14°C) were used to mimic a deep-subsurface sample contaminated with drilling fluid. After DNA extraction and PCR amplification of the 16S rRNA genes from individual samples separately, the amplified products from river water were observed to be denatured at 82°C and completely digested by exonuclease I (Exo I), while the amplified products from hot spring fluid remained intact after denaturation at 84°C and enzyme digestion with Exo I. DNAs extracted from the two samples were mixed and used as a template for amplification of the 16S rRNA genes. The amplified rRNA genes were denatured at 84°C and digested with Exo I before clone library construction. The results indicated that the 16S rRNA gene sequences from the river water were almost completely eliminated, whereas those from the hot spring fluid remained.
In recent years, international drilling projects, such as the Integrated Ocean Drilling Program (http://www.iodp.org/) and the International Continental Scientific Drilling Program (http://icdp.gfz-potsdam.de/), have been expanded for the purpose of scientific study. The goal of the Integrated Ocean Drilling Program is to drill 7,000 m below the seafloor in the near future. The International Continental Scientific Drilling Program, on the other hand, has already drilled to a depth of more than 9,000 m during the KTB Continental Deep Drilling Project (5, 6). The temperature of the crust increases steadily with depth and at a fairly uniform rate. The subsurface thermal gradients are between 15°C and 30°C per km in nonvolcanic regions (10). The deep-subsurface crust is therefore considered to be a high-temperature environment. Many microbiologists have become very interested in this vast subsurface environment because of the number and variety of unknown thermophilic microbes that it harbors (9). Thermophiles and hyperthermophiles inhabiting subsurface environments beneath active deep-sea hydrothermal fields and continental hot springs have been studied extensively (11, 18, 22, 31, 33).
A major problem in obtaining core samples by drilling for the study of deep-subsurface microbial communities is contamination of the samples by mesophilic microbes in the drilling fluid, which consists primarily of surface seawater or river water. This contamination by mesophilic microbes invalidates culture-independent phylogenetic investigations that are based on molecular evolutionary markers, such as the 16S rRNA gene sequence, because PCR tends to amplify, without discrimination, genes from indigenous thermophiles as well as genes from contaminating mesophiles. To date, tracer monitoring with perfluorocarbon chemicals or fluorescent microspheres has been used to test for microbial contamination (27, 28). However, this type of tracer monitoring is both costly and time-consuming and is applicable only to investigation of complete and dense cores. Since the materials in deep-subsurface geothermal environments are denatured and cracked by heat from magma, tracer monitoring is difficult to use for determining microbial contamination by deep drilling. Two novel drilling systems to avoid contamination of core samples have been proposed, one using riser pipes (excavation with drilling fluid in a closed circulation system) and the other using filter-sterilized drilling fluid. However, both of these approaches require expensive equipment. For reliable investigations of deep-subsurface microbial communities by culture-independent techniques, it is necessary to develop new methods for eliminating mesophilic microbes contaminating the cores recovered from deep drilling.
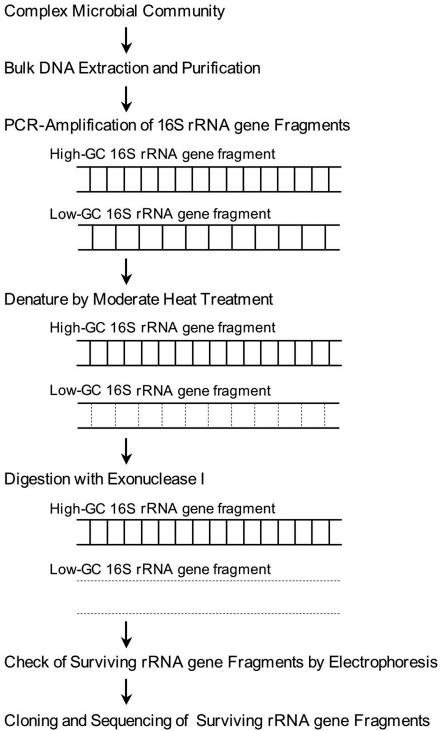
Here, we describe a simple method involving moderate heat treatment and digestion with exonuclease I (Exo I), which is known to be specific for single-stranded DNA (ssDNA) (4). This method is based on the following findings regarding the presence of rRNA genes in prokaryotes: the G+C contents of the rRNA genes are strongly correlated with the optimal growth temperatures of prokaryotes (8); the rRNA genes of thermophiles and hyperthermophiles have high G+C contents; and these high-G+C-content rRNA genes are indicative of melting temperatures (Tm) that are higher than those of mesophiles. Because of the differences in Tm values, 16S rRNA genes of mesophilic origin can be selectively denatured by moderate heat treatment, followed by digestion with Exo I. In this study, we demonstrated for the first time that there is a strong correlation between the G+C contents of 16S rRNA genes and their actual Tm values by using several bacterial strains with different optimal growth temperatures. Here we also describe a test using a mixture of hot spring water, simulating a deep-subsurface sample and containing thermophilic bacteria, and river water, simulating drilling fluid used in deep drilling and containing mesophilic bacteria, that was used to determine whether we eliminated 16S rRNA gene fragments from mesophiles by heat treatment and digestion with Exo I.
MATERIALS AND METHODS
Bacterial strains and environmental samples.
Seven bacterial strains, Psychrobacter okhotskensis JCM11840, Escherichia coli JCM5491, Anaerolinea thermoplia JCM11388, Thermoanaerobacter thermocopriae JCM7501, Thermus filiformis JCM11600, Thermotoga maritima JCM10099, and Aquifex pyrophilus JCM9492, were obtained from the Japan Collection of Microorganisms (http://www.jcm.riken.jp/). Each strain has a different optimal growth temperature in the range from 25 to 85°C. These strains were cultured under suitable conditions and were used for experimental measurement of the Tm values of PCR-amplified 16S rRNA gene fragments. In addition, the 16S rRNA gene sequences of these strains were obtained from the DDBJ/EMBL/GenBank database, and the G+C content of each of the sequences was calculated (Table 1).
TABLE 1.
Characterization of 16S rRNA genes, growth temperature ranges, and optimal growth temperatures of seven strains obtained from the Japan Collection of Microorganisms
| Type strain | 16S rRNA gene
|
Growth temp range (°C) | Topt (°C) | Reference | ||
|---|---|---|---|---|---|---|
| Accession no. | Length (bp) | G+C content (%)a | ||||
| Psychrobacter okhotskensis JCM11840 | AB094794 | 1,492 | 53.0 | 0-35 | 25 | 37 |
| Escherichia coli JCM5491 | J01859 | 1,541 | 54.4 | 8-48 | 37 | 3 |
| Anaerolinea thermoplia JCM11388 | AB046413 | 1,457 | 57.1 | 50-60 | 55 | 25 |
| Thermoanaerobacter thermocopriae JCM7501 | L09167 | 1,522 | 59.7 | 47-74 | 60 | 16 |
| Thermus filiformis JCM11600 | L09667 | 1,475 | 62.6 | 37-80 | 73 | 15 |
| Thermotoga maritima JCM10099 | M21774 | 1,562 | 63.6 | 55-90 | 80 | 13 |
| Aquifex pyrophilus JCM9492 | M83548 | 1,564 | 64.9 | 67-95 | 85 | 14 |
The G+C contents of 16S rRNA genes were calculated based on sequences obtained from the DDBJ/EMBL/GenBank database.
Two environmental samples, hot spring fluid (76°C) and river water (14°C), were collected at Nakabusa hot spring (36°23′20″N, 137°44′53″E), located in Nagano Prefecture, Japan. The sampling sites were very close to each other in the same hot spring field (the distance between the two sites was approximately 3 m).
DNA extraction, real-time PCR, and melting curve analysis.
To extract the genomic DNAs from the seven strains, a conventional extraction protocol was used (12). Cells of each strain were lysed with a lysozyme and proteinase K solution, and then the genomic DNAs were extracted with phenol-chloroform-isoamyl alcohol and precipitated with ethanol. The bulk DNAs in the two environmental samples were obtained by the following procedure. Exactly 2 liters of hot spring fluid and 2 liters of river water were each aseptically filtered with a Millipore (Bedford, MA) Sterivex-GV filter unit (pore size, 0.22 μm) using a tubing pump. The DNA from the microorganisms trapped by each of the two filter units was extracted by the method previously described by Somerville et al. (29).
The bacterial 16S rRNA genes from the DNAs of the seven reference strains and the two environmental samples were PCR amplified using a universal primer set, Bac27F and Uni1492R (19), and real-time PCR reagents (SYBR Green PCR master mixture; Applied Biosystems, Foster City, CA). After PCR amplification, the Tm values of the PCR products were immediately determined by melting curve analysis. Fluorescence signal monitoring for both the real-time PCR and the melting curve analyses was performed by using a 7300 real-time PCR system (Applied Biosystems, Foster City, CA).
PCR, cloning, and restriction fragment length polymorphism (RFLP).
The bacterial 16S rRNA genes from bulk DNAs from the hot spring fluid and the river water were PCR amplified using primers Bac27F and Uni1492R and KOD DNA polymerase (TOYOBO, Osaka, Japan). The resultant PCR products were cloned using a Zero Blunt TOPO PCR cloning kit (Invitrogen, Carlsbad, CA); 16S rRNA gene clone libraries were constructed separately.
Plasmid DNAs were isolated from single colonies from each of the two libraries originating from the two environmental samples. For this isolation we used a QIAprep Spin miniprep kit (QIAGEN, Valencia CA). The 16S rRNA gene inserts were PCR amplified using the following vector-specific primers: T7 (5′-TAA TAC GAC TCA CTA TAG GG-3′) and M13 reverse (5′-CAG GAA ACA GCT ATG A-3′). To group similar clones into operational taxonomic units (OTUs), the PCR products were divided based on RFLP analysis with restriction enzyme HaeIII (Promega, Madison, WI), which recognizes 4-bp restriction sites. The resulting RFLP was visualized by electrophoresis with a 2% agarose gel. The PCR products that had the same restriction patterns were analyzed again with another 4-bp recognizant restriction enzyme, RsaI (Promega, Madison, WI), and further divided into groups. This analysis was repeated once more with another 4-bp recognizant restriction enzyme, MspI (Promega, Madison, WI).
Sequencing and homology search.
The sequences of representative 16S rRNA gene clones in each OTU were determined with a capillary DNA sequencer (CEQ 8000 genetic analysis system; Beckman Coulter, Fullerton, CA). For the sequencing reactions, vector-specific primers T7 and M13 reverse and the following internal 16S rRNA gene-specific primers were used: 324R, 357F, 519R, 530F, 1114F, 1100R, 1389R (19), and 803R (30). The sequences were checked for chimera formation by using the Chimera check program of the Ribosomal Database Project II database (21). The OTUs that survived the Chimera check analysis were homology searched using the program FASTA (20) in the DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp/).
Heat treatment and digestion with Exo I.
Bacterial 16S rRNA genes from bulk DNAs in hot spring fluid and river water were PCR amplified using primers Bac27F and Uni1492R. In addition, a DNA mixture was prepared by blending equal amounts of bulk DNA extracted from the two environmental samples. The bacterial 16S rRNA genes in this DNA mixture were then amplified. The products from each of the three PCRs were denatured for 3 min at various temperatures between 80°C and 86°C and immediately chilled on ice in order to prevent rehybridization of denatured 16S rRNA genes. Exo I (TaKaRa, Kyoto, Japan) was then added to the chilled reaction tubes, and this was followed by incubation at 37°C for 30 min to allow sufficient digestion of the denatured rRNA genes. Since Exo I is a 3′-to-5′ exonuclease specific for ssDNA (4), all ssDNAs in a reaction tube were completely digested, leaving any double-stranded DNA intact (details of the procedure are shown in Fig. 1). Prior to cloning of the surviving 16S rRNA gene fragments, the Exo I was inactivated by heating the sample at 80°C for 15 min. Bacterial 16S rRNA gene fragments that survived the heat treatment were cloned and grouped by RFLP analysis. A representative clone in each OTU was sequenced using the universal forward primer Bac27F, which determined approximately 600 bp. The resulting sequences were homology searched in the DNA Data Bank of Japan using FASTA and were compared with sequences obtained from the original hot spring fluid and river water samples.
FIG. 1.
Flow chart showing the concept of the approach used for elimination of 16S rRNA genes with low G+C contents, generally found in mesophiles, and for selective analysis of 16S rRNA gene fragments with higher G+C contents derived from thermophiles and hyperthermophiles.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences determined in this study have been deposited in the DDBJ/EMBL/GenBank database under the following accession numbers: AB199562 to AB199567 (hot spring fluid) and AB199568 to AB199581 (river water).
RESULTS AND DISCUSSION
Correlation between optimal growth temperatures of various prokaryotes and the G+C contents of their 16S rRNA gene sequences.
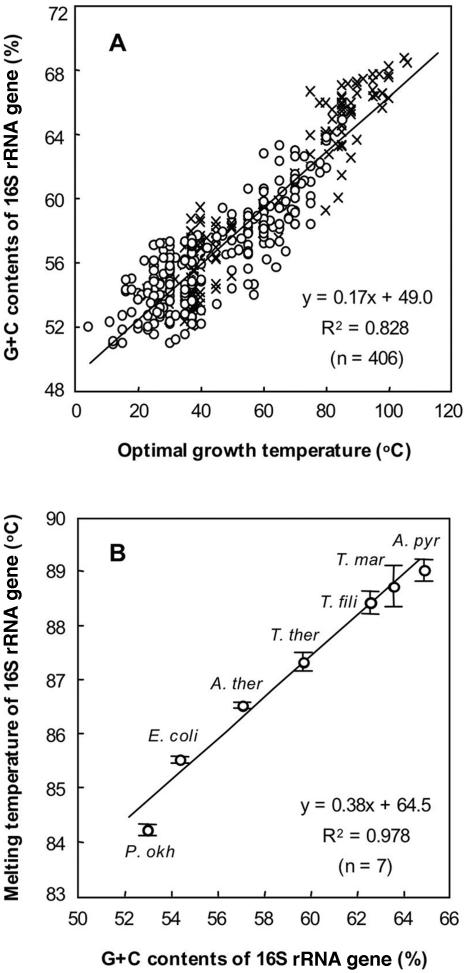
Until now, no correlation has been found between the growth temperature ranges of prokaryotes and their genomic G+C contents. However, we found that there is a correlation between the G+C contents of the 16S rRNA genes of different prokaryotes and their optimal growth temperatures. We plotted the optimal growth temperatures against the G+C contents of the 16S rRNA genes of 406 prokaryotes (254 bacterial strains and 152 archaeal strains) (Fig. 2A). The data plot shows that the higher the optimal growth temperature (Topt) of a prokaryote, the higher its G+C content (PGC) (PGC = 0.17Topt + 49.0; R2 = 0.828), indicating that there is a direct correlation between these values and suggesting that the 16S rRNA genes of thermophiles and hyperthermophiles generally have higher G+C contents than the 16S rRNA genes of mesophiles or psychrophiles.
FIG. 2.
Correlations between the optimal growth temperatures of prokaryotic strains and the G+C contents of their 16S rRNA genes (A) and between the G+C contents and the actual melting temperatures of 16S rRNA gene fragments amplified from seven bacterial strains (B). (A) Bacterial strains (○) and archaeal strains (×) were randomly chosen from various publications reviewing the phylogeny, metabolism, and taxonomy of microorganisms (2, 24). (B) Seven strains were selected from the bacterial strains, and the plot indicates data for the 16S rRNA gene fragments amplified from Psychrobacter okhotskensis (P. okh), Escherichia coli (E. coli), Anaerolinea thermoplia (A. ther), Thermoanaerobacter thermocopriae (T. ther), Thermus filiformis (T. fili), Thermotoga maritima (T. mar), and Aquifex pyrophilus (A. pyr). The 16S rRNA gene sequences of the strains were obtained from the DDBJ/EMBL/GenBank database. The G+C contents were calculated with GENETYX-MAC. The error bars indicate the standard deviations for triplicate determinations.
Although an obvious correlation between the G+C contents and optimal growth temperatures was recognized (Fig. 2A), a few exceptions were also found, including some strains of the genera Deinococcus and Halomonas in the domain Bacteria and some strains of the genera Halococcus and Halorubrum in the domain Archaea. All of these organisms are mesophilic strains but have relatively high 16S rRNA gene G+C contents (56.6 to 59.4%). These exceptional strains were halophilic or radiation-tolerant microorganisms that live in extreme environments, which appears to be reflected in the high G+C contents of their 16S rRNA genes. Furthermore, we also found thermophiles that have relatively low 16S rRNA gene G+C contents, including some strains of the genus Methanothermus in the domain Archaea and some strains of the genera Caminicella, Thermoanaerobacter, and Desulfotomaculam in the domain Bacteria. These thermophilic strains had relatively low 16S rRNA gene G+C contents (54.1 to 60.0%). Aside from these few exceptions, the majority of prokaryotes were in line with the correlation.
It is well established that the G+C content is closely correlated with the Tm of DNA fragments. In order to determine whether there is also a correlation for PCR-amplified 16S rRNA gene fragments, the 16S rRNA gene of each of the seven randomly selected bacterial strains was amplified. The Tm of the 16S rRNA gene fragments from each of the strains was determined by melting curve analysis. The correlation between the Tm values and the G+C contents of the 16S rRNA genes is shown in Fig. 2B. The results showed that there was a strong correlation between the Tm values and the G+C contents within the 99% confidence range (Tm = 0.38PGC + 64.5; R2 = 0.978). There was also a strong correlation between the Topt values of the representative strains and their Tm values within the 99% confidence range (Tm = 0.08Topt + 82.3; R2 = 0.989).
These mutual relationships suggest that the PCR-amplified 16S rRNA gene fragments of thermophiles and hyperthermophiles have higher Tm values than the PCR-amplified 16S rRNA gene fragments of mesophiles or psychrophiles. Because the 16S rRNA gene fragments from mesophiles or psychrophiles have Tm values that are lower than those of the 16S rRNA gene fragments from thermophiles and hyperthermophiles, it is reasonable to expect that 16S rRNA gene fragments from mesophiles and psychrophiles can be completely eliminated by first denaturing them at a temperature below the lowest Tm at which the 16S rRNA gene fragments from the thermophiles and hyperthermophiles can be denatured and then digesting them with an ssDNA-digesting enzyme (Fig. 1). Our proposed method makes culture-independent analysis of the microflora in a sample from a geothermal environment easily possible, even when there is sample contamination by drilling fluid containing a large number of mesophiles.
Melting temperatures of 16S rRNA genes obtained from environmental samples.
The bacterial 16S rRNA genes from the bulk DNAs in hot spring fluid (76°C) and river water (14°C) were PCR amplified, and their Tm values were determined by melting curve analysis. The average Tm for the 16S rRNA gene fragments from the hot spring fluid was 86.8°C, whereas the average Tm for the 16S rRNA gene fragments from the river water was lower, 84.5°C. The mean G+C contents, inferred from the regression analysis (Fig. 2B), were approximately 59% in the case of the fragments from the hot spring fluid and 53% in the case of the fragments derived from river water. The actual measurements clearly indicated that microbes containing 16S rRNA genes with high Tm values and therefore with high G+C contents were dominant and that microbes in the river water had 16S rRNA gene sequences with low G+C contents.
Phylogenetic analysis of a bacterial community in hot spring fluid.
Approximately 1.5-kb portions of bacterial 16S rRNA gene fragments from the bulk DNAs in the hot spring fluid were amplified. A clone library was constructed, and 57 clones were analyzed (Table 2). The 16S rRNA gene clones were divided into six OTUs based on RFLP (NHS-01 to NHS-06). The phylogenetic analysis revealed that these OTUs belonged to the classes Aquificae, Thermodesulfobacteria, Gammaproteobacteria, and Betaproteobacteria. NHS-01, NHS-02, and NHS-03 were phylogenetically included in a cluster of Aquificae that was primarily composed of a number of authentic thermophilic strains and environmental clones obtained from hot springs and subsurface geothermal water (26, 32, 36). Clone sequences belonging to these Aquificae-related OTUs (NHS-01, NHS-02, and NHS-03) accounted for 70% of the clones in the library. NHS-04 was closely related to Geothermobacterium ferrireducens, which is known to be a hyperthermophile that is able to use Fe(III) as an electron acceptor (17). The clones in NHS-04 accounted for 12% of the clones. NHS-05 exhibited the highest level of homology to Acidithiobacillus ferrooxidans in the Gammaproteobacteria and accounted for 11% of the entire library. The closest relative, A. ferrooxidans, is a mesophile. However, NHS-05 appeared to be a clone that originated from a thermophilic bacterium, since the G+C content of the NHS-05 fragments was clearly high. NHS-06 showed the closest match to an environmental clone from a hot spring in Yellowstone National Park (SM2G09). The clones in NHS-06 accounted for 7% of all clones.
TABLE 2.
16S rRNA gene sequences obtained from hot spring fluid (76°C) and river water (14°C)
| Library | OTU | No. of clones | 16S rRNA gene
|
Bacterial division | Closest database match (%) | |
|---|---|---|---|---|---|---|
| Length (bp) | G+C content (%) | |||||
| Hot spring | NHS-01 | 18 | 1,484 | 57.1 | Aquificae | SRI-40 (97-98) |
| NHS-02 | 11 | 1,506 | 57.1 | Aquificae | NAK14 (96-97) | |
| NHS-03 | 11 | 1,519 | 60.9 | Aquificae | Hydrogenobacter subterraneus (95-96) | |
| NHS-04 | 7 | 1,534 | 62.2 | Thermodesulfobacteria | Geothermobacterium ferrireducens (96-97) | |
| NHS-05 | 6 | 1,522 | 58.3 | Gammaproteobacteria | Acidithiobacillus ferrooxidans (90-91) | |
| NHS-06 | 4 | 1,504 | 57.1 | Betaproteobacteria | SM2G09 (88-89) | |
| Total | 57 | |||||
| River water | RVW-01 | 18 | 1,492 | 53.8 | Betaproteobacteria | LO13-5 (95-96) |
| RVW-02 | 13 | 1,498 | 53.3 | Betaproteobacteria | KD3-141 (97-98) | |
| RVW-03 | 4 | 1,497 | 53.5 | Gammaproteobacteria | NB0.1-H (99-100) | |
| RVW-04 | 4 | 1,490 | 49.8 | Bacteroidetes | Dysgonomonas capnocytophagoides (92-93) | |
| RVW-05 | 3 | 1,487 | 50.6 | Bacteroidetes | PHOS-HE21 (85-86) | |
| RVW-06 | 3 | 1,490 | 55.7 | Betaproteobacteria | Leptothrix discophora (96-97) | |
| RVW-07 | 2 | 1,501 | 52.2 | Betaproteobacteria | ARKMP-77 (93-94) | |
| RVW-08 | 1 | 1,500 | 50.8 | Bacteroidetes | Flavobacterium ferrugineum (91-92) | |
| RVW-09 | 1 | 1,489 | 50.7 | Bacteroidetes | MBIC4147 (90-91) | |
| RVW-10 | 1 | 1,473 | 50.6 | Bacteroidetes | BIjii 32 (84-85) | |
| RVW-11 | 1 | 1,511 | 52.9 | Gammaproteobacteria | MS-81-1c (86-87) | |
| RVW-12 | 1 | 1,502 | 51.1 | Betaproteobacteria | HOClCi44 (84-85) | |
| RVW-13 | 1 | 1,495 | 53.7 | Betaproteobacteria | Rhodoferax ferrireducens (97-98) | |
| RVW-14 | 1 | 1,493 | 50.1 | Candidate division TM6 | NMW3.210WL (84-85) | |
| Total | 54 | |||||
The phylogenetic analysis indicated that almost all clones were closely related to clusters primarily composed of thermophilic microbes and/or thermal environment-related clone sequences. These findings suggest that all of the 16S rRNA gene fragments originated from thermophilic microbes inhabiting the hot spring. The G+C contents of the 16S rRNA gene fragments ranged from 57.1% (NHS-01, NHS-02, and NHS-06) to 62.2% (NHS-04), and the overall average was 58.6% (Table 2); the mean unexpectedly agreed with the G+C content inferred from the actual Tm of the total rRNA gene fragments.
Phylogenetic analysis of a bacterial community in river water.
A 16S rRNA gene clone library for the river water was constructed by using the same method that was used in the analysis of the hot spring fluid, and 54 clones in the library were analyzed (Table 2). The clones were divided into 14 OTUs based on RFLP (RVW-01 to RVW-14). The phylogenetic analysis indicated that these 14 OTUs belonged to clusters of the bacterial divisions Betaproteobacteria, Gammaproteobacteria, and Bacteroidetes and candidate division TM6 (34). All OTUs were closely related to mesophilic bacteria and environmental clones obtained from nongeothermal fields, including peat soils (23), forest soils (1), industrial bioreactor samples (7), and drinking water (35).
The G+C contents of these 16S rRNA gene fragments ranged from 49.8% (RVW-04) to 55.7% (RVW-06), and the overall average was 52.9% (Table 2). The average G+C content also agreed with that inferred from the actual Tm values of the total rRNA gene fragments.
Heat treatment, digestion, and cloning of surviving 16S rRNA genes.
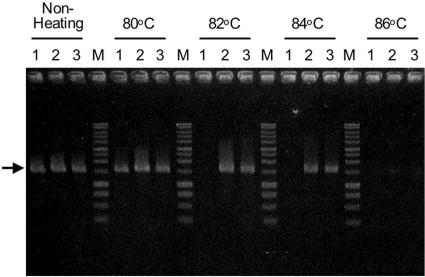
The bacterial 16S rRNA genes from bulk DNAs in hot spring fluid and river water were again amplified. In addition, a DNA mixture containing equal amounts of bulk DNA from hot spring fluid and bulk DNA from river water was prepared, and the 16S rRNA genes in the mixture were amplified by PCR under the conditions described above. The PCR products from these three amplifications were heat denatured at several temperatures and digested by Exo I. The extent of survival of each 16S rRNA gene was visualized by electrophoresis (Fig. 3). The PCR products obtained from the river water sample were completely denatured by heat treatment at 82°C, and consequently, all products disappeared after subsequent digestion with Exo I. In contrast, the 16S rRNA gene fragments from the hot spring survived heat treatment even at 84°C, as well as subsequent digestion, since their Tm values were clearly higher than those of the rRNA gene fragments that originated from the river water sample.
FIG. 3.
Heat denaturation and digestion with exonuclease I of bacterial 16S rRNA genes, as visualized in a 1.0% agarose gel. Lanes 1, 16S rRNA genes amplified from bulk DNA derived from river water; lanes 2, 16S rRNA genes amplified from bulk DNA derived from hot spring fluid; lanes 3, 16S rRNA gene fragments amplified from a DNA mixture that was a blend of equal amounts of bulk DNAs extracted from hot spring fluid and river water; lanes M, DNA marker (1-kb DNA ladder; Promega, Madison, WI). The temperatures of the heat treatments are indicated at the top. The size of each amplified 16S rRNA gene fragment was approximately 1,500 bp (arrow).
After heat treatment and digestion of PCR products derived from the DNA mixture, the survival pattern was almost identical to the pattern for the PCR products derived from the hot spring sample (Fig. 3). A clone library was constructed from the rRNA gene fragments derived from the DNA mixture that survived heat treatment at 84°C, as well as subsequent digestion. Fifty-nine clones in the library were analyzed and divided into seven OTUs (MIX-01 to MIX-07) based on RFLP (Table 3). The phylogenetic analysis suggested that these OTUs belonged to the classes Aquificae, Thermodesulfobacteria, Gammaproteobacteria, and Betaproteobacteria. The composition of these OTUs was very similar to that of the OTUs derived from the hot spring sample, both in terms of homology and in terms of frequency. Six of the seven OTUs exhibited significant similarity to the rRNA genes from the hot spring fluid, and almost all of the clones (93% of all clones) clearly originated from thermophilic microbes. However, one OTU, MIX-06, was closely related to RVW-06, which was found in the clone library created from the river water sample, and this OTU was considered a 16S rRNA gene of mesophilic origin. Because the sequence of RVW-06 had the highest G+C content (55.7%) among the 16S rRNA genes from river water, it was also able to survive heat treatment at 84°C.
TABLE 3.
Frequency and homology of 16S rRNA gene fragments that were amplified from a DNA mixture (DNAs extracted from hot spring fluid and river water) and that survived heat treatment at 84°C and subsequent digestion by Exo I
| OTU | No. of clones | Bacterial division | Closest database match (%) |
|---|---|---|---|
| MIX-01 | 13 | Aquificae | NHS-01 (99-100) |
| MIX-02 | 12 | Aquificae | NHS-02 (99-100) |
| MIX-03 | 10 | Thermodesulfobacteria | NHS-04 (99-100) |
| MIX-04 | 9 | Aquificae | NHS-03 (98-99) |
| MIX-05 | 6 | Gammaproteobacteria | NHS-05 (99-100) |
| MIX-06 | 5 | Betaproteobacteria | RVW-06 (99-100) |
| MIX-07 | 4 | Betaproteobacteria | NHS-06 (98-99) |
| Total | 59 |
Applicability of the method for selective phylogenetic analysis of archaea.
In this study, we developed a new method of selective phylogenetic analysis, and we demonstrated that the approach which we used was effective when a PCR primer set for bacterial 16S rRNA genes was used. To test the applicability of the this method for phylogenetic analysis of archaea, PCR amplification with a primer set for archaeal 16S rRNA genes was also performed. Whereas the archaeal rRNA genes derived from the fluid of the hot spring were easily amplified by PCR, no PCR product was obtained from the river water, in spite of repeated trials with various primer sets. Therefore, in this paper, we limit our discussion to the bacterial clone analysis. However, it should be noted that there was an obvious correlation between the optimal growth temperatures and the G+C contents of the 16S rRNA gene sequences of archaea (PGC = 0.20Topt + 47.8; R2 = 0.901; n = 152) (Fig. 2A). Thus, this method ought to be applicable in a selective phylogenetic analysis of thermophilic and hyperthermophilic archaea.
Conclusions.
In this paper we demonstrate that 16S rRNA gene fragments can be efficiently removed from a mixture of 16S rRNA genes derived from a microbial community of mesophiles and thermophiles by a new method based on differences between Tm values of 16S rRNA genes from mesophiles and thermophiles. This method is useful for phylogenetic analyses targeted at thermophilic or hyperthermophilic bacteria (and probably also archaea) in geothermal or hydrothermal samples, e.g., deep-subsurface cores obtained in the process of deep drilling that may be contaminated with abundant mesophilic microorganisms in the drilling fluid. The new method has the following advantages: (i) the experimental procedures are very simple to perform and can be completed within 1 h; (ii) the method does not require costly and bulky equipment to monitor samples for microbial contamination, nor does it require a novel drilling system with riser pipes and filter-sterilized drilling fluid; and (iii) the method frees microbiologists from cumbersome aseptic treatment of core samples at drilling sites. Our proposed method presents a solution to dealing with the problem of possible microbial contamination of samples obtained by deep drilling.
On the other hand, significant amounts of rRNA gene of mesophilic origin have been detected in geothermal cores or hot groundwater samples, and several researchers believe that some kinds of mesophiles may survive even in deep-subsurface geothermal environments. Although whether these mesophiles are indigenous to geothermal environments or just introduced during sampling is still controversial, the possibility of their presence in such environments must be considered. If they do exist in such environments, our method risks missing the presence of indigenous mesophiles because it automatically eliminates rRNA genes with low G+C contents. This needs to be kept in mind when our method is used. In deep drilling, however, the cores recovered from the deep-subsurface environments are often contaminated with abundant mesophiles via the drilling fluid. The cell density of the mesophiles in the drilling fluid is much higher than the cell density of thermophiles or hyperthermophiles that inhabit deep-subsurface environments. In this case, our method is exceedingly effective for phylogenetic analysis, because it can efficiently eliminate very large numbers of clone sequences introduced from the contaminating mesophiles. This approach is thus a time-saving and cost-effective technique for clone analyses of deep-subsurface cores contaminated with massive amounts of mesophiles via drilling fluid.
Acknowledgments
This work was supported in part by a Special Coordination Fund, “International Research Project on the Interaction between the Sub-Vent Biosphere and Geo-Environment (Archaean Park Project),” from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
REFERENCES
- 1.Axelrood, P. E., M. L. Chow, C. C. Radomski, J. M. McDermott, and J. Davies. 2002. Molecular characterization of bacterial diversity from British Columbia forest soils subjected to disturbance. Can. J. Microbiol. 48:655-674. [DOI] [PubMed] [Google Scholar]
- 2.Boone. D. R., and R. W. Castenholz. 2001. Bergey's manual of systematic bacteriology 2nd ed., vol. 1. Springer-Verlag, New York, N.Y.
- 3.Brenner, D. J., B. R. Davis, A. G. Steigerwalt, C. F. Riddle, A. C. McWhorter, S. D. Allen, J. J. Farmer III, Y. Saitoh, and G. R. Fanning. 1982. Atypical biogroups of Escherichia coli found in clinical specimens and description of Escherichia hermannii sp. nov. J. Clin. Microbiol. 15:703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brody, R., K. Doherty, and P. Zimmerman. 1986. Processivity and kinetics of the reaction of exonuclease-I from Escherichia coli with polydeoxyribonucleotides. J. Biol. Chem. 261:7136-7143. [PubMed] [Google Scholar]
- 5.Drescher, J., T. Kirsten, and K. Schafer. 1998. The rate gas inventory of the continental crust, recovered by the KTB continental deep drilling project. Earth Planet. Sci. Lett. 154:247-263. [Google Scholar]
- 6.Emmermann, R. 1995. Abenteuer Tiefbohrung. Geowissenschaften 13:114-128. [Google Scholar]
- 7.Friedrich, U., K. Prior, K. Altendorf, and A. Lipski. 2002. High bacterial diversity of a waste gas-degrading community in an industrial biofilter as shown by a 16S rDNA clone library. Environ. Microbiol. 4:721-734. [DOI] [PubMed] [Google Scholar]
- 8.Galtier, N., N. Tourasse, and M. Gouy. 1999. A nonhyperthermophilic common ancestor to extant life forms. Nature 283:220-221. [DOI] [PubMed] [Google Scholar]
- 9.Gold, T. 1992. The deep, hot biosphere. Proc. Natl. Acad. Sci. USA 89:6045-6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gold, T. 1999. Life at borders, p. 11-36. In T. Gold (ed.), The deep hot biosphere: the myth of fossil fuels. Springer-Verlag, New York, N.Y.
- 11.Higashi, Y., M. Sunamura, K. Kitamura, K.-I. Nakamura, Y. Kurusu, J.-I. Ishibashi, T. Urabe, and A. Maruyama. 2004. Microbial diversity in hydrothermal surface to subsurface environments of Suiyo Seamount, Izu-Bonin Arc, using a catheter-type in situ growth chamber. FEMS Microbiol. Ecol. 47:327-336. [DOI] [PubMed] [Google Scholar]
- 12.Hiraishi, A. 1992. Direct automated sequencing of 16S rDNA amplified by polymerase chain reaction from bacterial cultures without DNA purification. Lett. Appl. Microbiol. 15:210-213. [DOI] [PubMed] [Google Scholar]
- 13.Huber, R., T. A. Langworthy, H. König, M. Thomm, C. R. Woese, U. B. Sleytr, and K. O. Stetter. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90oC. Arch. Microbiol. 144:324-333. [Google Scholar]
- 14.Huber, R., T. Wilharm, D. Huber, A. Trincone, S. Burggraf, H. König, R. Rachel, I. Rockinger, H. Fricke, and K. O. Stetter. 1992. Aquifex pyrophilus gen. nov. sp. nov., represents a novel group of marine hyperthermophilic hydrogen-oxidizing bacteria. Syst. Appl. Microbiol. 15:340-351. [Google Scholar]
- 15.Hudson, J. A., H. W. Morgan, and R. M. Daniel. 1987. Thermus filiformis sp. nov., a filamentous caldoactive bacterium. Int. J. Syst. Bacteriol. 37:431-436. [Google Scholar]
- 16.Jin, F., K. Yamasato, and K. Toda. 1988. Clostridium thermocopriae sp. nov., a cellulolytic thermophile from animal feces, compost, soil, and a hot spring in Japan. Int. J. Syst. Bacteriol. 38:279-281. [Google Scholar]
- 17.Kashefi, K., D. E. Holmes, A.-L. Reysenbach, and D. R. Lovley. 2002. Use of Fe(III) as an electron acceptor to recover previously uncultured hyperthermophiles: isolation and characterization of Geothermobacterium ferrireducens gen. nov., sp. nov. Appl. Environ. Microbiol. 68:1735-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura, H., R. Asada, A. Masta, and T. Naganuma. 2003. Distribution of microorganisms in the subsurface of the Manus Basin hydrothermal vent field in Papua New Guinea. Appl. Environ. Microbial. 69:644-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.) Nucleic acid techniques in bacterial systematics. John Willy and Sons, New York, N.Y.
- 20.Lipman, D. J., and W. R. Person. 1985. Rapid and sensitive protein similarity searches. Science 227:1435-1441. [DOI] [PubMed] [Google Scholar]
- 21.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marteinsson, V. T., S. Hauksdóttir, C. F. V. Hobel, H. Kristmannsdóttir, G. O. Hreggvidsson, and J. K. Kristjánsson. 2001. Phylogenetic diversity analysis of subterranean hot springs in Iceland. Appl. Environ. Microbiol. 67:4242-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris, S. A., S. Radajewski, T. W. Willison, and J. C. Murrell. 2002. Identification of the functionally active methanotroph population in a peat soil microcosm by stable-isotope probing. Appl. Environ. Microbiol. 68:1446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reysenbach, A.-L., D. Götz, and D. Yernool. 2002. Microbial diversity of marine and terrestrial thermal springs, p. 345-421. In J. T. Staley and A.-L. Resysenbach (ed.), Biodiversity of microbial life. Foundation of Earth's biosphere. Wiley-Liss, Inc., New York, N.Y.
- 25.Sekiguchi, Y., T. Yamada, S. Hanada, A. Ohashi, H. Harada, and Y. Kamagata. 2003. Anaerolinea thermophila gen. nov., sp. nov. and Caldinea aerophila gen. nov., sp. nov., novel filamentous thermophiles that represent a previous uncultured lineage of the domain Bacteria at the subphylum level. Int. J. Syst. Evol. Microbiol. 53:1843-1851. [DOI] [PubMed] [Google Scholar]
- 26.Skirnisdottir, S., G. O. Hreggvidsson, S. Hjörleifsdottir, V. T. Marteinsson, S. K. Petursdottir, O. Holst, and J. K. Kristjansson. 2000. Influence of sulfide and temperature on species composition and community structure of hot spring microbial mats. Appl. Environ. Microbiol. 66:2835-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith, D. C., A. J. Spivack, M. R. Fisk, S. A. Haveman, H. Staudigel, and the Leg 185 Shipboard Scientific Party. 2000. Methods for quantifying potential microbial contamination during deep ocean coring. Ocean Drilling Program Technical Note 28. [Online.] http://www-odp.tamu.edu/publications/tnotes/tn28/INDEX.HTM.
- 28.Smith, D. C., A. J. Spivack, M. R. Fisk, S. A. Haveman, H. Staudigel, and Ocean Drilling Program Leg 185 Shipboard Scientific Party. 2000. Tracer-based estimates of drilling-induced microbial contamination of deep-sea crust. Geomicrobiol. J. 17:207-219. [Google Scholar]
- 29.Somerville, C. C., I. T. Knight, W. L. Straube, and R. R. Colwell. 1989. Simple, rapid method for direct isolation of nucleic acids from aquatic environments. Appl. Environ. Microbiol. 55:548-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stackebrandt, E., and O. Charfreitag. 1990. Partial 16S rRNA primary structure of five Actimomyces species: phylogenetic implications and development of an Actinomyces israelii-specific oligonucleotide probe. J. Gen. Microbiol. 136:37-43. [DOI] [PubMed] [Google Scholar]
- 31.Summit, M., and J. A. Baross. 2001. A novel microbial habitat in the midocean ridge subseafloor. Proc. Natl. Acad. Sci. USA 98:2158-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takai, K., T. Komatsu, and K. Horikoshi. 2001. Hydrogenobacter subterraneus sp. nov., an extremely thermophilic, heterotrophic bacterium unable to grow on hydrogen gas, from deep subsurface geothermal water. Int. J. Syst. Evol. Microbiol. 51:1425-1435. [DOI] [PubMed] [Google Scholar]
- 33.Takai, K., T. Gamo, U. Tsunogai, N. Nakayama, H. Hirayama, K. N. Nealson, and K. Horikoshi. 2004. Geochemical and microbiological evidence for a hydrogen-based, hyperthermophilic subsurface lithoautotrophic microbial ecosystem (HyperSLiME) beneath an active deep-sea hydrothermal field. Extremophiles 8:269-282. [DOI] [PubMed] [Google Scholar]
- 34.von Wintzingerode, F., O. Landt, A. Ehrlich, and U. B. Göbel. 2000. Peptide nucleic acid-mediated PCR clamping as a useful supplement in the determination of microbial diversity. Appl. Environ. Microbiol. 66:549-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams, M. M., J. W. Domingo, M. C. Meckes, C. A. Kelty, and H. S. Rochon. 2004. Phylogenetic diversity of drinking water bacteria in a distribution system simulator. J. Appl. Microbiol. 96:954-964. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto, H., A. Hiraishi, K. Kato, H. X. Chiura, Y. Maki, and A. Shimizu. 1998. Phylogenetic evidence for the existence of novel thermophilic bacteria in hot spring sulfur-turf microbial mats in Japan. Appl. Environ. Microbiol. 64:1680-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yumoto, I., K. Hirota, Y. Sogabe, Y. Nodasaka, Y. Yokota, and T. Hoshino. 2003. Psychrobacter okhotskensis sp. nov., a lipase-producing facultative psychrophile isolated from the coast of the Okhotsk Sea. Int. J. Syst. Evol. Microbiol. 53:1985-1989. [DOI] [PubMed] [Google Scholar]