Abstract
The cyanobacterium Prochlorococcus numerically dominates the photosynthetic community in the tropical and subtropical regions of the world's oceans. Six evolutionary lineages of Prochlorococcus have been described, and their distinctive physiologies and genomes indicate that these lineages are “ecotypes” and should have different oceanic distributions. Two methods recently developed to quantify these ecotypes in the field, probe hybridization and quantitative PCR (QPCR), have shown that this is indeed the case. To facilitate a global investigation of these ecotypes, we modified our QPCR protocol to significantly increase its speed, sensitivity, and accessibility and validated the method in the western and eastern North Atlantic Ocean. We showed that all six ecotypes had distinct distributions that varied with depth and location, and, with the exception of the deeper waters at the western North Atlantic site, the total Prochlorococcus counts determined by QPCR matched the total counts measured by flow cytometry. Clone library analyses of the deeper western North Atlantic waters revealed ecotypes that are not represented in the culture collections with which the QPCR primers were designed, explaining this discrepancy. Finally, similar patterns of relative ecotype abundance were obtained in QPCR and probe hybridization analyses of the same field samples, which could allow comparisons between studies.
The unicellular cyanobacterium Prochlorococcus is remarkable not only for its high population density (concentrations often exceed 100,000 cells ml−1 [4, 7, 21]) but also for its vast horizontal and vertical habitat range in the oceans, characteristics that together make it the most abundant marine oxyphototroph (4, 7, 21). Prochlorococcus has been found ubiquitously in the oligotrophic oceans between 40°N and 40°S latitude. The abundance is maximal in the stratified, nutrient-poor waters during the summer but is also significant in the deeply mixed, (relatively) nutrient-rich waters during the winter and spring (4, 7, 21). In the oligotrophic oceans, the euphotic zone (which is about 200 m deep) is characterized by steep vertical gradients of light, temperature, and nutrients, which can vary significantly by season (15, 24). Remarkably, the photosynthetic community of the entire euphotic zone at such sites is typically dominated by Prochlorococcus (4, 7, 21).
The Prochlorococcus lineage is genetically and physiologically diverse (17, 18, 20, 22), which is a likely key to its numerical dominance. Prochlorococcus is composed of at least six distinct lineages, or ecotypes, and early studies of the physiological properties of these lineages suggested that they partition the niche vertically. Closely related ecotypes eMED4 and eMIT9312 (the prefix “e” distinguishes an ecotype from the type strain that it was named after, as described by Ahlgren et al. (1]) were predicted to dominate the upper euphotic zone, as they display optimal growth rates at higher light intensities than the other four ecotypes. The other four ecotypes (eMIT9211, eNATL2A, eSS120, and eMIT9313) were predicted to dominate the deeper waters, based on their lower light optima and the ability of some of them to utilize nitrite, which in stratified waters is found deeper in the euphotic zone (17-19). While not quantitative, sequence analysis of Prochlorococcus petB/D genes in the Atlantic Ocean (26) and of rpoC genes in the Pacific Ocean (10) provided the first evidence that supported the following predictions: surface “types” are phylogenetically distinct from deep “types,” and the frequency of “high-light” ecotypes is highest in surface waters.
To test these predictions for Prochlorococcus population structure in a quantitative manner, two different molecular methods have recently been developed, probe hybridization with 16S rRNA genes as the target (28) and quantitative (real-time) PCR with the internal transcribed spacer (ITS) or 23S rRNA gene as the target (1). Both of these methods were designed to target individual ecotypes, although currently the probes cannot distinguish some of the known low-light ecotypes. Probe hybridization studies of a stratified water column in the eastern North Atlantic Ocean showed that there was complete domination of the upper euphotic zone by the high-light eMED4 ecotype, while the lower euphotic zone contained only low-light ecotypes (28). In sharp contrast, quantitative PCR (QPCR) analysis of a stratified water column in the western North Atlantic showed that the high-light eMIT9312 ecotype dominated the upper euphotic zone, while the eMED4 ecotype was present but the eMED4 counts were only a fraction of the eMIT9312 counts (1). The lower euphotic zone was dominated by two low-light ecotypes, eNATL2A and eMIT9313, but interestingly, the eMIT9312 ecotype was also present at significant levels at these depths. This population structure resembled that of a stratified site in the Red Sea, as determined by probe hybridization, except that the eMED4 ecotype was not detected at the latter site (29).
The general prediction that the high-light and low-light ecotypes vertically partition the water column is thus supported by both of these field studies, which employed different methods of analysis. However, the degree of partitioning and the identities of the dominant ecotypes appear to vary from site to site. Direct comparisons between probe hybridization and QPCR methods for Prochlorococcus quantification have not been done. Because the two methods target different loci, it is possible that the differences observed in the eastern and western North Atlantic are due to the methods used to detect different populations. In this study we addressed this question directly by applying the two methods to the same samples.
Flow cytometry (FCM) identifies Prochlorococcus by its distinct forward light scatter (an indicator of cell size) and chlorophyll autofluorescence signal and is used to quantify the abundance of the total population in the field (5, 11). Thus, comparisons of FCM counts with the sum of the QPCR counts for the six known ecotypes provide a means to assess the ability of the current QPCR primers to target the majority of the cells in the population. In the western North Atlantic profile analyzed by Ahlgren et al. (1), such comparisons indicated that Prochlorococcus was well accounted for by QPCR in the upper euphotic zone, where the eMIT9312 ecotype was by far the dominant taxon. The six targeted ecotypes represented only a fraction of the total population in the lower euphotic zone, however, indicating that the deeper waters were dominated by novel ecotypes of Prochlorococcus. A second goal of this study was to describe these “missing” ecotypes, for future redesign of our suite of QPCR primer pairs to capture more of the Prochlorococcus population.
The analyses of Prochlorococcus ecotype distributions to date, i.e., those described by West and Scanlan (28), Ahlgren et al. (1), and here, provide a small snapshot of the distribution of ecotypes and the environmental variables with which they covary. A full understanding of the dynamics of this system globally requires analysis of many samples collected from a broad range of environmental gradients in time and space. To accomplish this, the third goal of this work was to modify our existing QPCR protocol (1) in order to improve its speed and sensitivity and provide a means to quantify novel ecotypes as they are discovered, even if they have no representatives in culture.
MATERIALS AND METHODS
Strains and culture conditions.
Prochlorococcus strains MIT 9312, MED4, MIT 9211, NATL2A, SS120, and MIT 9313 (22) were cultured for QPCR analysis. Cultures were grown at 22°C with a cycle consisting of 14 h of light and 10 h of darkness under cool white fluorescent lamps which provided approximately 20 μmol quanta m−2 s−1 of photosynthetically active radiation (measured with a QSL-100 quantum scalar irradiance meter [Biospherical Instruments, San Diego, CA]). Cultures were grown in Pro99 natural seawater-based medium (described but not named by Moore et al. [19]) in glassware that was soaked overnight in 1 N HCl and then rinsed in 18-MΩ water (Milli-Q Biocel system; Millipore, Billerica, MA).
Field sample collection and storage.
Water samples were collected on two cruises in the North Atlantic Ocean. Samples from the western Sargasso Sea were collected on 29 August 2002 (CTD 29; 33.223°N, 64.879°W) aboard the R/V Endeavor (EN375). Samples from the eastern North Atlantic were collected on 19 September 2003 (CTD 15; 34.683°N, 22.99°W) aboard the R.R.S. James Clark Ross (AMT13). CTD measurements (conductivity, temperature, barometric pressure, chlorophyll a fluorescence, and photosynthetically active radiation) were obtained as the samples were collected with a Niskin rosette. Water was transferred from Niskin bottles to amber collection bottles before processing. Filtration of EN375 samples was performed with plastic filter funnels washed between samples with Milli-Q water and a small volume of seawater from the sample about to be filtered. Filtration of AMT13 samples was performed with glass filter funnels cleaned between samples by soaking in 10% (vol/vol) bleach, followed by rinsing with 18-MΩ water. Samples used for QPCR analysis were collected by filtering 100 ml of seawater onto 25-mm-diameter, 0.2-μm-pore-size polycarbonate filters under a low vacuum (less than 10 mm Hg). The filters were washed with 3 ml preservation solution (10 mM Tris [pH 8.0], 100 mM EDTA, 0.5 M NaCl [28]) and placed into 2-ml polypropylene screw-cap tubes (Sarstedt Inc., Newton, NC). The tubes were stored in liquid nitrogen (EN375) or at −80°C in a low-temperature freezer (AMT13) until analysis. Duplicate 1-ml aliquots of seawater were also prepared for flow cytometry (see below).
Flow cytometry.
Duplicate 1-ml samples (laboratory and natural seawater samples) were prepared for FCM by glutaraldehyde fixation. Cells were fixed in 0.125% (vol/vol) glutaraldehyde during a 10-min incubation in the dark and then frozen in liquid nitrogen until analysis. Cell concentrations were measured with an EPICS flow cytometer or a FACScan (Becton Dickinson) by using red fluorescence and forward angle light scattering properties to identify the cells, as described previously (5, 8).
Preparation of QPCR standards from Prochlorococcus cultures.
QPCR standards were prepared by filtration from laboratory cultures having known cell concentrations (measured by flow cytometry) as described previously (1), with two modifications. First, the filtration funnels used to collect the cells were soaked for 2 h in 10% (vol/vol) bleach and then rinsed with 18-MΩ water, rather than just rinsed. Second, the filters with cells were stored at −80°C rather than −20°C.
Preparation of QPCR standards from ITS sequences.
Plasmid clones containing the 16S/ITS/23S rRNA gene regions of the six type strains were generated for use as alternative QPCR standards. The 16S/ITS/23S rRNA gene regions of strains MIT 9312, MED4, MIT 9211, NATL2A, SS120, and MIT 9313 were amplified by PCR, using the cyanobacterium-specific primers 16S-1247f and 23S-1608r (22). The PCR was performed with a PTC-100 (MJ Research, Inc., Waltham, MA) as described by Rocap et al. (22). Products from these amplifications were then cloned directly into the pCR4-TOPO vector using a TOPO TA cloning kit for sequencing and introduced into Escherichia coli TOP10 cells by following the manufacturer's instructions (Invitrogen Corp., Carlsbad, CA). DNA sequencing of the inserts was performed using primers targeting the vector, as well as internal primers generated from the information obtained from sequencing from the ends. All DNA sequencing reported in this work was performed at Davis Sequencing, Inc. (Davis, CA) or the Harvard Medical School Biopolymers Facility (Boston, MA). For all six QPCR standard clones, perfect matches with the corresponding QPCR primers were found at the priming sites.
Clones of the 16S/ITS/23S rRNA gene regions were prepared for use as standards for QPCR by linearizing the plasmids with the PstI restriction enzyme. Clones were first purified from the TOP10 cells (Invitrogen Corp., Carlsbad, CA) using a QIAprep Spin Miniprep kit (QIAGEN, Valencia, CA) by following the manufacturer's instructions and eluted in 50 μl of buffer EB. Purified clones were stored at −80°C until digestion. PstI digestion was performed in 30-μl mixtures with 3 μl of a purified clone, 2 μl of PstI, 1× buffer 3, and 1× bovine serum albumin (New England Biolabs, Inc., Beverly, MA). Incubation was performed at 37°C for 3 h. Products were stored at −80°C until analysis. For each reaction, single bands were observed on 1% (wt/vol) agarose gels, indicating that the clones contained a single PstI restriction site (located in the vector sequence).
Concentrations of the PstI digests of the ITS clones were determined with the PicoGreen double-stranded DNA quantitation reagent (Molecular Probes, Inc., Eugene, OR) by following the manufacturer's instructions. PicoGreen-stained DNA was quantified with a Synergy HT multidetection microplate reader (Bio-Tek Instruments, Inc., Winooski, VT), with excitation set at 485 nm and emission set at 528 nm. Five replicates were measured for each digest, and the mean values are reported below.
QPCR template DNA preparation.
Extraction of genomic DNA by chemical and enzymatic lysis was performed as described by Ahlgren et al. (1), except that the DNA template was stored at −80°C rather than −20°C. Preparation of the DNA template by our new heat lysis method involved four steps. First, frozen samples in screw-cap tubes were thawed on ice, and 650 μl of 10 μM Tris (pH 8.0) was added. Second, the tubes were placed in a mini-bead beater (BioSpec Products, Bartlesville, OK) and vortexed for 2 min at the maximum speed (4,800 rpm) to resuspend the cells. Third, using a narrow-gauge 1-ml pipette tip, 500 μl of the resuspended cells was transferred from the filter shreds into a new microcentrifuge tube. Finally, the tube was incubated at 95°C for 15 min to inactivate DNases and lyse the cells. Cell lysates were aliquoted and stored at −80°C, and aliquots were never thawed and refrozen more than four times during analysis.
Quantitative PCR.
To determine the abundance of each of the ecotypes in the samples, QPCR, using ecotype-specific primers (1), was performed with replicate filtered samples for each depth. Reactions were performed in duplicate for each standard, environmental sample, and negative control lacking template DNA (10 mM Tris [pH 8]). For all field data presented in this work, standards were prepared by heat lysis of cell cultures. QPCR was performed by using 25-μl mixtures at the DNA Engine Opticon (MJ Research, Inc., Waltham, MA). QPCR was performed with 10 μl of each lysate supplemented with the Quantitect SYBR Green PCR master mixture (QIAGEN Inc., Valencia, CA). Individual QPCR were performed for each ecotype primer set, and the appropriate standards were analyzed for each QPCR. The ecotype cell concentration for a given sample was determined by comparing the CT value (the PCR cycle when the fluorescence value [an indicator of amplicon concentration] crosses a designated threshold value) of the sample template with the CT values of the standards. The R2 values for the fluorescence-versus-CT curves for the standards were typically 0.99 or higher. The threshold value was set manually as the lowest fluorescence value exhibiting the greatest R2 value for the standard set. The primers used to specifically amplify each ecotype were the primers described by Ahlgren et al. (1). The primers were used at a final concentration of 500 nM, except for the MIT 9312 ecotype reactions (300 nM) and the MIT 9313 ecotype reactions (1,000 nM). All QPCR were initiated with 15 min of incubation at 95°C and were terminated with 5 min of incubation at 72°C. Forty or more cycles were used for each ecotype, with the following parameters: for the eMIT9312 ecotype, 95°C for 45 s, 56°C for 45 s, and 72°C for 10 s; for the eMED4 ecotype, 95°C for 45 s, 58°C for 45 s, and 72°C for 30 s; for the eMIT9211 ecotype, 95°C for 15 s, 56°C for 30 s, and 72°C for 10 s; for the eNATL2A ecotype, 95°C for 30 s, 56°C for 30 s, and 72°C for 5 s; for the eSS120 ecotype, 95°C for 15 s, 56°C for 45 s, and 72°C for 10 s; and for the eMIT9313 ecotype, 95°C for 45 s, 56°C for 5 s, and 72°C for 20 s.
For all QPCR measurements described in this work, levels of templates (standards and field samples) are expressed as concentrations (in cells ml−1) of Prochlorococcus in situ in the ocean. These concentrations were calculated by dividing the concentration of cells on a filter (determined by FCM for the cultures used as standards) by the volume of seawater collected in the field per filter (100 ml). In this work, the standard set concentrations for field sample analyses ranged from a few cells ml−1 to more than 1,000,000 cells ml−1, which allowed quantification by interpolation within this concentration range. For this study, the most dilute standard concentrations used for the ecotypes were 3.39, 3.06, 3.02, 1.62, 5.31, and 2.73 cells ml−1 for eMED4, eMIT9312, eMIT9211, eNATL2A, eSS120, and eMIT9313, respectively. Measurements of template concentrations were valid if they met two criteria: (i) their CT values were lower than those of the negative controls, which lacked template DNA, and (ii) the melting curve analysis of the products showed an absence of nonspecific amplification. Sample template concentrations lower than the concentration of the most dilute standard were also reported in the data, as long as they had CT values that were lower than those of the no-template negative controls (10 mM Tris [pH 8]), with the caveat that they were extrapolated values beyond the concentrations for the standard set. The theoretical limit of detection in this assay is 0.65 cells ml−1 or 1 cell per PCR. For purposes of graphic presentation, values that were below this limit were changed to this limit, as were values for samples that showed that nonspecific products were amplified (these values were only for a few eMIT9313 reactions of field samples and gave artifactual values of less than 10 cells ml−1).
QPCR product cloning and sequencing.
QPCR products for each of the six ecotype reactions for the EN375 CTD 29 profile were stored at −20°C until cloning. QPCR products for the eMIT9312, eMED4, and eNATL2A ecotype primer reactions were cloned directly using a TOPO TA cloning kit for sequencing into the pCR4-TOPO vector by following the manufacturer's instructions (Invitrogen Corp., Carlsbad, CA). QPCR products for the eMIT9313, eMIT9211, and eSS120 ecotype primer reactions could not be cloned directly, for reasons that were unknown but perhaps were related to their larger sizes relative to the sizes of eMIT9312, eMED4, and eNATL2A (226 to 419 versus 78 to 120 bp). Therefore, the QPCR products of eMIT9313, eMIT9211, and eSS120 were subjected to an additional five cycles of PCR in the Opticon using the same cycle parameters that were used for the QPCR, followed by 5 min of incubation at 72°C. These secondary reactions were performed in 50-μl mixtures with 1 μl of Taq polymerase (Promega, Madison, WI) in Taq buffer (Promega) supplemented with 3 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 0.5 mM, 5 μl of the QPCR product as the template, and the appropriate primers at the same concentration that was used for QPCR. Clones were prepared for sequencing with a QIAprep Spin miniprep kit (QIAGEN, Valencia, CA). Sequencing was performed on one strand for all clones and also on the reverse strand for all unique clones.
Construction of ITS libraries from field samples.
DNA from sample EN375 CTD 29 (depths, 0 m, 100 m, and 125 m) collected in the Sargasso Sea was used as a template for PCR, followed by cloning and sequencing. Primers targeting the full ITS region were modified from the primers described by Ernst et al. (9) to be specific for all Prochlorococcus ecotypes except the eNATL2A ecotype (two mismatches), which resulted in fragments between 703 and 993 bp long. Amplification was performed with primers ACMITS-2F (5′-GAAGTCGTTACTCCAACCC-3′) and ACMITS-3R (5′-TCATCGCCTCTGTGTGCC-3′), using the following thermal program: hot start at 95°C for 5 min, followed by 30 cycles of 95°C for 45 s, 63°C for 45 s, and 72°C for 2 min and a final extension step at 72°C for 10 min. Amplified DNA was gel purified and cloned using a TOPO TA 4.0 cloning kit (Invitrogen, Carlsbad, CA) by following the manufacturer's instructions and was subsequently sequenced using ACMITS-2F as the sequencing primer.
Phylogeny.
ITS sequences were aligned using the ARB software package (16) against a custom-built database containing all known Prochlorococcus ITS sequences. The sequences that were aligned were limited to the region between the ACMITS-2F and ACMITS-3R primers, and as described by Rocap et al. (22), the two tRNA sequences in the ITS region were excluded. A base frequency filter (16) excluding positions different in more than 50% of the selected sequences was calculated to ensure that homologous positions were compared, which resulted in a total of 310 bp for the analysis. The exact nucleotide positions used in this alignment differ from those used by Rocap et al. (22), as the suites of sequences aligned were different and base frequency filtering is dependent on the sequences analyzed. However, the overall tree topologies were essentially the same for the two alignments. Selected sequences were exported to PAUP* version 4b10 for a detailed phylogenetic analysis using neighbor-joining (minimum evolution as the criterion and paralinear [logdet] distance correction), maximum-parsimony, and maximum-likelihood (HKY) model parameters, gamma shape optimized by iterative likelihood searches starting from a maximum-parsimony tree. Bootstrap analysis (100 resamplings) was done with heuristic searches utilizing random addition and tree bisection reconnection branch swapping methods.
16S rRNA gene probe studies.
Aliquots of the AMT13 CTD 15 heat lysates prepared initially for QPCR were used as templates in the PCR using the oxygenic phototroph-specific 16S rRNA gene primers OXY107F and OXY1313R (27) as described previously (26). Dot blot hybridization with the 16S rRNA gene-specific oligonucleotide probes was performed as described previously (28) except that we used a modified, more specific HLII probe, S2PRO640R (12).
Nucleotide sequence accession numbers.
Environmental DNA sequences have been depsoted in the GenBank database under accession numbers DQ016306 to DQ016319 (ITS clones obtained with cyanobacterial primers ACMITS-2F and ACMITS-2R), DQ019002 (QPCR clone of the eMIT9211 ecotype), DQ019018 to DQ019030 (QPCR clones of the eNATL2A ecotype), DQ019031 and DQ019032 (QPCR clones of the eSS120 ecotype), and DQ019003 to DQ019017 (QPCR clones of the eMIT9313 ecotype). The sequences for QPCR clones of the eMED4 and eMIT9312 ecotypes, which are shorter than the minimum length for the GenBank database (50 bp), are as follows: eMED4 operational taxonomic unit 1 (OTU 1), TACATATATATAAAGAGGGAAATTGCTTTGAGTCGTGTCCTAATTT; eMED4 OTU 2, TACATATAGTTGAAGTGGGAAATTGTTTTGAGTCGTGTCCTAATTT; and eMIT9312 OTU 1, CTTTGATCCGGGAATTTCCGAATGGGGCAACCCC.
RESULTS AND DISCUSSION
Development of a streamlined QPCR assay.
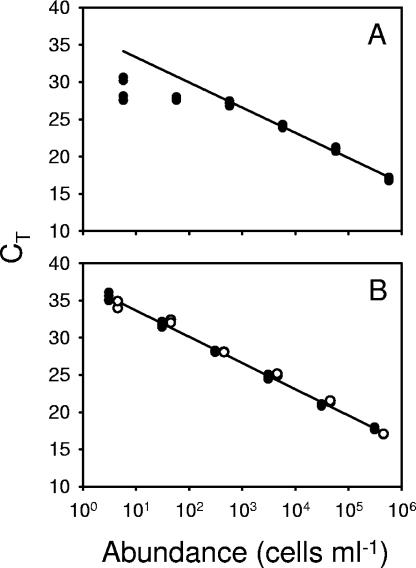
With the capacity to run 96 or even 384 reactions in parallel and with a reaction time on the order of several hours, QPCR has the potential for high-throughput quantification of environmental samples. This potential was limited in our earlier QPCR method for Prochlorococcus ecotypes (1), as well as in other systems (2, 3, 13, 25), by the time required to extract and purify the environmental DNA. By employing a new heat lysis protocol (see Materials and Methods), we were able to decrease the sample preparation times from hours to minutes and also to eliminate the use of hazardous materials, such as phenol and chloroform. For all strains tested, the streamlined technique was accurate over 6 orders of magnitude for template concentration (Fig. 1). Preparations of template from a culture dilution series using the original and streamlined methods yielded standard curves with statistically indistinguishable slopes and y intercepts (as determined by Student's two-tailed t tests, with a significance of α = 0.05) (data not shown). Any RNA remaining in the extracts did not interfere with the quantification (data not shown), even though Taq polymerase has been shown to have some reverse transcriptase activity in several studies (14, 23).
FIG. 1.
Generation of QPCR standards from laboratory cultures of Prochlorococcus strain MIT 9312, using the heat lysis method. (A) Standards generated from filtering a dilution series of cultured cells, using 18-MΩ water washes between filtrations. The first two points (∼10 and ∼100 cells ml−1) were excluded from the regression. Multiple points at each concentration represent replicate analyses of the same culture. (B) Standards generated from dilution series for two different cultures (culture 1, open symbols, culture 2, solid symbols), using a bleach soak and 18-MΩ rinse between filtrations. Regression for both cultures: y = −1.53 ln(x) + 37.14 (R2 = 0.994).
Because the environmental DNA is prepared as a cell extract in our heat lysis method, there is the potential for the extract to have long-term storage effects on the integrity of the DNA. Two tests confirmed that this is unlikely to be an issue for this method. First, a freshly prepared heat-treated lysate of strain MIT 9312 was subjected to 10 cycles of freezing at −80°C and thawing and compared by QPCR to a replicate lysate that was never frozen. The CT values were not significantly different after this treatment (data not shown). Second, we observed that QPCR analysis of the same DNA extract from field samples analyzed a year apart gave the same result (the R2 was 0.996 for a comparison of results obtained with all six primer sets for 11 samples, including levels that spanned 5 orders of magnitude) (data not shown). This is a particularly important feature of the system as we will undoubtedly want to reexamine samples as we refine our primers (see below).
Improving the limits of detection.
The second modification of our earlier protocol was made to increase the sensitivity of measurement. Routinely in biological oceanography, field samples are collected via filtration, and the filtration unit is typically cleaned between samples by rinsing it with seawater and/or 18-MΩ water. Through a series of control experiments, we learned that carryover contamination from one sample to another on our filter rigs could not be completely eliminated by rinsing with water. This was shown by lower-than-expected CT values for the most dilute standards, indicating greater-than-expected template concentrations (Fig. 1A). We therefore added a bleach soak cleaning step for the filtration units (2 h of soaking in 10% [vol/vol] bleach), which lowered the limit of detection from 10 to 100 cells ml−1 to a few cells ml−1 (Fig. 1B), which is near the theoretical limit of detection (1 cell per PCR) but above any background detected in control reactions without template addition. This represents a 1- to 2-order of magnitude improvement in the method (Fig. 1A) (1). With the QIAGEN Quantitect SYBR Green PCR master mixture, control reactions without a DNA template rarely showed product at the end of the QPCR. When they did, the concentration was on the order of a few cells ml−1 (data not shown), and these values were used as the limit of detection for the reactions run in parallel with template DNA added.
Development of culture-independent QPCR standards: ITS clones.
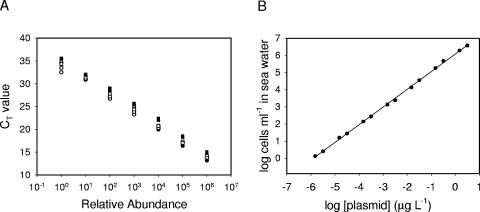
In our original protocol, standards for QPCR quantification were derived from genomic DNA prepared from Prochlorococcus cultures (1; this study). Given the challenge of culturing and quantifying Prochlorococcus in the laboratory, we generated an alternative, more accessible set of standards consisting of cloned ITS fragments. A set of clone standards was generated by PCR amplification and cloning of the ITS of the six ecotype type strains (Table 1). For eMED4 (Fig. 2A) and the other five ecotypes (Table 1), abundance-versus-CT curves of both types of standards had the same slope. For the cell culture-based standards, dilutions were prepared before extraction, while for the clones they were prepared after extraction. The agreement of the slopes of the standard curves demonstrates that the efficiency of template extraction for the culture-based method is independent of the cell concentration (Fig. 2 and Table 1).
TABLE 1.
Characteristics of ITS clones used as QPCR standards for the six Prochlorococcus ecotypes
| Ecotype | ITS clone | ITS insert size (bp) | Cells and ITS clones used as standards have equivalent slopes?a | ITS clone calibrationb
|
||
|---|---|---|---|---|---|---|
| Slope | y intercept | R2 | ||||
| eMED4 | pEZ10 | 2,482 | Yes (0.878) | 1.03 | 6.01 | 0.997 |
| eMIT9312 | pEZ11 | 2,489 | Yes (0.645) | 0.998 | 6.07 | 0.999 |
| eMIT9211 | pEZ12 | 2,635 | Yes (0.570) | 1.04 | 6.47 | 0.987 |
| eNATL2A | pEZ13 | 2,575 | Yes (0.487) | 1.04 | 6.16 | 0.998 |
| eSS120 | pEZ14 | 2,605 | Yes (0.349) | 1.05 | 6.25 | 0.998 |
| eMIT9313 | pEZ15 | 2,772 | Yes (0.948) | 0.971 | 6.23 | 0.988 |
| Avg | 6.20 | |||||
| SD | 0.16 | |||||
Comparisons of slopes for abundance-versus-CT value curves for cell culture-derived and ITS clone QPCR standards. See Fig. 2A for an example of a slope comparison (for MED4). The values in parentheses are results from two-tailed t tests with a significance of α = 0.05.
Calibration of ITS clone standards was performed by comparison to standards derived from Prochlorococcus cell cultures. See Results and Discussion for details and Fig. 2B for an example of a calibration curve (for MED4).
FIG. 2.
Calibration of ITS clones for use as QPCR standards in absolute quantification of field samples. (A) Dilution series of templates derived from cell cultures (circles) and clones of the ITS region (squares) of MED4. The open and solid symbols indicate replicate cultures or plasmid preparations. The relative abundance indicates the abundance within each dilution series (the most dilute concentration for cells was 3.02 cells ml−1, and most dilute concentrations for plasmid preparations were 3.17 and 1.5 pg liter−1 for preparations 1 and 2, respectively). (B) Cross-calibration of ITS clone standards with cell culture standards. Other ecotypes produced similar curves with similar slopes (Table 1).
ITS clones were calibrated for the absolute quantification of ecotypes in the field by first determining the concentrations (in μg liter−1) of the plasmid clones and then converting the values to numbers of cells ml−1 in seawater by comparing the CT values with the values for cell culture standards. For eMED4 (Fig. 2B) and the five other ecotypes (Table 1), the slopes of the linear regression for plasmid concentration versus cell abundance were approximately 1, and all had high R2 values (Table 1). Because the slopes were 1, the y intercept became the factor for converting the concentration of the plasmid to cell abundance in the field: log10 cell abundance (in cells ml−1) = log10 plasmid concentration (in μg liter−1) + y intercept. The y intercept value for the log-log plots of the six ecotypes was 6.20 ± 0.16 (average ± standard deviation) (Table 1).
Since the y intercepts are so similar for the six known ecotypes, it is reasonable to assume that any novel ecotypes would have comparable values and thus any new ecotypes that are discovered (see below) could be quantified without having to culture them. Clones of their ITS regions could be used as standards for QPCR with a reasonable degree of confidence; the y intercept for these clones would be assigned a value of 6.20, the average for all Prochlorococcus ecotypes.
Field application of the improved QPCR method.
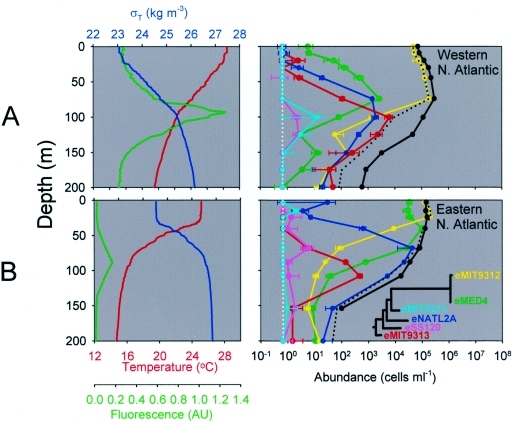
Field samples were collected in late August 2002 in the western North Atlantic Ocean near Bermuda. At this time of year at this site the water column was well stratified with a shallow (10-m), warm (>28°C) mixed layer and a deep chlorophyll maximum at about 100 m (Fig. 3A, left panel). Using the QPCR method described here, we found all six ecotypes in the water column, although the levels were vastly different. The eMIT9312 ecotype dominated the surface waters, and the concentrations were more than 100,000 cells ml−1 at the subsurface maximum (Fig. 3A, right panel). Although eMIT9312 was at least 2 orders of magnitude more abundant than the other ecotypes, the other ecotypes were still very easily quantified using our methods. The eMED4, eNATL2A, and eMIT9313 ecotypes displayed well-defined concentration maxima with increasing depth, which resulted in a “stacked” appearance in the water column (Fig. 3A). Finally, the maximum concentrations of eMIT9211 and eSS120 were only 10 cells ml−1 or less.
FIG. 3.
Hydrographic features (left panels) and depth profiles of Prochlorococcus (right panels) in the western North Atlantic on 29 August 2002 (A) and in the eastern North Atlantic on 19 September 2003 (B). In the left panels, hydrographic features are indicated by different colors, as follows: water temperature, red; density (σT), blue; and fluorescence, green (values in arbitrary units [AU]). In the right panels, the six ecotype QPCR values are indicated by different colors, as follows: eMIT9312, yellow; eMED4, green; eNATL2A, blue; eMIT9313, red; eMIT9211, light blue; and eSS120, pink. An idealized phylogenetic tree is shown in panel B (based on the study of Rocap et al. [22]). The solid black lines indicate the total counts as determined by flow cytometry. The dotted black lines indicate the sum of the QPCR counts of all six ecotypes. The error bars indicate one standard deviation of the mean for replicate filters. The white dotted vertical line at 0.65 cells ml−1 indicates the theoretical limit of detection (1 cell per PCR). QPCR values less than or equal to this limit are placed on this line.
Ahlgren et al. (1) studied the relative abundance of ecotypes at a similar site in the Atlantic Ocean a year earlier using the original QPCR methodology. Their water column had a deeper (40-m) and colder (∼26°C) mixed layer, but the deep chlorophyll maximum was at about the same depth as in our profile (100 m). Ahlgren et al. found relative abundance patterns for ecotypes that were very similar to the patterns which we found. Although total Prochlorococcus abundance undergoes significant seasonal variation in this region, the year-to-year variation during a given season is small (7). It is perhaps not surprising that ecotype abundances appear to be relatively stable as well, but only longer time series could validate this.
Sequence analysis of the QPCR amplicons.
Since the QPCR primer sets were designed to capture the “cluster” of strains represented in a particular group, it is likely that they amplify a number of different ribotypes. To study this and to verify that primers were indeed specific for a particular cluster, we cloned and sequenced the QPCR products from selected depths. For all six primer sets, all sequences most closely resembled those of the ecotypes targeted; the levels of identity to known cultured ecotype representatives ranged from 80% to 100% (Table 2). In contrast, the levels of sequence similarity to members of nontarget ecotypes were less than 75%. The exception, as expected, was the results for the eMIT9312 primer set because the region between the primers is the same for strains in the eMIT9312, eMED4, and eMIT9313 clusters, although there is significant divergence between the ecotypes in the sequence targeted by the primers (1). We concluded that the primers were able to amplify specifically DNA from the target ecotypes. Most of the clone sets contained one or two OTUs (an OTU was defined as any clone with a unique sequence). Interestingly, both the eNATL2A and eMIT9313 clone sets had a high number of OTUs. In fact, all 15 clones sequenced for the eMIT9313 ecotype primer reactions were unique. This provides preliminary evidence that for these two ecotypes at least, the microdiversity within the clades can be very high, even in a relatively small sample of water (100 ml).
TABLE 2.
Properties of cloned QPCR amplicons from the western North Atlantic Ocean
| Ecotype primer set | Sample depth (m) | Between-primer read length (bp) | Total no. of clones | No. of OTUs | % Clone identity to known ecotype members | Highest % identity to nonmembers |
|---|---|---|---|---|---|---|
| eMIT9312 | 74 | 34 | 10 | 1 | 100 | 100a |
| eMED4 | 100 | 46 | 9 | 2 | 80.4-91.3 | 52.1-54.3 |
| eMIT9211 | 100 | 215 | 10 | 1 | 99.5 | 55.4 |
| eNATL2A | 100 | 69 | 16 | 13 | 92.7-100 | 62.5-66.6 |
| eSS120 | 100 | 177 | 8 | 2 | 99.4-100 | 74.4 |
| eMIT9313 | 100 | 377 | 15 | 15 | 94.4-99.4 | 61.2-62.9 |
MED4 and MIT 9313 have perfect matches to the eMIT9312 ecotypes sequences between the primers but differ in the primer regions, where PCR specificity is conferred.
One caveat with the sequence data concerns the two low-abundance ecotypes, eMIT9211 and eSS120. The maximum concentration for both of these ecotypes was 10 cells ml−1 or less in the depth profile. The sequences of the QPCR amplicons were very similar (99 to 100%) to the sequences of the type strains, MIT 9211 and SS120, suggesting that there was possible contamination from our cultured strains. We think that it is more likely, however, that these signals were real as this and other depth profiles (Chisholm et al., unpublished data) always showed that the maximum abundance of eMIT9211 and eSS120 was in the subsurface. If PCR well contamination were the source of the signal, we would not have seen these consistent (and logical) patterns with depth.
QPCR analysis of an eastern North Atlantic site.
Examination of a second depth profile from a different oceanic region provided further validation of the QPCR technique and insights into the geographical variability of ecotype distributions (Fig. 3B). Like the profile shown in Fig. 3A, this profile was obtained in the summer (19 September 2003) when the water column was highly stratified, with the deep chlorophyll maximum concentration at approximately 100 m (Fig. 3B, left panel). However, significant differences between the two sites were observed with respect to the relative abundances of the ecotypes at different depths. While the eMIT9312 ecotype dominated the surface mixed layer at both stations, its abundance decreased dramatically below the mixed layer (30 m) at the eastern site (Fig. 3B, right panel), whereas its abundance remained high down to 100 m at the western site (Fig. 3A). eMED4 was relatively rare at the surface at the western site, and the peak abundance was at the depth where the chlorophyll concentration was maximum. In contrast, at the eastern site the maximum eMED4 abundance was just below the surface mixed layer, where this ecotype was the most abundant ecotype in the water column. In fact, its integrated abundance in the upper 50 m was roughly equal to that of eMIT9312. The eNATL2A ecotype was also present in the eastern profile, but the concentration was 1 order of magnitude higher than the maximum concentration in the west (∼40,000 versus ∼2,000 cells ml−1). For eMIT9313 there was a deep subsurface maximum concentration at both sites, while there was little or no eMIT9211 and eSS120.
In contrast to the western profile, the total abundance of all ecotypes measured by QPCR at the eastern site closely matched the abundance measured by flow cytometry at all depths. Hence, the degree to which the current suite of QPCR primer sets can account for the total Prochlorococcus populations appears to vary not only with respect to depth but also with respect to geographical location (see below).
Direct comparisons of QPCR and 16S rRNA gene probe methods.
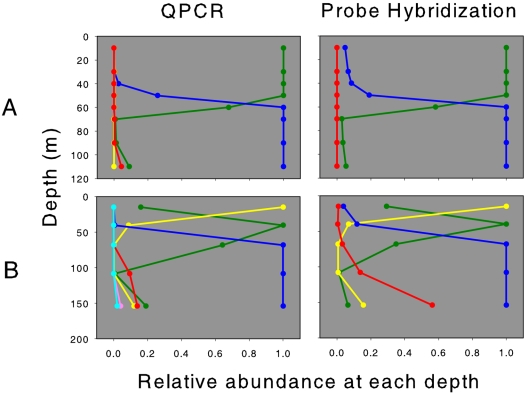
Seven years before our study (July 1996) West and Scanlan (28) used a probe method to analyze the relative abundance of Prochlorococcus ecotypes at a site very close to our eastern North Atlantic site. In their study, ecotype-specific oligonucleotide probes, which targeted particular regions of the 16S rRNA gene, were hybridized to field samples of DNA and quantified in dot blots (28) or by fluorescence microscopy (29). In sharp contrast to our analysis of these waters (Fig. 3B), West and Scanlan found that eMIT9312 (desiganted HLII in their work) and eMIT9313 were not detectable (28) (Fig. 4A, right panel). The eMED4 (HLI) ecotype was the only ecotype detected in the upper 50 m, and the waters below this were dominated by the “low-light” ecotypes (which include eNATL2A and eMIT9313, although eMIT9313-specific probes showed no detectable counts).
FIG. 4.
QPCR and probe hybridization analyses of two field stations in the eastern North Atlantic: relative abundances of the six ecotypes of Prochlorococcus at each depth for profile 2 of the July 1996 PRIME cruise (A) and for the 19 September 2003 eastern North Atlantic profile (B). For QPCR data, the colors indicate different ecotypes, as described in the legend to Fig. 3. For probe hybridization data, the colors indicate relative hybridizations for the different probes, as follows: HLI (targets the eMED4 ecotype), green; HLII (targets the eMIT9312 ecotype), yellow; LL (targets both the eNATL2A and eMIT9313 ecotypes), blue; and MIT1023 (targets the eMIT9313 ecotype), red. For ease of comparison, ecotype abundances at each depth were normalized to the most abundant type at that depth. Probe hybridization data for the PRIME cruise are from the study of West and Scanlan (28).
We wondered whether these differences were indeed real or whether the two methods, targeting different DNA loci, differentially detect and quantify the Prochlorococcus ecotypes. The ITS and the 16S rRNA gene can evolve at dramatically different rates, and the ITS evolves much faster. Therefore, these two techniques could target different groups or subgroups of natural populations. To address this question, a direct comparison of the QPCR and probe hybridization methods was performed with samples from the two eastern North Atlantic profiles, the profile reported here (Fig. 3B) and the profile reported by West and Scanlan (28). The two methods use different normalization methods to obtain absolute quantification of the ecotypes (28; this study). Therefore, to perform a direct comparison, the relative abundances of the ecotypes were examined at each depth. For both depth profiles, there was close agreement between the two methods for quantification of the Prochlorococcus ecotypes (Fig. 4). In the July 1996 profile, the QPCR results closely matched the results of the probe hybridization method: partitioning of the water column by eMED4 and eNATL2A, with eMIT9312 and eMIT9313 making insignificant contributions to the total population (Fig. 4A). Likewise, for the September 2003 profile, the probe hybridization results closely matched the results of the QPCR method (Fig. 4B). The only difference of note is the larger relative contribution of eMIT9313 at 150 m measured by the probe hybridization method.
The fact that the two methods gave very similar results for both depth profiles indicates that the different molecular methods appear to target similar groups and the contrasting results of the two studies are indeed real. One significant difference between the two profiles that may account for the differences in population structure is water temperature. The surface mixed layers of the two profiles were similar depths (30 m), but the temperatures were different: 21°C in July 1996 versus 25°C in September 2003 (28; this study). The thermocline of the September 2003 profile was also significantly steeper than that of the July 1996 profile. It is possible that temperature, either directly or indirectly (by its influence on nutrient upwelling, for example), plays a significant role in establishing the distribution patterns of the ecotypes.
Identification of ecotypes not detected by the QPCR primers.
One of the advantages of working with Prochlorococcus is that the abundance of the total population can be measured using FCM. We take this as a robust measure of the total number of Prochlorococcus cells because simultaneous measurements of total Prochlorococcus numbers and divinyl chlorophyll a and b concentrations (pigments unique to Prochlorococcus) from field samples yield values that correspond to those measured in cultures (6, 18). Thus, by comparing the flow cytometry counts with the total number of cells targeted with the QPCR primers (the sum of all ecotype abundances), one can determine whether the QPCR method can account for the majority of the cells. Indeed, at the eastern North Atlantic site (Fig. 3B) these numbers agreed reasonably well throughout the depth profile. They also agreed well in the upper 75 m at western North Atlantic site (Fig. 3A), where the eMIT9312 ecotype is orders of magnitude more abundant than the other ecotypes. This result provides reasonable confidence that the eMIT9312 primers were able to catch the majority of the eMIT9312 members; if the QPCR counts represent only a fraction of the true counts for the ecotype, then flow cytometry also missed the vast majority of total Prochlorococcus cells, which, as described above, is highly unlikely.
In contrast to the upper 75 m there is a dramatic discrepancy between the flow cytometry and summed QPCR counts for the deeper water at the western North Atlantic site (Fig. 3A), which is consistent with the results of Ahlgren et al. (1). Thus, the QPCR primers currently in use are inadequate to target most of the Prochlorococcus cells at these depths at this time of year; i.e., they are biased toward the lineages found at the surface and the lineages that are readily cultured.
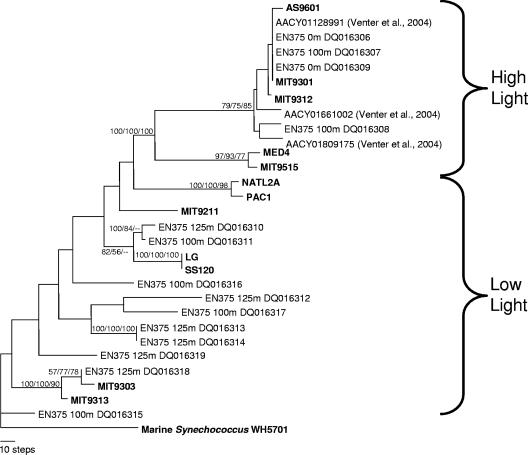
To begin exploration of the missing diversity at this site, a small, pilot clone library was constructed and analyzed. Using primers that target Prochlorococcus ITS (except for the eNATL2A ecotype [see Materials and Methods]), clone libraries were generated from surface and deep waters. In agreement with the QPCR and FCM data (Fig. 3A), clones from 25 m were dominated by the Prochlorococcus eMIT9312 ecotype (18 of 18 clones). Also in agreement with the QPCR and FCM data, at 100 m and 125 m, only 3 of 11 clones sequenced matched any of the known ecotypes; two at 100 m matched MIT 9312, while one at 125 m matched MIT 9313. The remaining eight clones appear to represent novel lineages of Prochlorococcus, and the sequences were not recognized by the six current primer sets. A phylogenetic analysis showed that these new sequences fall within the “low-light” cluster (Fig. 5) but are distinct from the four known “low-light” lineages. Except for two clones that appear to form an individual lineage within the SS120 ecotype, the relative positions of the individual low-light clades are not well resolved (this is due to the high level of divergence between ITS sequences from the new “low-light” clades and the low number of sequence entries available). Although this was only a pilot study, this analysis is entirely consistent with the relative values for total Prochlorococcus observed by FCM and QPCR; where there is a close match between total counts determined by QPCR and FCM results, the clone library is dominated by known ecotypes, and when there is not a close match, the clone libraries are dominated by unknown ecotypes. Future large-scale library analysis should be performed to thoroughly search for new ecotypes and facilitate generation of new primer sets that can quantify these new groups.
FIG. 5.
Maximum-parsimony tree of 16-23S ITS sequences from Prochlorococcus. Sequences generated in this study from PCR amplification of field samples, followed by cloning and sequencing, are indicated by EN375, followed by the depth from which the sample was collected (0, 100, or 125 m) and the GenBank accession number. Sequences from cultures are indicated by boldface type (22), and sequences from shotgun clones from the Bermuda Atlantic Time Series are indicated by “Venter et al., 2004” (27). The bootstrap values at the nodes are based on neighbor-joining, maximum-parsimony, and maximum-likelihood algorithms (in that order).
Conclusions.
While the field studies described here were intended primarily to validate the streamlined QPCR approach, we nevertheless drew some significant conclusions about the Prochlorococcus population structure in the North Atlantic, especially by comparing our results with those of West and Scanlan (28) for the eastern North Atlantic and those of Ahlgren et al. (1) for the western North Atlantic. We observed that the relative abundance of Prochlorococcus ecotypes varies with region and depth in the North Atlantic. Dominance can shift from one ecotype to another, although we did find that some ecotypes (eMIT9211 and eSS120) were always rare. The eMIT9312 ecotype dominated the upper euphotic zone in the summer in the western North Atlantic, whereas the upper water column was dominated by eMED4 in one of the eastern profiles and by both eMIT9312 and eMED4 in the other. Most likely, differences in environmental parameters, such as temperature and nutrients, play a role in establishing the different community structures observed in these three profiles. Geographical differences in ecotype genome composition (and emergent physiology) could also contribute and cannot be ruled out at this time.
The challenge now is to use the methods reported here to measure the dynamics of these populations over extensive temporal and spatial scales, with more exhaustive sampling, in order to better understand what regulates their relative abundance. Since we clearly cannot account for all of the Prochlorococcus ecotypes using existing QPCR primers and probes, we anticipate an iterative process; as more sequence information about the Prochlorococcus lineages becomes available, we can refine our primers and probes to account for more of the population as determined by flow cytometry. We are optimistic that ultimately, the sum of the ecotypes that we detect will closely approximate the flow cytometrically determined numbers in all samples.
Acknowledgments
We thank the captains and crews of the R/V Endeavor and the R.R.S. James Clark Ross, as well as B. Binder (chief scientist, Endeavor cruise). We also thank M. Polz, D. Lindell, M. Sullivan, R. Sarma-Rupavtarm, G. Rocap, and N. Ahlgren for valuable discussions.
In this study we used CTD/sea surface data from the Atlantic Meridional Transect Consortium (NER/0/5/2001/00680), provided by the British Oceanographic Data Centre and supported by the Natural Environment Research Council. This work was supported by NSF postdoctoral research fellowship DBI-0102109 to E.R.Z., by grants from NSF, DOE, and the Gordon and Betty Moore Foundation to S.W.C, and by NERC grant NER/T/S/2000/00621 to D.J.S.
Footnotes
AMT contribution no. 108.
REFERENCES
- 1.Ahlgren, N. A., G. Rocap, and S. W. Chisholm. Measurement of Prochlorococcus ecotypes using real-time PCR reveals different abundances of genotypes with similar light physiologies. Environ. Microbiol., in press. [DOI] [PubMed]
- 2.Becker, S., M. Fahrbach, P. Boger, and A. Ernst. 2002. Quantitative tracing, by Taq nuclease assays, of a Synechococcus ecotype in a highly diversified natural population. Appl. Environ. Microbiol. 68:4486-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinkman, N. E., R. A. Haugland, L. J. Wymer, M. Byappanahalli, R. L. Whitman, and S. J. Vesper. 2003. Evaluation of a rapid, quantitative real-time PCR method for enumeration of pathogenic Candida cells in water. Appl. Environ. Microbiol. 69:1775-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell, L., H. Liu, H. A. Nolla, and D. Vaulot. 1997. Annual variability of phytoplankton and bacteria in the subtropical North Pacific Ocean at Station ALOHA during the 1991-1994 ENSO event. Deep-Sea Res. Part I 44:167-192. [Google Scholar]
- 5.Cavender-Bares, K. K., S. L. Frankel, and S. W. Chisholm. 1998. A dual sheath flow cytometer for shipboard analyses of phytoplankton communities from the oligotrophic oceans. Limnol. Oceanogr. 43:1383-1388. [Google Scholar]
- 6.Chisholm, S. W., R. J. Olson, E. R. Zettler, R. Goericke, J. B. Waterbury, and N. A. Welschmeyer. 1988. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature 334:340-343. [Google Scholar]
- 7.DuRand, M. D., R. J. Olson, and S. W. Chisholm. 2001. Phytoplankton population dynamics at the Bermuda Atlantic Time-series Station in the Sargasso Sea. Deep-Sea Res. Part II 48:1983-2003. [Google Scholar]
- 8.Dusenberry, J. A., and S. L. Frankel. 1994. Increasing the sensitivity of a FACScan flow cytometer to study oceanic picoplankton. Limnol. Oceanogr. 39:206-209. [Google Scholar]
- 9.Ernst, A., S. Becker, U. I. A. Wollenzien, and C. Postius. 2003. Ecosystem-dependent adaptive radiations of picocyanobacteria inferred from 16S rRNA and ITS-1 sequence analysis. Microbiology 149:217-228. [DOI] [PubMed] [Google Scholar]
- 10.Ferris, M. J., and B. Palenik. 1998. Niche adaptation in ocean cyanobacteria. Nature 396:226-228. [Google Scholar]
- 11.Frankel, S. L., B. J. Binder, H. M. Shapiro, and S. W. Chisholm. 1990. A high-sensitivity flow cytometer for studying picoplankton. Limnol. Oceanogr. 35:1164-1169. [Google Scholar]
- 12.Fuller, N. J., N. J. West, D. Marie, M. Yallop, T. Rivlin, A. F. Post, and D. J. Scanlan. 2005. Dynamics of community structure and phosphate stature of picocyanobacterial populations in the Gulf of Aqaba, Red Sea. Limnol. Oceanogr. 50:363-375. [Google Scholar]
- 13.Harms, G., A. C. Layton, H. M. Dionisi, I. R. Gregory, V. M. Garrett, S. A. Hawkins, K. G. Robinson, and G. S. Sayler. 2003. Real-time PCR quantification of nitrifying bacteria in a municipal wastewater treatment plant. Environ. Sci. Technol. 37:343-351. [DOI] [PubMed] [Google Scholar]
- 14.Jones, M. D., and N. S. Foulkes. 1989. Reverse transcription of messenger-RNA by Thermus aquaticus DNA-polymerase. Nucleic Acids Res. 17:8387-8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karl, D. M. 1999. A sea of change: biogeochemical variability in the North Pacific Subtropical Gyre. Ecosystems 2:181-214. [Google Scholar]
- 16.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, G. A. W., O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore, L. R., and S. W. Chisholm. 1999. Photophysiology of the marine cyanobacterium Prochlorococcus: ecotypic differences among cultured isolates. Limnol. Oceanogr. 44:628-638. [Google Scholar]
- 18.Moore, L. R., R. Goericke, and S. W. Chisholm. 1995. Comparative physiology of Synechococcus and Prochlorococcus: influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Mar. Ecol. Prog. Ser. 116:259-275. [Google Scholar]
- 19.Moore, L. R., A. F. Post, G. Rocap, and S. W. Chisholm. 2002. Utilization of different nitrogen sources by the marine cyanobacteria, Prochlorococcus and Synechococcus. Limnol. Oceanogr. 47:989-996. [Google Scholar]
- 20.Moore, L. R., G. Rocap, and S. W. Chisholm. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464-467. [DOI] [PubMed] [Google Scholar]
- 21.Partensky, F., W. R. Hess, and D. Vaulot. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63:106-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rocap, G., D. L. Distel, J. B. Waterbury, and S. W. Chisholm. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 68:1180-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaffer, A. L., W. Wojnar, and W. Nelson. 1990. Amplification, detection, and automated sequencing of gibbon interleukin-2 messenger RNA by Thermus aquaticus DNA-polymerase reverse transcription and polymerase chain-reaction. Anal. Biochem. 190:292-296. [DOI] [PubMed] [Google Scholar]
- 24.Steinberg, D. K., C. A. Carlson, N. R. Bates, R. J. Johnson, A. F. Michaels, and A. H. Knap. 2001. Overview of the US JGOFS Bermuda Atlantic Time-series Study (BATS): a decade-scale look at ocean biology and biogeochemistry. Deep-Sea Res. Part II 48:1405-1447. [Google Scholar]
- 25.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urbach, E., and S. W. Chisholm. 1998. Genetic diversity in Prochlorococcus populations flow cytometrically sorted from the Sargasso Sea and Gulf Stream. Limnol. Oceanogr. 43:1615-1630. [Google Scholar]
- 27.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y.-H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 28.West, N. J., and D. J. Scanlan. 1999. Niche-partitioning of Prochlorococcus populations in a stratified water column in the eastern North Atlantic Ocean. Appl. Environ. Microbiol. 65:2585-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.West, N. J., W. A. Schonhuber, N. J. Fuller, R. I. Amann, R. Rippka, A. F. Post, and D. J. Scanlan. 2001. Closely related Prochlorococcus genotypes show remarkably different depth distributions in two oceanic regions as revealed by in situ hybridization using 16S rRNA-targeted oligonucleotides. Microbiology 147:1731-1744. [DOI] [PubMed] [Google Scholar]