Abstract
The HilC and HilD proteins of Salmonella enterica serovar Typhimurium are members of the AraC/XylS family of transcription regulators. They are encoded on Salmonella pathogenicity island 1 (SPI1) and control expression of the hilA gene, which encodes the major transcriptional activator for many genes encoded on SPI1 and elsewhere that contribute to invasion of host cells. Gel electrophoretic shift and DNase footprinting assays revealed that purified HilC and HilD proteins can bind to multiple regions in the hilA and hilC promoters and to a single region in the hilD promoter. Although both HilC and -D proteins can bind to the same DNA regions, they showed different dependencies on the sequence and lengths of their DNA targets. To identify the binding-sequence specificity of HilC and HilD, a series of single base substitutions changing each position in a DNA fragment corresponding to positions −92 to −52 of the hilC promoter was tested for binding to HilC and HilD in a gel shift DNA-binding assay. This mutational analysis in combination with sequence alignments allowed deduction of consensus sequences for binding of both proteins. The consensus sequences overlap but differ so that HilC can bind to both types of sites but HilD only to one. The hilA and hilC promoters contain multiple binding sites of each type, whereas the hilD promoter contains a site that binds HilC but not HilD without additional binding elements. The HilC and HilD proteins had no major effect on transcription from the hilA or hilD promoters using purified proteins in vitro but changed the choice of promoter at hilC. These results are consistent with a model derived from analysis of lacZ fusions stating that HilC and HilD enhance hilA expression by counteracting a repressing activity.
Salmonella enterica serovars are prevalent bacterial pathogens that cause diseases ranging from localized gastroenteritis to disseminated enteric fevers in humans and animals. Salmonellae express a variety of virulence factors, including very polymorphic surface carbohydrates, multiple fimbrial adhesins, phase-variable flagella, and mechanisms for invasion and survival in host macrophages and other cells (see review in references 13 and 44). Because Salmonella is acquired mainly by oral ingestion of contaminated materials, a key step of infection is its passage across the intestinal epithelium by invasion of M cells in Peyer's patches (7, 20, 39) or of enterocytes (46). Many of the genes required for intestinal penetration and invasion of host cells are carried on the 40-kb region at centisome 63, which is called Salmonella pathogenicity island 1 (SPI1) (reviewed in reference 10). Genes of SPI1 encode a type III secretion system (20, 33), which can inject into host cells various effector proteins, including those encoded by sptP in SPI1, sopB in SPI5, sopD at centisome 64, and sopE2 at centisome 40. These injected proteins elicit cytoskeletal changes in host cells that lead to bacterial internalization (8, 14, 40). The SPI1 locus and many of these effector genes are highly conserved in Salmonella lineages (34). SPI1 function contributes to cell invasion, intestinal colonization, destruction of M cells in Peyer's patches, activation of cytokine secretion, and triggering of neutrophil migration (reviewed in reference 31). Genetic disruption of SPI1 functions typically reduces infectivity by the oral route (15) (also see reference 35).
Expression of SPI1 invasion and effector genes responds to multiple environmental signals and is decreased under conditions of high oxygen, low osmolarity, low pH, and stationary-phase growth (5, 11, 16, 25). Many regulatory proteins influence invasion gene expression (31, 32), but the key regulator is the SPI1-encoded HilA protein, which can activate their transcription directly (1, 4, 28) or by increasing expression of the activator InvF (9). The N-terminal region of HilA carries a DNA-binding and transcription-activating helix-loop-helix motif typical of OmpR/ToxR family members (4). In laboratory culture hilA-lacZ fusions respond to numerous regulatory genes, including the PhoP/PhoQ, BarA/SirA, and EnvZ/OmpR two-component regulatory systems and the CsrAB, FliZ, and FadD regulatory proteins (2, 30). Of the small nucleoid-binding proteins, H-NS and Hha can repress hilA expression, but FIS and HU may activate (12; citation in references 30 and 47). Action of these factors and the repression of hilA by low osmolarity or high oxygen require DNA sequences within positions −332 to −39 upstream of the hilA transcription start site (42). Deletion of this upstream region results in high and unregulated hilA expression. A model suggests that hilA promoter activity is blocked by a negative factor until it is counteracted under inducing conditions by specific regulatory protein(s) (43).
Two SPI1-encoded regulators of hilA expression are the AraC/XylS family members, HilC (also called SirC or SprA) and HilD. Lucas and Lee (30) proposed that HilD is required for hilA expression under normal conditions and that HilC mediates only the action of EnvZ/OmpR. However, overexpression of either HilC or HilD resulted in constitutive activation of hilA expression. Expression of a hilC-lac reporter is regulated by the environmental conditions that modulate hilA expression, but hilD-lac expression is little affected.
Understanding the action of HilC and HilD in hilA expression will require knowledge of their binding sites at target promoters and of their effect on transcription with purified components. To this end, we purified HilC and HilD proteins that were active for DNA binding and transcriptional regulation. By using gel shift, DNase protection, and scanning mutagenesis procedures, the sites of binding of HilC and HilD in the hilA, hilC, and hilD promoters were characterized. Both proteins were found to bind to the same target sequences in the various promoters, but they differed in the number of binding sites and in their sequence requirements for binding. The effect of HilC and HilD proteins on transcription of these promoters indicated that all three promoters are active in the absence of other cellular proteins.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this work are listed in Table 1. DNA isolation and recombinant DNA manipulations were carried out using standard methods (41) or as previously described (37). Plasmids pINO3, pCP9, and pDP14 were used for in vitro transcription experiments and were constructed as follows. The promoter regions for the hilA, hilC, and hilD genes were amplified from S. enterica serovar Typhimurium SL1344 chromosomal DNA by PCR using Vent polymerase (New England Biolabs) using conditions specified by the manufacturer. The following primers were used to create PCR amplimers flanked by EcoRI and HindIII sites (introduced sites are underlined): HilApr5Ec (5′-CCGGAATTCACGCTTGTTAGCTTTCTGCCAG) and HilApr3Hi (5′-TCGAATGGAAGCTTCCGTATATCCTGGT) for the hilA promoter; 5prHilC (5′-CCGGAATTCATTACAAAATTGTGCATAAAG) and HilC + 88 (5′-GCTGTTGAAGCTTATTATTGCTAATGGCCT) for the hilC promoter; and 5prHilD (5′-CCGGAATTCATATATACTGTTAGCGATGTCTG) and 3prHilD (5′-TACTTACAAAGCTTACATTTTCCATAT) for the hilD promoter.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| Serovar-Typhimurium | ||
| SL1344 | rpsL hisG | 19 |
| SD11 | SL1344 ΔSP11 | 42 |
| CL4419 | metA22 metE551 trpE2 ilv-452 xyl-404 vsp2120 hsdSA hsdT flaA66 | O. Colson |
| E. coli | ||
| MC4100 | ΔlacU169 araD139 deoC1 flbB5301 ptsF25 rbsR rpsL 150 | 6 |
| BL21 (DE3) | ompT hsdSB gal dcm (λDE3) | Novagen |
| Plasmids | ||
| pSR | Ap, rmB t1+2 terminators | 22 |
| pINO3 | pSR containing −367 to +90 of hilA promoter | This work |
| pCP9 | pSR containing −157 to +75 of hilC promoter | This work |
| pDP14 | pSR containing 283 to +48 of hilD promoter | This work |
| pET15b | Ap, T7 promoter | Novagen |
| pHilC14 | pET15b expressing His6-HilC | This work |
| pHilD21 | pET15b expressing His6-HilD | This work |
| pRS415 | Ap, lacZYA transcriptional fusion plasmid | 45 |
| pTL61T | Ap, lacZYA transcriptional fusion plasmid | 27 |
| pRS415P | pRS415 with polylinker from pTL61T | This work |
| philCL | pRS415P with hilC promoter and variants driving lacZ | This work |
| philDL | pRS415P with hilD promoter driving lacZ | This work |
The PCR products were cloned as EcoRI-HindIII fragments by ligation into plasmid pSR (22), which contains the rrnB Rho-independent transcription terminator downstream of the HindIII site, to form plasmids pINO3, pCP9, and pDP14.
Plasmids pHilC14 and pHilD21, in which the hilC and hilD genes are expressed under the control of the T7 promoter, were constructed by cloning the PCR-amplified hilC and hilD genes from SL1344 chromosomal DNA into vector pET15b (Novagen, Inc.). The following primers were used to create PCR fragments flanked by NdeI -BamHI sites, with introduced sites underlined:HilC5Nde (5′-GGGAATTCCATATGGTATTGCCTTCAATGAATAAATCAG) and HilC3Bam (5′-CGCGGATCCTCAATGGTTCATTGTACGCATAAAGC) for the hilC gene; and HilD5Nde (5′-GGGAATTCCATATGGAAAATGTAACCTTTGTAAGTAATA) and HilD3Bam (5′-CGCGGATCCTTAATGGTTCGCCATTTTTATGAA) for the hilD gene.
These constructs allowed overexpression of the HilC and HilD proteins with N-terminal His6 tag extensions to facilitate their purification. All DNA inserts were verified by automated DNA sequencing at the Biomolecular Research Facility at the University of Virginia School of Medicine.
Fusions of hilC or hilD promoter regions to lacZYA were constructed as follows: the polylinker region from the plasmid vector pTL61T (27) was cloned as an EcoRI-BamHI fragment into the corresponding sites of the lacZ transcriptional reporter plasmid pRS415 (45) to yield pRS415P. The EcoRI-HindIII fragments carrying the wild-type hilC and hilD promoters from plasmids pCP9 and pDP14, respectively, were ligated into EcoRI-HindIII-digested pRS415P, to generate plasmids philCL and philDL. The hilC promoter variants were introduced in similar manner. The reporter plasmids were passaged in the restriction-negative modification-positive serovar Typhimurium strain CL4419 prior to transformation into SL1344 derivatives.
Purification of HilC and HilD proteins.
Plasmids pHilC14 and pHilD21 were introduced into Escherichia coli strain BL21(DE3) by transformation. Isolates were grown in Luria broth (LB) supplemented with ampicillin (0.2 mg/ml) at 37°C. Cultures of E. coli BL21(DE3)/pHilC14 were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) in the early log phase. The cells were allowed to grow to an A600 of 1.0 and were harvested by centrifugation. The IPTG induction was omitted in the case of E. coli BL21(DE3)/pHilD21 because a high level of HilD expression inhibited cell growth
The N-terminal His-tagged HilC and HilD proteins were purified from cell extracts by Ni2+ affinity chromatography as described in the Novagen standard protocol. In brief, cells grown in 2 liters of LB were washed and suspended in 60 ml of binding buffer (20 mM Tris-HCl, pH 7.9, 0.5 M NaCl, 5 mM imidazole, and 10% [vol/vol] glycerol) and disrupted by sonication of 20-ml portions for three 3-min periods, with 10-min intervals for cooling. Unbroken cells and particulate material were removed by centrifugation at 20,000 × g for 30 min, and the cell extract was applied onto an Ni-nitrilotricetic acid agarose (Qiagen) column with 2 ml of resin volume. The resin was washed with 10 volumes of wash buffer (binding buffer + 55 mM imidazole), and adsorbed His-tagged proteins were eluted with 5 ml of binding buffer + 100 mM imidazole. Eluted proteins were concentrated using Centriplus YM-10 Centrifugal Filter Devices (Amicon, Inc., Beverly, Mass.), dialyzed against 500 ml of storage buffer (40 mM HEPES, pH 7.8, 40 mM KCl, 1 mM EDTA, and 10% [vol/vol] glycerol), and stored in aliquots at −70°C. Yields of HilC and HilD proteins were in the range of 0.8 to 1 mg per liter of culture, and a purity of 90 to 95% was estimated by Coomassie brilliant blue staining following sodium dodecyl sulfate-polyacrylamide gel electrophoresis (24).
Gel mobility shift assay.
DNA fragments carrying portions of the hilA, hilC, and hilD promoters were generated by PCR using plasmids pINO3, pCP9, and pDP14 as templates, respectively. Mutant DNA templates carrying single base substitutions in the hilC promoter were prepared using plasmid pCP9 as template. Mutations in the upstream half of the C2 region were incorporated into a fragment spanning residues −92 to −20, and mutations in the downstream half were in fragments spanning residues −162 to −52. The sequences of primers used for production of labeled fragments and for mutagenesis are available upon request. The amplified DNA fragments were purified from agarose gels using QIAquick Gel Extraction Kit (Qiagen) and were 5′ end labeled by incubation with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (3,000 Ci/mmol; ICN). Labeled DNA fragments were separated from unincorporated nucleotide by gel filtration through Sephadex G-25 or G-50 Quick Spin columns (Boehringer Mannheim). Radiolabeled DNA fragments (ca. 10,000 cpm per reaction) were incubated with HilC or HilD protein at 37°C for 15 min in binding buffer (40 mM Tris-HCl pH 8.0, 50 mM KCl, 10 mM MgCl2, 10 mM dithiothreitol, 5% glycerol, and 2 ng of poly d[I-C]/μl). Samples were resolved by electrophoresis in 1.5-mm-thick, nondenaturing 6% polyacrylamide gels containing Tris-glycine (5 mM Tris, 38 mM glycine, pH 8.6) at 20 mA for 45 min at room temperature. The positions of radioactive DNA fragments in the gels were detected using a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) and the ImageQuant program for quantitative comparisons.
DNase I protection assay.
DNase I footprinting reactions were performed as described by Galas and Schmitz (17, 37). The 5′ 32P-end-labeled DNA fragments contained the hilA promoter from position −286 to +30, the hilC promoter from position −157 to +75, and the hilD promoter from position −161 to +48. The top strand primers were 5′ end labeled by incubation with T4 polynucleotide kinase and [γ-32P]ATP (3,000 Ci/mmol) prior to use in PCR, as described above. The labeled PCR products were resolved on nondenaturing polyacrylamide gels, detected by autoradiography, and recovered by crush-soak elution and binding to Quick Spin PCR columns (Qiagen).
Promoter-containing DNA fragments (ca. 50,000 cpm) at a final concentration of 1 nM were incubated with specified amounts of purified HilC or HilD protein in 20 μl of TXN buffer (40 mM Tris-HCl, pH 8.0, 50 mM KCl, 10 mM MgCl2, and 10 mM dithiothreitol). After 15 min at 37°C, digestion was begun by addition of 2 μl of TXN buffer containing 25 mM CaCl2, 25 mM MgCl2, and 0.5 μg of DNase I/ml. After 30 s at ambient temperature, 4 μl of stop solution (0.18 M EDTA, 0.34 μg/ml of poly d[I-C], and 30% [vol/vol] glycerol) was added. The DNA was precipitated with 95% (vol/vol) ethanol, washed with 70% (vol/vol) ethanol, dried under vacuum, dissolved in loading buffer, and resolved by electrophoresis on 5% sequencing gels (41). DNA size markers were generated by Maxam-Gilbert sequencing reactions (41) on the same DNA fragment. The gels were dried, and the radioactive DNA fragments were detected by PhosphorImager (Molecular Dynamics).
In vitro transcription.
Transcription assays were performed at 37°C in TXN buffer (see above), using plasmids pINO3, pCP9, and pDP14 as DNA templates. Indicated amounts of HilC or HilD protein and DNA template were incubated for 10 min in 10 μl of TXN buffer prior to the addition of 5 μl of E. coli RNA polymerase (RNAP) holoenzyme (USB). After 15 min at 37°C, 5 μl of TXN buffer containing nucleoside triphosphate substrates was added to yield the following final concentrations: 1 nM DNA template; 0 to 50 nM HilC or HilD; 35 nM RNAP; ATP, CTP, and GTP at 200 μM; and 40 μM [α32P]UTP (2.5 Ci/nmol). After 15 min, the reaction was terminated by addition of 5 μl of transcription stop solution (7 M urea, 0.1 M EDTA, 0.4% [wt/vol] sodium dodecyl sulfate, 40 mM Tris-HCl [pH 8.0] 0.5% bromophenol blue, and 0.5% xylene cyanol). Products were resolved by electrophoresis in 5% polyacrylamide-7 M urea gels in Tris-borate-EDTA buffer and detected by autoradiography.
RNA isolation and primer extension.
RNA was isolated from cultures of serovar Typhimurium SL1344 and SD11 grown to an optical density at 600 nm (OD600) of 1.0 in permissive conditions of limited oxygen and high osmolarity (LB + 0.3 M NaCl, without aeration). Cells from 100 ml of culture were collected by centrifugation, and RNAs were isolated using the RNAgents kit (Promega). Traces of DNA were removed using DNase I (RNase free; Boehringer Mannheim GmbH, Mannheim, Germany). RNA (17 μg) was hybridized to 5′ 32P-end-labeled oligonucleotide (5′-ACATAATAGTCTCTTA CGTCAGCTAA) complementary to the hilC coding strand or to 5′ 32P-end-labeled oligonucleotide (5′-CTGCTGAGTCTGACTTTTAATTTGCT) complementary to the hilD coding strand. Primer extension was performed as described by Ausubel et al. (3) using avian myeloblastosis virus reverse transcriptase (Promega). Transcripts were resolved by electrophoresis as described above, next to a sequencing ladder generated using the same labeled oligonucleotides and PCR-generated fragments of the corresponding genes as templates. For identification of the in vitro transcription start sites, the in vitro RNA synthesis was scaled up using nonradioactive nucleoside triphosphates and a reaction volume of 100 μl. DNase I (RNase free) was added to a final concentration of 20 μg/ml, and incubation was continued for 10 min at 37°C. Reaction mixtures were extracted twice with equal volumes of phenol-chloroform (1:1) and once with chloroform. The RNA transcripts were annealed to 5′ 32P-end-labeled oligonucleotide IO770 (5′-GATGCCTGGCAGTTCCCTACTCTCGC) complementary to part of the vector pSR (22), which was transcribed from the cloned hilC or hilD promoters. Reverse transcription was carried out as described above. Dideoxy sequencing standards were prepared using the same labeled oligonucleotide and pCP9 or pDP14 plasmid DNAs as templates.
β-Galactosidase assay.
Cells carrying the transcriptional fusion plasmids were grown in LB with aeration or in LB + 1% NaCl in capped, static culture tubes to provide repressing or inducing conditions, respectively. Assay of β-galactosidase was performed as previously described (38) by continuous determination at 415 nm of the rate of o-nitrophenyl-galactopyranoside hydrolysis in a microplate reader (Molecular Dynamics).
RESULTS
Purification and activity of recombinant HilC and HilD proteins.
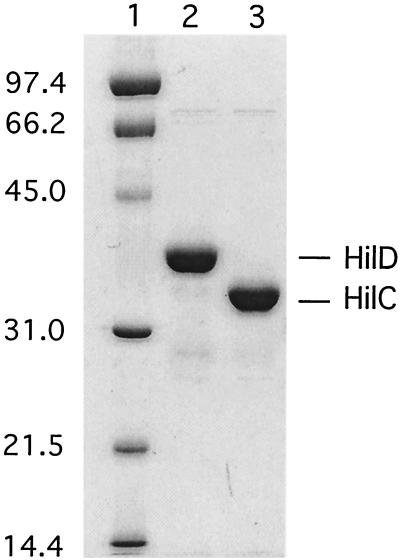
The serovar Typhimurium HilC and HilD proteins were expressed with an N-terminal hexa-histidine (His6) adduct to facilitate purification. The choice of His6 tag was fortuitous in light of subsequently reported difficulties in purification of an active form of HilC-Myc with a Myc epitope tag (43). The His6-tagged forms of HilC and HilD proteins were produced in E. coli and purified to >90% homogeneity by elution from an Ni matrix column (Fig. 1). The DNA-binding activities of the His-tagged proteins at the hilA promoter region (positions −286 to +30; all coordinates are relative to the transcription start sites) were examined by DNase protection assay. Both proteins protected sites within region I (−230 to −187) and region II (−95 to −60) (data not shown), as described for the HilD-Myc protein by Schechter and Lee (43). This result suggested that the His-tagged proteins are suitable for further analysis of their DNA-binding properties. Owing to the high expression of these proteins from plasmids, it was not meaningful to compare their in vivo regulatory properties with those of their wild-type counterparts.
FIG. 1.
Purification of HilC and HilD. Protein samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue. Lane contents follow: 1, molecular weight standards, as indicated on the left; 2, HilD; and 3, HilC. The HilC and HilD protein samples were purified by elution from an Ni matrix column and retain the His6 tag. Kilodaltons are given on left.
HilC- and HilD-binding sites in the hilA promoter.
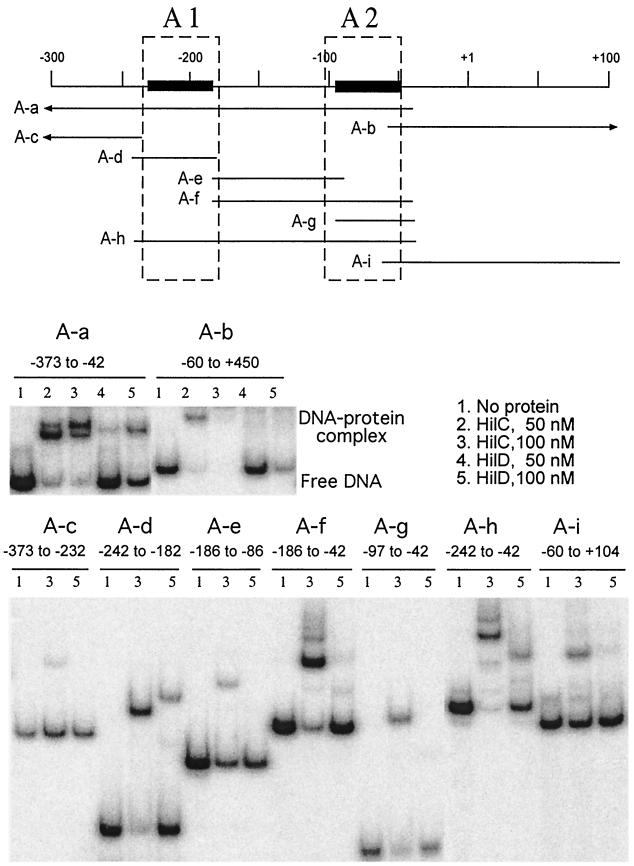
To delineate the contribution of portions of the hilA promoter for binding of the HilC and HilD proteins, fragments of the hilA promoter were constructed by PCR and tested by gel shift assay (Fig. 2). The regions that were protected by HilC and HilD against DNase cleavage were designated RI and RII by Schechter and Lee (43), but we use the designations A1 and A2 to distinguish them from the related protein-binding regions in the other promoters studied here. There were notable differences between the binding of HilC and that of HilD to the hilA promoter. DNA fragment A-a, which carries the upstream regulatory sequences from −373 to −42, formed two major shifted complexes with HilC but only a single complex with HilD. Fragment A-b, which carries the promoter and transcribed sequences (−60 to +450), bound HilC well and HilD less well, as shown by the loss of the band corresponding to free DNA, but most of the bound complexes remained in the wells. The behavior of fragment A-b suggests that some HilC- and HilD-binding sites are present in the transcribed region beyond +100.
FIG. 2.
Electrophoretic mobility shift assay analysis of binding of HilC and HilD to portions of the hilA promoter. The top panel is a schematic diagram of the hilA promoter and the regions carried on the DNA fragments used in this assay. Fragment names are identified in the top panel, and their end points are given above each data set. Arrowheads indicate that the end point extends beyond the region shown. The dashed boxes represent the protein-binding positions assigned by Schechter and Lee (43), and the solid lines indicate the protein-binding regions determined here, which are designated A1 and A2. Fragments were labeled with 32P on the 5′ end of the top strand. The lower panels show the gel shift results. The added proteins in each lane are as follows: lanes 1, no added protein; lanes 2, 50 nM HilC; lanes 3, 100 nM HilC; lanes 4, 50 nM HilD, and lanes 5, 100 nM HilD.
Fragments A-c (−373 to −232), A-e (−186 to −86), and A-i (−60 to +104), which do not carry region A1 or A2, showed no binding of HilD and weak binding of HilC. This weak binding of HilC could be sequence independent, because HilC gave a similar shift at the unrelated but A+T-rich uhpT promoter (not shown). Fragment A-d (−242 to −182) formed a single shifted complex with HilC and HilD, showing that region A1 alone could be stably bound by either protein. The affinity for HilC was higher than for HilD. The lower mobility of the HilD-DNA complex than of the HilC-DNA complex could reflect the larger size of HilD or differences in DNA bending. The isolated region A2 carried on fragment A-g (−97 to −42) gave a single shifted complex with HilC but no apparent binding of HilD. Fragment A-h, which carries both A1 and A2 (−242 to −42), showed behavior similar to that of fragment A-a, namely, multiple retarded species with HilC but a single shifted species with HilD. Finally, fragment A-f, which carries region A2 and the sequences up to A1 (−186 to −42), formed multiple retarded species with HilC but only weak binding of HilD. These results indicated that the A1 region (−242 to −182) is an independent binding site for both proteins but that the promoter-proximal A2 region (−85 to −61) is capable of independent binding of HilC but not of HilD. DNase protection assays confirmed the binding of both HilC and HilD to these two ca.-40-bp regions when both regions were present on the same DNA fragment. Weaker protection of the intervening region from −110 to −140 was seen for HilC but not for HilD (data not shown). Features of these binding sequences are discussed below.
Effect of HilC and HilD on transcription at hilC and hilD promoters.
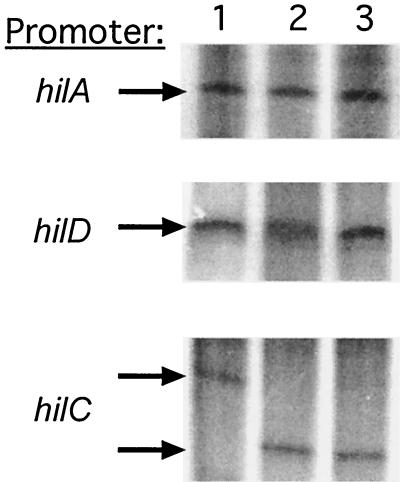
Expression of hilA-lacZ fusions displays complex regulation and can be activated by the HilC or HilD protein (42). Since some transcriptional regulators exhibit autogenous control, we examined the effect of the HilC and HilD proteins on in vitro transcription by E. coli RNAP Eσ70 holoenzyme of the hilA, hilC, and hilD promoters. Transcription of supercoiled plasmid DNAs carrying each hil promoter gave rise to a single major transcript specific for each promoter (Fig. 3, lane 1). The level of the vector-encoded RNA-I transcript was used to normalize for RNAP activity and sample loading (not shown). The addition of 50 nM HilC (lane 2) or HilD (lane 3) had no obvious effect on the size or amount of the hilA and hilD transcripts. In contrast, both HilC and HilD blocked formation of the hilC transcript that was made in their absence and activated the synthesis of a transcript that was ca. 70 nucleotides shorter. This result indicated that hilC has two promoters and that the upstream promoter is repressed by HilC and HilD while the downstream one is activated. The consequences of this change in transcription start site for gene expression are not yet known. However, these results indicate that the purified proteins are active for both DNA binding and transcriptional control.
FIG. 3.
In vitro transcription of the hilA, hilC, and hilD promoters. The promoter fragments used as templates for in vitro transcription reactions are indicated on the left of each panel and are identified in Materials and Methods. Reactions for all lanes contain 50 nM E. coli RNAP Eσ70 holoenzyme. Additional proteins present during transcription are depicted as follows: lanes 1, none; lanes 2, 50 nM HilC; and lanes 3, 50 nM HilD. Transcription was carried out and products were resolved as described in Materials and Methods.
hilC and hilD start sites.
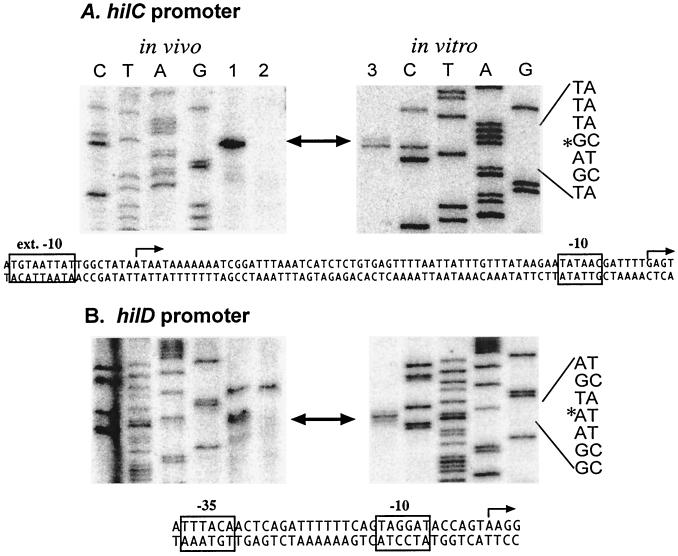
The hilC and hilD start sites used during cellular and in vitro transcription were localized by primer extension (Fig. 4). RNA was extracted from wild-type serovar Typhimurium SL1344 (Fig. 4, lane 1) and from an isogenic mutant deleted for SPI1 (Fig. 4, lane 2) grown under inducing conditions for hilA expression, i.e., high salt and low aeration (32). A single SPI1-dependent transcript was detected for hilC and hilD. The identical start sites were found for the in vitro transcripts synthesized in the presence of HilC (Fig. 4, lane 3). The 5′ end of the in vitro hilC transcript produced in the absence of HilC was mapped to position −71, relative to the start used in the presence of HilC (data not shown).
FIG. 4.
Identification of the in vivo and in vitro transcription start sites of the hilC and hilD promoters. The 5′ ends of the transcripts specific for hilC (A) and hilD (B) were identified by primer extension, as described in Materials and Methods. Primer extension products were electrophoresed alongside products of dideoxy sequencing reactions using the same primers and DNA template, CTAG. The RNA samples used as template for primer extension were as follows: lanes 1, RNA from serovar Typhimurium SL1344; lanes 2, RNA from strain SD11 (ΔSPI1); and lanes 3, RNA synthesized during in vitro transcription in the presence of HilC protein. The promoter-specific transcripts are indicated with arrows, and the sequence of the start site is marked with an asterisk on the sequence on the right. Below each set of panels is the nucleotide sequence of the promoter region, where the start site is indicated by an arrow and the promoter elements are boxed. In the hilC promoter, the transcription start site in the presence of HilC or in cells is on the right, and the in vitro start site in the absence of HilC is on the left.
Sequences upstream of the start sites of hilC and hilD were examined for potential promoter elements (Fig. 4). The hilD start is preceded by the sequence TTTACA-N16-TAGGAT, which matches the Eσ70 promoter consensus at 9 of 12 positions and is expected to specify an effective promoter. The corresponding sequence of the HilC/D-dependent hilC promoter, TTTAAT-N16-TATAAC, matches the consensus at 8 of 12 positions but should specify a weak promoter, owing to the absence of the highly conserved T at position −7 and the poor match of the −35 element. The hilC start site used in the absence of HilC and HilD is associated with the sequence TGTAATTAT, which matches at six of eight positions the extended −10 promoter element that allows transcription initiation in the absence of a −35 element (23). The extended −10 promoter element upstream of hilC is absent from the equivalent region of the hilD promoter, which differs at five of nine positions.
HilC- and HilD-binding sites in the hilC promoter.
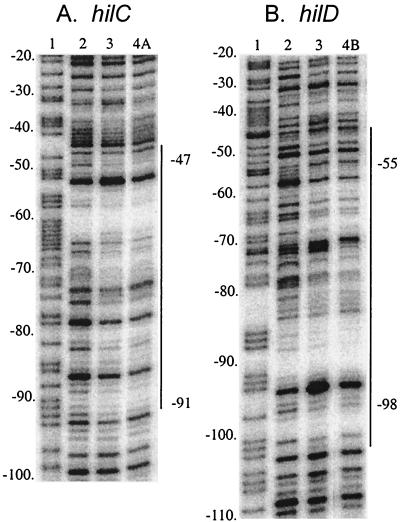
Since HilC and HilD affected the choice of hilC transcription start sites, their binding to the hilC promoter was determined. DNase I protection assay of a DNA fragment carrying residues −157 to +75 of the hilC promoter and labeled on the 5′ end of the top strand showed that both proteins gave the same pattern of protection of sites between positions −91 and −47 (Fig. 5). As occurred at the hilA promoter, these sites were only partially protected and were interspersed with sites of little or no protection and a few DNase-hypersensitive sites. At protein concentrations higher than used for Fig. 5, the region of protection was extended in both directions.
FIG. 5.
DNase I protection assay of HilC and HilD binding to the hilC and hilD promoters. The hilC (A) and hilD (B) promoter fragments indicated on the top of the figure and defined in the text were subjected to DNase I digestion in the presence of no added protein (lanes 2), 50 nM HilC (lanes 3), 200 nM HilD (lane 4A), or 400 nM HilD (lane 4B). In lanes 1 are the products of the A+G sequencing reactions of the same DNA fragments. The regions of sites protected from DNase digestion are indicated on the right of each panel.
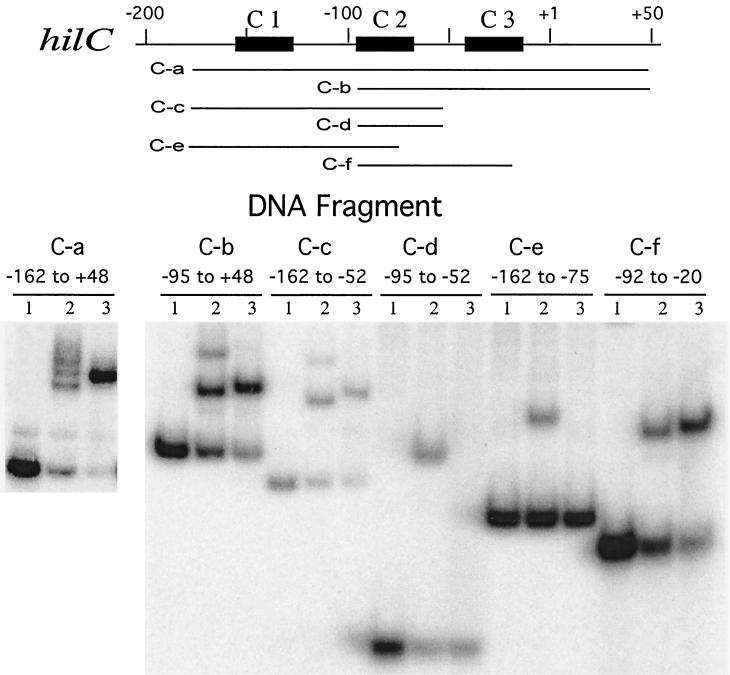
To identify the sequences that contribute to protein binding at the hilC promoter, DNA fragments carrying segments between −162 and +48 were analyzed for gel shift activity with the HilC and HilD proteins (Fig. 6). Binding of HilC to fragment C-a (−162 to +48) revealed at least three closely spaced, retarded species. Under other electrophoretic conditions, only a single HilC-retarded species was seen (data not shown). The multiple retarded species could reflect DNA bending or looping of HilC molecules between different pairs of sites. HilC binding to the DNA fragments C-b (−95 to +48) and C-c (−162 to −52) resulted in formation of two retarded bands, suggesting that each of these fragments contains two HilC-binding sites, i.e., one each in addition to the shared region of −91 to −47 identified by DNase footprinting, here called region C2. Binding of HilC to DNA fragments C-d (−95 to −52) and C-f (−92 to −20), which carry the C2 region, yielded a single shifted complex. Fragment C-e (−162 to −75), which lacks part of the protected region, also formed a single bound complex, but a fragment carrying only the region −83 to −39 showed no binding of HilC (not shown). These results indicate that HilC can bind to multiple sites in the hilC promoter besides the C2 region (residues −92 and −52) (Fig. 6). At least two other HilC-binding regions lie on either side of C2 and are designated C1 and C3 (Fig. 6), but their locations have not been determined.
FIG. 6.
Electrophoretic mobility shift assay analysis of HilC and HilD binding to the hilC promoter region. DNA fragments containing portions of the hilC promoter region, as indicated schematically in the top panel, were synthesized by PCR and used in gel shift analysis, as described for Fig. 2. Proteins incubated with the DNA fragments prior to electrophoresis are depicted as follows: lanes 1, no added protein; lanes 2, 150 nM HilC; and lanes 3, 150 nM HilD.
As was found for the hilA promoter, binding of HilD to the hilC promoter resulted in a single retarded complex (Fig. 6). HilD also formed a single complex with fragments C-a, C-b, C-c, and C-f and a fragment carrying regions −92 to −39 (not shown). Binding of HilD was not detected to fragment C-d or C-e or a fragment carrying −83 to −39 (not shown). These results indicated that HilD binds only to the C2 region, as expected from DNase footprinting (Fig. 5) and not to the C1 and C3 regions. However, this binding of HilD requires a larger target than does binding of HilC, as seen by comparing the binding to fragments C-d and C-f.
Mutational analysis of HilC- and HilD-binding site C2 in the hilC promoter.
The nucleotide positions that determine binding of HilC and HilD were investigated by analysis of base substitutions within the 39-bp C2 region of the hilC promoter. To enhance binding of HilD, flanking sequences from one or the other side were incorporated in fragments carrying the variant sequences. Substitutions were made at each position throughout the C2 region to make the most extreme base change. Thus, a purine was converted to the pyrimidine that normally pairs with the other purine; e.g., each G was converted to T. At several positions, all three possible base substitutions were made.
A panel of 51 variant C2 fragments was analyzed for binding to HilC and HilD in the gel shift assay. Representative results are shown in Fig. 7. DNA-binding activity was calculated by measuring the fraction of each labeled DNA fragment that was shifted in the presence of 100 to 300 nM HilC and HilD. The PhosphorImager output measured for the shifted complex was divided by the sum of the volumes of the shifted and unshifted bands and was then expressed relative to the value of the wild-type fragment normalized to 100%. Each binding reaction was assayed at least three times, and average values are presented (Fig. 8).
FIG. 7.
A representative electrophoretic mobility shift assay analysis of protein binding to hilC promoter variants. The 5′ 32P-labeled DNA fragments carrying single base substitutions in the C2 region of the hilC promoter from positions −92 to −54 were incubated in the presence of no added protein (lanes 1), 100 nM HilC (lanes 2), or 100 nM HilD (lanes 3). Samples were subjected to electrophoretic mobility shift assay, and the distribution of radioactivity in the free DNA and protein-bound complexes was detected and quantified by PhosphorImager. The base changes in these representative samples are indicated along the top. WT, wild type.
FIG. 8.
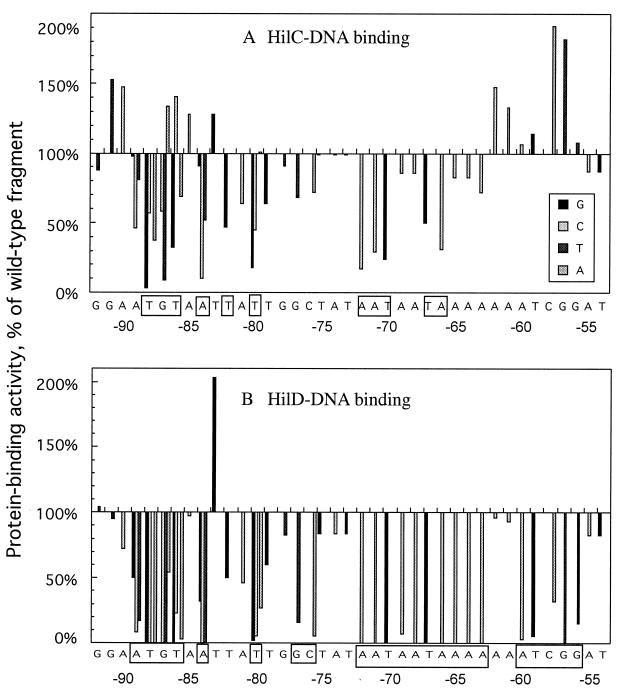
Summary of effects of base substitutions in the 39-bp hilC fragment on binding of HilC and HilD measured by electrophoretic mobility shift assay. Results compiled for experiments similar to the example in Fig. 7 were averaged and expressed relative to the binding of HilC (A) or HilD (B) to the fragment with the wild-type sequence as 100%. Binding was determined with protein concentrations of 100 and 200 nM, and values are averaged. At each position, the base in the wild-type sequence was changed to its extreme alternative. At several positions all base changes were made and identified in the insert. At the bottom of each panel are the nucleotide coordinates relative to the transcription start site. Those residues at which substitution strongly decreased protein binding are indicated in boxes.
The base substitutions had strikingly different effects on binding of the two Hil proteins. For HilC, no base substitution abolished binding and important positions were located in two short blocks (Fig. 8A). The degree of impairment of binding depended on the nature of the base substitution, and transition substitutions, i.e., purine for purine, were usually much less detrimental than were transversion changes at the six sites tested in detail. For example, replacement of −88T by G, A, or C decreased HilC-binding activity to 4, 33, or 53% of the result for the wild type, respectively. Similarly, replacement of −87G by T or C decreased HilC binding to 9 or 55% of that for the wild type, respectively, whereas −87G to A increased binding. The sequences important for HilC binding are TGT starting at −88, −84A, −82T, and −80T; AAT starting at −72; and TA starting at −67.
Different specificity was found for binding of HilD. Substitutions at 15 of the 39 positions eliminated detectable binding by HilD, and changes at six other positions decreased binding by >90% (Fig. 8B). All positions that were important for HilC binding were also important for HilD binding. In addition, the blocks from −72 to −63 and from −60 to −56 were critical for HilD binding but had little if any effect on HilC binding. Furthermore, substitutions at 10 positions allowed increased binding of HilC, but changes at only one position, −83T to G, showed increased binding of HilD. Thus, although HilC and HilD proteins can bind to the same DNA target, they have different sequence requirements for DNA recognition.
HilC- and HilD-binding sites in the hilD promoter.
DNase I protection assays with the DNA fragment carrying residues −161 to +48 of the hilD promoter showed that HilC and HilD protected the same sites in the hilD promoter between positions −98 and −47, termed here the D region (Fig. 5). Comparable protection of the D region required about twice the amount of HilD that was effective at the C2 region.
Various regions of the hilD promoter were synthesized by PCR and tested by gel shift assay (not shown). DNA fragments that carried the D region bound HilC. Only the fragment carrying −283 to +48 bound HilD, whereas a fragment carrying −169 to +7 did not. The longer fragment carries the promoter for prgH, and it is possible that stable HilD binding to the hilD promoter requires the presence of this divergent upstream promoter. Taken together, HilD binding requires a larger target than HilC at the hilC and hilD promoters.
Effect of HilC- and HilD-binding sites on hilC expression.
The potential role of the HilC- and HilD-binding sites in expression of the hilC and hilD promoters was examined by in vitro transcription and lac fusions driven by promoter variants. Previous studies found that hilD-lacZ expression is unaffected by growth conditions or by the presence of factors encoded on SPI1 (30). We too found (Table 2) that β-galactosidase expression from a hilD-lacZ transcriptional fusion, carried on a moderate-copy-number plasmid in serovar Typhimurium, was not substantially affected by the presence of SPI1. Growth under conditions that induce hilA expression resulted in a modest increase in hilD-lac expression. In contrast, expression of a hilC-lacZ fusion increased about threefold under inducing conditions and decreased about threefold in the absence of SPI1. The wide range of values for this strain reflected the substantial differences in β-galactosidase activity throughout the growth cycle. The extent of regulation by these factors could be greater than observed because the reporters were carried on multicopy plasmids.
TABLE 2.
Effect of SPI1 and mutations in region C2 of the hilC promoter on β-galactosidase expression from hilC-lac and hilD-lac fusion strains grown under repressing and inducing conditions
| Promotera | SPI1 genotype | β-Galactosidase activity following growth underb:
|
|
|---|---|---|---|
| Repressing conditions | Inducing conditions | ||
| hilC | + | 2,125 ± 1,050 | 5,960 ± 1,180 |
| Δ | 660 ± 230 | 3,380 | |
| hilC wild type | + | 2,005 ± 941 | 6,283 ± 377 |
| hilC −88T to G | + | 2,625 ± 286 | 4,935 ± 697 |
| hilC −59T to G | + | 2,325 ± 191 | 4,700 ± 480 |
| hilD | + | 5,715 ± 640 | 9,980 |
| Δ | 5,275 ± 780 | 7,134 | |
Plasmid philCL, encoding the hilC-lacZ transcriptional fusion or its derivatives carrying the T-88G or the T-59G substitution, or plasmid philDL, encoding the hilD-lacZ transcriptional fusion, was introduced into serovar Typhimurium strain SL1344 (SDI1+) or SDI1 (ΔSDI1).
Cells were inoculated into LB with 0.5% NaCl and grown with vigorous aeration (repressing conditions) or inoculated into LB + 1% NaCl and incubated in static culture in filled sealed tubes (inducing conditions). Samples were withdrawn at various stages through the culture growth curve. β-Galactosidase activity was measured in triplicate by continuous assay in microplate reader. Activity is expressed as ΔOD410 × min−1 × OD650−1.
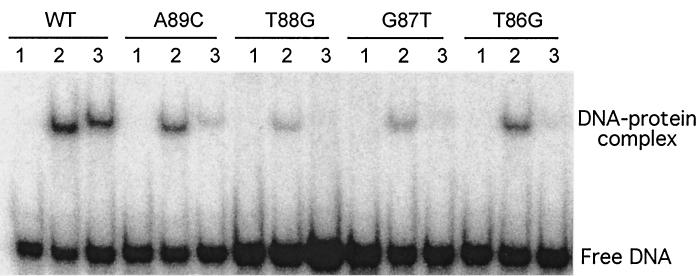
When the hilC-lac reporter plasmid carried the T-88G or the T-59G mutations, there was little change in β-galactosidase expression relative to that of the wild-type promoter, although the induction response was dampened (Table 2). This minimal effect upon changes in the C2 protein-binding region was consistent with the in vitro results that expression of the hilC promoter was comparable in the absence or presence of HilC and HilD, even though a different start site was used. However, in vitro transcription was strongly decreased by the T-88G substitution, which should impair both HilC and HilD binding and the activity of the upstream extended −10 promoter element. Transcription was not affected by the T-59G substitution, which should affect only HilD binding.
DISCUSSION
Expression of the hilA gene appears to be a key step in the regulatory cascade controlling the invasion of host cells by Salmonella. Analysis of hilA-lacZ transcriptional fusions by Lee and colleagues (30-32, 42), among others, led to a model proposing that hilA transcription is repressed by a negative factor acting within the promoter region between positions −300 and −100. The HilD and perhaps the HilC proteins play central roles in controlling hilA transcription and may counteract this repressor activity. The lack of appreciable effect of HilC or HilD protein on transcription of the hilA promoter in vitro found here is consistent with this model. Identification of the repressor and demonstration that HilC and HilD can overcome its action are necessary for definition of the regulatory cascade.
Although overexpression of the HilC or HilD protein results in high and unregulated expression of hilA (30), the relative contribution of changes in the level or activity of either protein in control of hilA expression is not clear. Analysis of lac fusions suggested that transcription of hilC was regulated by the same environmental conditions that influence hilA expression but that of hilD was little affected (30). In addition to their binding to multiple sites in the hilA promoter, the HilC and HilD proteins were shown here to bind to the hilC and hilD promoters. They occupy roughly 34-bp sites between positions −91 and −47 at the hilC promoter and between −98 and −55 at the hilD promoter. Neither protein had much effect on the amount of transcripts produced during in vitro transcription at the three hil promoters, but both proteins determined the choice of promoter for hilC transcription. The HilC and HilD proteins repressed the upstream promoter, which uses an extended −10 element at positions −88 to −80, and activate the silent downstream promoter. Reverse transcriptase PCR analysis suggested that hilC is also transcribed from even farther upstream (21). These results indicate that HilC and HilD can function as direct transcription activators at the hilC promoter, unlike their proposed role at the hilA promoter to relieve repression. The hilD promoter is active in vitro and unaffected by the HilC and HilD proteins or other SPI1 products.
The HilC and HilD proteins are members of the AraC/XylS family of transcription activators, which typically bind to a 34-bp DNA target (18, 36, 42). The DNA-binding activity resides in the C-terminal domain and possesses two helix-turn-helix motifs, so that a protein monomer binds to two successive DNA helical turns. AraC family members often exist as dimers, joined by a dimerization surface in the N-terminal domain, whose activity may be regulated by ligand recognition. It remains to be seen whether the activities of HilC or HilD are affected by a small molecule ligand or other form of posttranscriptional control. Some members of the AraC/XylS family have difficult solubility properties, and the HilC and HilD proteins lost DNA-binding activity upon prolonged storage. This instability complicated comparison of the binding affinities of the two proteins for their DNA targets, but HilC routinely had higher affinity than did HilD when both proteins were prepared simultaneously.
DNA-binding activity.
The HilC and HilD proteins bind to the same regions of each other's promoter, as well as to the same regions of the hilA promoter. Nonetheless, the binding process and recognition determinants at these sites were quite different. HilC binds to at least three sites in the hilC promoter. The binding to the C2 region was localized to positions −96 to −47 by DNase footprinting, whereas a gel shift showed requirement for positions −95 to −52, and a gel shift of sequence variants identified important nucleotides for binding between positions −89 and −63. The two flanking binding sites, C1 and C3, which contribute to formation of multiple retarded electrophoretic species with HilC, lie between positions −162 and +48 but were not precisely localized. The multiple electrophoretic species could reflect binding of additional molecules of HilC or formation of DNA loops by dimeric HilC between binding regions. The C1 and C3 regions bind HilC but not HilD in gel shift assays. The D region of the hilD promoter, which is equivalent to the C2 region, bound HilC but not HilD. Similarly, HilC and HilD bound in different manners to the hilA promoter. HilC can bind to fragments carrying only the upstream A1 or only the downstream A2 site, but HilD bound well only to the A1 region and not to A2 alone. Binding of HilD to the A2 region required the presence of flanking sequences. In summary, sites A1 and C2 can bind both HilC and HilD in the absence of flanking sequences, whereas sites A2 and D can bind HilC but not HilD, without additional flanking sequences. Even at the C2 site, binding of HilD required a longer stretch of specific base sequences than did binding of HilC.
These different binding specificities of HilC and HilD seen in the gel shift assays were reflected in the sequence dependence for binding to the C2 region. Base substitutions at 12 of the 39 positions studied caused a substantial decrease in HilC binding (>50% decrease). In contrast, substitutions at 25 of the 39 positions strongly reduced HilD binding. Substitutions that depressed HilC binding by >10% also depressed HilD binding, usually to a much greater degree. Substitutions unimportant for HilC binding generally had little effect on HilD binding, except for the segment from residues −60 to −56. These results show that HilC and HilD use overlapping binding sites and depend on the same nucleotide positions for DNA recognition. The greater dependency of HilD for the wild-type hilC sequence could reflect its lower affinity for this DNA sequence, such that any change in residues in its DNA target has a strong impact, whereas the higher affinity of HilC for DNA allows its binding to altered DNA sequences.
HilC- and HilD-binding sites.
Comparison of the four known binding site sequences revealed that the C2 region and the A1 region, which bind HilC and HilD, are closely related. Likewise, the D region resembles the A2 region, and both bind HilC but not HilD without flanking sequences. Sequence similarity extends for a considerable distance on either side of the D and A2 regions (not shown). Alignment of the four binding regions (Fig. 9) reveals some relevant features. Out of 34 positions, 8 are invariant and 11 more are present in three of the four regions. Residues at nine positions discriminate between the two types of binding sites, where the same base is present in the HilC- and HilD-binding A1 and C2 sites and a different base is present in the HilC-binding A2 and D sites. The A1 and C2 sites match at 23 of 34 positions and are very A+T rich (88 and 82%, respectively). Search for repeated sequence motifs in the HilC- and HilD-binding regions showed only the palindromic repeats of the sequence AT-T—TATT-N9-AATA—A-AT. Changes at any of these positions depressed binding of HilD, and all but the last three positions contributed to binding of HilC. Combination of the shared residues in the two HilC- and HilD-binding regions with the results of the mutagenesis analysis on protein binding yielded the consensus sequence: a/g T a/g TN a/t N3 TN t/g t/g c/g N3 AA a/t A2TA a/t A2N2AT a/c a/g G. Query with this consensus of the available genomic sequences for Salmonella serovars Typhimurium, Typhi, and Paratyphi revealed that each genome had only two matches, which were each an exact match to the A1 and C2 sequences.
FIG. 9.
Comparison of aligned HilC- and HilD-binding sequences in the hilA, hilC, and hilD promoters. The protein-binding sequences defined by DNase footprinting and electrophoretic mobility shift assay are aligned. The invariant sequences present in all four regions are enclosed in boxes and are identified in the consensus on the bottom as capital letters. Those bases present in three of the four regions are indicated in the consensus in lowercase letters. Those positions that discriminate between the two types of protein-binding regions are indicated by shading and are identified at the bottom. Along the top are shown the regions of the hilC promoter that were proven by mutational analysis to be important for binding of HilC or HilD. The arrows indicate the sequences with dyad symmetry in the A1 and A2 regions.
The HilC-binding A2 and D sequences match at 25 of 34 positions and are not as A+T rich (68 and 62%, respectively). They show greater palindromic character than do the HilC- and HilD-binding regions, with two pairs of repeats of the consensus sequence ATGGT in dyad arrangement. The discrimination between HilC and HilD cannot yet distinguish the contributions of specific base sequence recognition, A+T-rich character, or DNA bending.
As members of the AraC family, HilC and HilD monomers are expected to bind to a 17-bp region, in which each helix-turn helix motif binds to 3- to 6-bp stretches on successive DNA helical turns (29). For comparison, the AraC-binding sites at the araBAD and araFGH promoters are 38 bp in length and are arranged as two 17-bp binding regions in direct orientation separated by 4 bp (29, 36). The SoxS and Rob proteins bind to the same pairs of 17-bp sequences, but they require different bases for optimal binding (26). The HilC- and HilD-binding sites are described as two 17-bp regions in dyad order (Fig. 9), although the extent of dyad symmetry is most obvious only for the A2 region. Further study is needed to determine the orientation of the protein monomers on the DNA.
Preliminary attempts to test the relevance of these protein-binding sites in gene expression were equivocal. As in previous studies (30), we found only marginal modulation of expression of a hilD-lacZ reporter by environmental conditions or the presence of SPI1. Hence, mutagenesis of HilC-binding site D in the hilD promoter was not performed. Expression of the hilC-lac reporter was increased by the presence of SPI1 and by environmental conditions reported to induce hilA expression. We constructed two single base substitutions in the hilC-lac promoter region. The T-88G substitution was designed to alter a position that is important both for the extended −10 region for the upstream hilC promoter and for HilC and HilD binding to the overlapping C2 region. HilC binding to this promoter region was not eliminated, owing to the presence on the DNA fragment of the C1 and C3 regions (not shown). This variant promoter had very low activity during in vitro transcription (not shown) but retained substantial β-galactosidase expression when expressed in serovar Typhimurium, albeit with decreased response to modulatory signals. The same behavior was seen for the T-59G substitution in the hilC-lac reporter, which was expected to reduce binding of HilD but not of HilC. These results suggest that the control of hilC expression is more complex than just occupancy of the C2 region. These studies should be extended with single-copy-number reporters and analysis of changes of the hilA promoter regions. However, it is clear from this study that protein occupancy of sites in the hilA and hilC promoters is complex. Further work will address the structure, orientation, and consensus elements of these binding sites and seek evidence for DNA looping. The major question regarding how HilC or HilD affects hilA transcription in the cell may be approached when the putative repressor protein is identified.
Acknowledgments
We are indebted to Catherine Lee and Thomas Linn for providing strains and plasmids and to Aaron Mackey for genome sequence analysis.
This work was supported by research grant GM38681 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Ahmer, B. M., M. J. van Reeuwijk, P. R. Watson, T. S. Wallis, and F. Heffron. 1999. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol. Microbiol. 31:971-982. [DOI] [PubMed] [Google Scholar]
- 2.Altier, C., M. Suyetomo, and S. D. Lawhon. 2000. Regulation of Salmonella enterica serovar Typhimurium invasion genes by csrA. Infect. Immun. 68:6790-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology, vol. 1. John Wiley & Sons, New York, N.Y.
- 4.Bajaj, V., C. Hwang, and C. A. Lee. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18:715-727. [DOI] [PubMed] [Google Scholar]
- 5.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703-714. [DOI] [PubMed] [Google Scholar]
- 6.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in E. coli using bacteriophage transposons. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 7.Clark, M. A., M. A. Jepson, N. L. Simmons, and B. H. Hirst. 1994. Preferential interaction of Salmonella typhimurium with mouse Peyer's patch M cells. Res. Microbiol. 145:543-552. [DOI] [PubMed] [Google Scholar]
- 8.Collazo, C. M., and J. E. Galan. 1997. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol. Microbiol. 24:747-756. [DOI] [PubMed] [Google Scholar]
- 9.Darwin, K. H., and V. L. Miller. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181:4949-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darwin, K. H., and V. L. Miller. 1999. Molecular basis for the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 12:405-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ernst, R. K., D. M. Domboski, and J. M. Merrick. 1990. Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella typhimurium. Infect. Immun. 58:2014-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fahlen, T. F., R. L. Wilson, J. D. Boddicker, and B. D. Jones. 2001. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 183:6620-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fierer, J., and D. G. Guiney. 2001. Diverse virulence traits underlying different clinical outcomes of Salmonella infection. J. Clin. Investig. 107:775-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu, Y., and J. E. Galan. 1998. The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Mol. Microbiol. 27:359-368. [DOI] [PubMed] [Google Scholar]
- 15.Galan, J. E., and R. I. Curtiss. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galan, J. E., and R. I. Curtiss. 1990. Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect. Immun. 58:1879-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galas, D., and A. Schmitz. 1978. DNase footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 5:3157-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 20.Jones, B. D., N. Ghori, and S. Falkow. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of Peyer's patches. J. Exp. Med. 180:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein, J. R., T. Fahlen, and B. D. Jones. 2000. Transcriptional organization and function of invasion genes within Salmonella enterica serovar Typhimurium pathogenicity island 1, including the prgH, prgI, prgJ, prgK, orgA, orgB, and orgC genes. Infect. Immun. 68:3368-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolb, A., D. Kotlarz, S. Kusano, and A. Ishihama. 1995. Selectivity of the Escherichia coli RNA polymerase E sigma 38 for overlapping promoters and ability to support CRP activation. Nucleic Acids Res. 23:819-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar, A., R. A. Malloch, N. Fujita, D. A. Smillie, A. Ishihama, and R. S. Hayward. 1993. The minus 35-recognition of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J. Mol. Biol. 232:406-418. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Lee, C. A., and S. Falkow. 1990. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc. Natl. Acad. Sci. USA 98:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Z., and B. Demple. 1996. Sequence specificity for DNA binding by Escherichia coli SoxS and Rob proteins. Mol. Microbiol. 20:937-945. [DOI] [PubMed] [Google Scholar]
- 27.Linn, T., and R. St. Pierre. 1990. Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J. Bacteriol. 172:1077-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lostroh, C. P., V. Bajaj, and C. A. Lee. 2000. The cis requirements for transcriptional activation by HilA, a virulence determinant encoded on SPI1. Mol. Microbiol. 37:300-315. [DOI] [PubMed] [Google Scholar]
- 29.Lu, Y., C. Flaherty, and W. Hendrickson. 1992. AraC protein contacts asymmetric sites in the Escherichia coli araFGH promoter. J. Biol. Chem. 267:24848-24857. [PubMed] [Google Scholar]
- 30.Lucas, R. L., and C. A. Lee. 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:2733-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lucas, R. L., and C. A. Lee. 2000. Unraveling the mysteries of virulence gene regulation in Salmonella typhimurium. Mol. Microbiol. 36:1024-1033. [DOI] [PubMed] [Google Scholar]
- 32.Lucas, R. L., C. P. Lostroh, C. C. DiRusso, M. P. Spector, B. L. Wanner, and C. A. Lee. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:1872-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mills, D. M., V. Bajaj, and C. A. Lee. 1995. A 40-kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol. Microbiol. 15:749-759. [DOI] [PubMed] [Google Scholar]
- 34.Mirold, S., K. Ehrbar, A. Weissmüller, R. Prager, H. Tschape, H. Rüssman, and W.-D. Hardt. 2001. Salmonella host cell invasion emerged by acquisition of a mosaic of separate genetic elements, including Salmonella pathogenicity island 1 (SPI1), SPI5, and sopE2. J. Bacteriol. 183:2348-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray, R. A., and C. A. Lee. 2000. Invasion genes are not required for Salmonella enterica serovar Typhimurium to breach the intestinal epithelium: evidence that Salmonella pathogenicity island 1 has alternative functions during infection. Infect. Immun. 68:5050-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niland, P., R. Hühne, and B. Müller-Hill. 1996. How AraC interacts specifically with its target DNAs. J. Mol. Biol. 264:667-674. [DOI] [PubMed] [Google Scholar]
- 37.Olekhnovich, I. N., J. L. Dahl, and R. J. Kadner. 1999. Separate contributions of UhpA and CAP to activation of transcription of the uhpT promoter of Escherichia coli. J. Mol. Biol. 292:973-986. [DOI] [PubMed] [Google Scholar]
- 38.Olekhnovich, I. N., and R. J. Kadner. 1999. RNA polymerase α and σ70 subunits participate in transcription of the Escherichia coli uhpT promoter. J. Bacteriol. 181:7266-7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Penheiter, K. L., N. Mathur, D. Giles, T. Fahlen, and B. D. Jones. 1997. Non-invasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal Peyer's patches. Mol. Microbiol. 24:697-709. [DOI] [PubMed] [Google Scholar]
- 40.Pfeifer, C. G., S. L. Marcus, O. Steele-Mortimer, L. A. Knodler, and B. B. Finlay. 1999. Salmonella typhimurium virulence genes are induced upon bacterial invasion into phagocytic and nonphagocytic cells. Infect. Immun. 67:5690-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Schechter, L. M., S. M. Damrauer, and C. A. Lee. 1999. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol. Microbiol. 32:629-642. [DOI] [PubMed] [Google Scholar]
- 43.Schechter, L. M., and C. A. Lee. 2001. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 40:1289-1299. [DOI] [PubMed] [Google Scholar]
- 44.Scherer, C. A., and S. I. Miller. 2001. Molecular pathogenesis of Salmonellae, p. 265-333. In E. A. Groisman (ed.), Principles of bacterial pathogenesis. Academic Press, San Diego, Calif.
- 45.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 46.Takeuchi, A. 1967. Electron microscope studies of experimental Salmonella infection. I. Penetration into the intestinal epithelium by Salmonella typhimurium. Am. J. Pathol. 50:109-136. [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson, R. L., S. J. Libby, A. M. Freet, J. D. Boddicker, T. F. Fahlen, and B. D. Jones. 2001. Fis, a DNA nucleoid-associated protein, is involved in Salmonella typhimurium SPI-1 invasion gene expression. Mol. Microbiol. 39:79-88. [DOI] [PubMed] [Google Scholar]