Abstract
AvrPtoB is a type III effector protein from Pseudomonas syringae pv. tomato that physically interacts with the tomato Pto kinase and, depending on the host genotype, either elicits or suppresses programmed cell death associated with plant immunity. We reported previously that avrPtoB-related sequences are present in diverse gram-negative phytopathogenic bacteria. Here we describe characterization of avrPtoB homologs from P. syringae pv. tomato T1, PT23, and JL1065, P. syringae pv. syringae B728a, and P. syringae pv. maculicola ES4326. The avrPtoB homolog from P. syringae pv. maculicola, hopPmaL, was identified previously. The four new genes identified in this study are designated avrPtoBT1, avrPtoBPT23, avrPtoBJL1065, and avrPtoBB728a. The AvrPtoB homologs exhibit 52 to 66% amino acid identity with AvrPtoB. Transcripts of each of the avrPtoB homologs were detected in the Pseudomonas strains from which they were isolated. Proteins encoded by the homologs were detected in all strains except P. syringae pv. tomato T1, suggesting that T1 suppresses accumulation of AvrPtoBT1. All of the homologs interacted with the Pto kinase in a yeast two-hybrid system and elicited a Pto-dependent defense response when they were delivered into leaf cells by DC3000ΔavrPtoΔavrPtoB, a P. syringae pv. tomato strain with a deletion of both avrPto and avrPtoB. Like AvrPtoB, all of the homologs enhanced the ability of DC3000ΔavrPtoΔavrPtoB to form lesions on leaves of two susceptible tomato lines. With the exception of HopPmaL which lacks the C-terminal domain, all AvrPtoB homologs suppressed programmed cell death elicited by the AvrPto-Pto interaction in an Agrobacterium-mediated transient assay. Thus, despite their divergent sequences, AvrPtoB homologs from diverse P. syringae pathovars have conserved avirulence and virulence activities similar to AvrPtoB activity.
Pseudomonas syringae infects a broad range of plant species and causes significant economic losses worldwide. Over 50 pathovars of P. syringae have been identified based largely on their host ranges. Among these pathovars, P. syringae pv. tomato, the causal agent of bacterial speck disease in tomato and a pathogen of Arabidopsis thaliana, has been established as a model system to study plant-pathogen interactions because of its economic importance and tractability in the laboratory. Genetic analyses of the tomato-Pseudomonas pathosystem have revealed that resistance to P. syringae pv. tomato race 0 strains is specified by a pathogen avirulence gene, avrPto, and a single resistance gene in tomato, Pto (33). However, disruption of the avrPto gene in P. syringae pv. tomato strain DC3000 or JL1065 does not eliminate the ability of the mutant pathogens to elicit Pto-specific plant resistance (34), suggesting that there is a second avr gene that is recognized by Pto. Indeed, using a cross-kingdom yeast two-hybrid screening method, a second avr gene, avrPtoB, was discovered in DC3000 (23). Mutation of either avrPto or avrPtoB individually did not abolish the ability of DC3000 to elicit Pto-dependent resistance, which suggested that avrPto and avrPtoB have redundant avirulence activities (23, 34).
Although AvrPto and AvrPtoB (also known as HopAB2 [27]) exhibit limited amino acid sequence similarity, they both interact with Pto in a yeast two-hybrid system and elicit Pto-mediated cell death in tomato when they are expressed via an Agrobacterium tumefaciens transient assay (33). An avrPto/avrPtoB deletion mutant of DC3000 causes disease on Pto-expressing tomato leaves, indicating that avrPto and avrPtoB have redundant avirulence activities and are the only two avirulence genes in DC3000 that are recognized by Pto. Moreover, deletion of either avrPto or avrPtoB from DC3000 results in a subtle decrease in bacterial virulence, whereas deletion of both avrPto and avrPtoB significantly reduces the disease-forming ability of this strain (26). These data demonstrate that these two genes contribute additively to the virulence of DC3000 by promoting lesion formation in susceptible tomato plants.
AvrPto and AvrPtoB have both distinct and common virulence functions that block plant defense responses and target pathways involved in symptom development. For example, it has been reported that AvrPto can act as a suppressor of cell wall-based plant defense in A. thaliana (15) and that AvrPtoB enhances host susceptibility by inhibition of programmed cell death (PCD) (1, 2, 20). Recently, by using cDNA microarray analysis, a set of tomato genes involved in ethylene biosynthesis (LeACO1 and LeACO2, encoding the ethylene-forming enzyme 1-aminocyclopropane-1-carboxylate oxidase) were found to be up-regulated in response to either AvrPto or AvrPtoB (7). In addition, tomato lines that are deficient in ethylene formation or perception showed markedly decreased symptoms of bacterial speck disease. Thus, AvrPto and AvrPtoB appear to promote disease symptoms, at least in part, by up-regulating the production of ethylene in tomato leaves (7).
avrPtoB-related genes are present in diverse phytopathogenic gram-negative bacteria (19, 23). AvrPtoB exhibits 52% amino acid sequence identity with VirPphA, the first virulence effector cloned from P. syringae pv. phaseolicola. Ectopic expression of avrPtoB, virPphA, or the homologs virPphAPgy and virPphAPsv restored the water-soaking ability of a plasmid-cured, nonpathogenic strain of P. syringae pv. phaseolicola (RW60) on bean pods, demonstrating that these genes contribute to pathogen virulence (19, 23).
Here we addressed the question of whether homologs of AvrPtoB present in diverse Pseudomonas pathovars have biological activity similar to that of AvrPtoB. We characterized five avrPtoB homologs cloned from three different P. syringae pathovars, P. syringae pv. tomato, P. syringae pv. syringae, and P. syringae pv. maculicola, which cause bacterial speck on tomato, brown spot on snap bean (Phaseolus vulgaris L.), and bacterial leaf spot on Brassica spp., respectively. We then examined the effects of ectopic expression of each of the avrPtoB homologs in DC3000ΔavrPtoΔavrPtoB (referred to below as ΔavrPtoΔavrPtoB) in susceptible tomato lines, as well as their ability to elicit plant resistance in tomato plants expressing Pto.
MATERIALS AND METHODS
Bacterial strains, plasmids, and DNA manipulation.
The bacterial strains and plasmids used in this study are listed in Table 1. All P. syringae strains were grown in King's B (KB) medium (24) at 30°C unless indicated otherwise. Escherichia coli DH5α and TOP10 (Invitrogen, Carlsbad, CA) were used for plasmid maintenance and were grown in LB medium (13) at 37°C. A. tumefaciens GV2260 (30) was grown in LB medium at 30°C. The concentrations of antibiotics used in the selective media were as follows: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; rifampin, 100 μg/ml; spectinomycin, 50 μg/ml; streptomycin, 50 μg/ml; and tetracycline, 10 μg/ml.
TABLE 1.
Bacterial strains and plasmids used for DNA manipulation
| Designation | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | Invitrogen | |
| Escherichia coli TOP10 | Invitrogen | |
| Pseudomonas syringae pv. tomato DC3000ΔavrPtoΔavrPtoB | ΔavrPto::ΩSp/Smr, ΔavrPtoB::nptII, Rifr Spr Kanr | 26 |
| DC3000ΔavrPtoΔavrPtoB(avrPtoB) | DC3000ΔavrPtoΔavrPtoB containing broad-host-vector pCPP45 expressing avrPtoB under native hrp promoter, Rifr Spr Kanr Tetr | 26 |
| T1(avrPto) | Broad-host-range vector pDSK519 carrying avrPto under native hrp promoter | 39 |
| Agrobacterium tumefaciens GV2260 | Rifr | 30 |
| pBTEX::Pto | pBTEX expressing Pto under the control of CaMV 35S promoter, Rifr Kanr | 39 |
| pBTEX::avrPto | pBTEX expressing avrPto under the control of CaMV 35S promoter, Rifr Kanr | 39 |
| pBTEX::avrPtoB | pBTEX expressing avrPtoB under the control of CaMV 35S promoter, Rifr Kanr | 23 |
| Plasmids | ||
| pBluescript II SK(−) | Stratagene | |
| pCR2.1 | TA cloning vector, Ampr | Invitrogen |
| pCPP45 | Broad-host-range vector with RP4 par region, Tcr | D. W. Bauer (Cornell University) |
| pBTEX | Derivative of binary vector pBI121 containing polylinker site between cauliflower mosaic virus 35S promoter and Nos terminator, Kanr | R. A. Bressan (Purdue University) |
| pCPP2318 | Broad-host-range vector carrying β-lactamase gene, Tcr | 6a |
Amp, ampicillin; Kan, kanamycin; Tc, tetracycline; Sp, spectinomycin; Sm, streptomycin; Rif, rifampin.
DNA manipulation and PCR were performed using standard protocols (36). Primers used in this study are listed in Table 2. DNA and amino acid sequence analyses were performed using the DNASTAR software package (Madison, WI), and ClustalW was used for multiple-sequence alignment for AvrPtoB and AvrPtoB homologs (40). A neighbor-joining tree of AvrPtoB with nine other members of the AvrPtoB/VirPphA family was constructed using MEGA, version 2.1 (25).
TABLE 2.
Primers used to amplify avrPtoB homologs in different assays
| Primer | Sequence (5′→3′) | Nucleotide positiona |
|---|---|---|
| avrPto2-30 | ATGGCGGGTATCAA(C/T)(A/G)GAGC(A/G)G | 1-22 |
| avrPto2-3 | GACCTTCGAAGTGGCAGTGA | |
| avrPtoB-hrp | GGTGTGATGGAACTCTTTCCTGCTCb | |
| avrPtoBseq-1 | TATCGTTCAGCAATTGGTCAGTG | 393-415 |
| avrPtoBseq-2 | CCCCGGGTTCAGGTTAA | 1230-1224 |
| avrPtoBH-F | ATGGCGGGTATCAATAGAGCGG | 1-22 |
| avrPtoBH-R | TCAGGGGACTATTCTAAAAGC | 1740-1720 |
| avrPtoBHRT-F | GCCACGCGATAGCTCTTCCTTCTC | 509-532 |
| avrPtoBHRT-R | AACAACCGCCTGCCGCTCGTAAC | 1377-1355 |
| B728a-hrp | ACGCATCAGTGGAACCTTTb | |
| B728a-F | ATGTCGGGTATCAACGGAGCAG | 1-22 |
| B728a-R | TCAGTCAGGAACGACTCTAAAG | 1551-1530 |
| B728aseq-1 | ATGCCTCAGAAATGTAATGGATAA | 452-464 |
| B728aseq-2 | CTCCGCTTCAGGGTTCAG | 1214-1241 |
| hopPmaL-hrp | GATTTGCCGGGAGAAGTAAGGc | |
| hopPmaL-F | ATGGTAGGGATCAGTGGTAGAGCAG | 1-25 |
| hopPmaL-R | TCAAAATGCCTATGCATCAAGCC | 1158-1145 |
| hopPmaL-RT-R | GGCGAACCGGCCTAATAAT | 1029-1011 |
| 16SrRNA-F | GCGGCAGGCCTAACACAT | |
| 16SrRNA-R | GTTCCCCTACGGCTACCTT | |
| GWApBF | GGGGACAAGTTTGTACAAAAAAGCAGGCTTTGAGAATCTTTATTTTCAGGGCATGGCGGGTATCAATAGAGC | |
| GWApBR | GGGGACCACTTTGTACAAGAAAGCTGGGTTTCAGGGGACTATTCTAAAAGC |
Nucleotide position of avrPtoB or avrPtoB homolog.
The hrp box is underlined.
Upstream of hrp box.
Cloning of avrPtoB homologs.
Primers avrPto2-30 and avrPto2-3 generated from nucleotide sequences of avrPtoB and virPphA were used to PCR amplify the open reading frames of avrPtoB homologs in P. syringae pv. tomato T1, PT23, and JL1065 using Turbo Pfu polymerase (Stratagene, La Jolla, CA). Amplified fragments were cloned into pCR2.1 for sequencing. Two or three clones of each homolog were sequenced from independent PCRs to verify there were no PCR-induced errors. The sequences obtained were then used to design internal primers for sequencing the upstream and downstream regions of each avrPtoB homolog using genomic DNA as the template. The sequence of the avrPtoB homolog in P. syringae pv. syringae B728a was retrieved from a draft sequence of the P. syringae pv. syringae B728a genome available at www.jgi.doe.gov. The hopPmaL sequence was retrieved from the NCBI GenBank database (accession no. AF458050). Primer sets were designed using sequences retrieved from the GenBank database to amplify the two homologs from the genomic DNA of P. syringae pv. syringae B728a and P. syringae pv. maculicola ES4326.
Preparation of bacterial cells and supernatant for RNA and protein assays.
Bacteria grown overnight in 25 ml of KB medium at 30°C were washed twice with hrp-inducing minimal medium (HrpMM) (17) and resuspended in HrpMM. Each bacterial suspension was diluted to an optical density at 600 nm (OD600) of 0.15 to 0.2 in 100 ml of HrpMM and grown at 18°C until the OD600 was 0.4 to 0.5. Separation of bacterial cultures into cell and supernatant fractions was performed using the protocol described previously (12). Cell pellets from each bacterial strain were separated into two parts. One part was concentrated 100-fold and 10 μl was used for protein assays, and the other part was precipitated and resuspended in 200 μl RNAProtect bacterial reagent (QIAGEN, Valencia, CA) for RNA isolation. The supernatant proteins were precipitated with 20% trichloroacetic acid, dissolved in 20 μl 1× sodium dodecyl sulfate (SDS) sample buffer, and boiled at 100°C.
Reverse transcriptase PCR (RT-PCR).
Total bacterial RNA was isolated using an RNeasy mini kit and an RNase-free DNase set (QIAGEN, Valencia, CA). One microgram of total RNA from each sample was used for first-strand cDNA synthesis with random primers and SuperScript II RNase H− reverse transcriptase (Invitrogen, Carlsbad, CA). PCR was performed using the following gene-specific primers listed in Table 2: for avrPtoB, avrPtoBseq-1 and avrPtoBseq-2; for avrPtoBT1, avrPtoBPT23, and avrPtoBJL1065, avrPtoBHRT-F and avrPtoBHRT-R; for avrPtoBB728a, B728aseq-1 and B728aseq-2; and for hopPmaL, hopPmaL-F and hopPmaL-RT-R. Primers 16SrRNA-F and 16SrRNA-R were used for amplification of 16S rRNA as an internal control.
Generation of AvrPtoB antibody.
Recombinant AvrPtoB was expressed as an N-terminal glutathione S-transferase fusion with a tobacco etch virus (TEV) protease cleavage site directly upstream of the AvrPtoB start codon. The expression construct was generated by Gateway cloning the avrPtoB gene into the pDEST15 expression vector (Invitrogen, Carlsbad, CA) by following the manufacturer's protocol. Gateway-compatible avrPtoB was generated by PCR using the GWApBF and GWApBR primers. pDEST15::AvrPtoB was expressed in Escherichia coli BL21 Star(DE3) (Invitrogen, Carlsbad, CA) and was purified using standard protein expression methods (14). Briefly, glutathione S-transferase-AvrPtoB was purified on a column containing glutathione Sepharose 4 fast flow resin (Amersham-Pharmacia, Piscataway, NJ). The protein was cleaved from the column using 100 U of TEV protease (Invitrogen, Carlsbad, CA) and incubating the sample overnight at 4°C with gentle shaking. The cleaved protein was boiled in SDS sample buffer, separated by SDS-polyacrylamide gel electrophoresis, and stained with Coomassie blue, and AvrPtoB was excised from the gel. Polyclonal AvrPtoB antibody was generated in rabbits by Covance Research Products Inc. (Denver, PA) using acrylamide gel slices containing 2 mg of denatured AvrPtoB protein. Polyclonal AvrPtoB antiserum was affinity purified from the crude antiserum using recombinant AvrPtoB electrophoretically transferred onto polyvinylidene difluoride membranes (31).
Immunoblot assay.
Protein samples from cell and supernatant fractions were electrophoresed on 8% denaturing SDS-polyacrylamide gels and electrotransferred to polyvinylidene difluoride membranes (Immobilon R; Millipore, Bedford, MA) according to the manufacturer's protocol (Bio-Rad, Hercules, CA). AvrPtoB and AvrPtoB homolog proteins were detected with ECL Plus Western blotting detection reagents (Amersham-Pharmacia, Piscataway, NJ) using purified anti-AvrPtoB primary antibodies and horseradish peroxidase-conjugated anti-rabbit immunoglobulin G secondary antibody (Promega Co., Madison, WI). Broad-host-range vector pCPP2318, which carries a constitutively expressed β-lactamase gene, was introduced into each Pseudomonas strain tested, and β-lactamase was used as the cytoplasmic marker. AvrPto and β-lactamase were detected using anti-AvrPto and anti-β-lactamase antibodies, respectively. Hemagglutinin (HA)-tagged AvrPtoB was detected using anti-HA primary antibodies (Roche Applied Science, Indianapolis, IN) and peroxidase-conjugated anti-rat immunoglobulin G antibody (Roche Applied Science, Indianapolis, IN).
Yeast two-hybrid interactions.
A LexA yeast two-hybrid system was used to test the interaction between Pto and AvrPtoB homologs. Bait construct pEG202::Pto and prey constructs pJG4-5::AvrPto and pJG4-5::AvrPtoB were generated in previous studies (23, 39). avrPtoB homologs were PCR amplified using the following primer sets (Table 2): for avrPtoBT1 and avrPtoBPT23, avrPtoBH-F and avrPtoBH-R; for avrPtoBB728a, B728a-F and B728a-R; and for hopPmaL, hopPmaL-F and hopPmaL-R. PCR products were cloned into pCR2.1 for sequencing, and the EcoRI fragments were subcloned into an EcoRI-digested pJG4-5 vector. The pJG4-5 constructs were introduced into a yeast strain containing pEG202::Pto and a reporter vector (10). Yeast cells containing both prey and bait proteins were streaked on medium containing galactose and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Photographs were taken after 2 days of incubation of the plates at 30°C.
Construction of ΔavrPtoΔavrPtoB expressing avrPtoB homologs.
DNA fragments containing the native hrp box and the open reading frames of individual avrPtoB homologs were PCR amplified using the primers listed in Table 2 and were cloned into pCR2.1 for sequencing. Primers avrPtoB-hrp and avrPtoBH-R were used to amplify avrPtoBT1 and avrPtoBPT23. avrPtoBB728a was amplified using primers B728a-hrp and B728a-R, and hopPmaL was amplified using primers hopPmaL-hrp and hopPmaL-R. The EcoRI fragments from pCR2.1 containing individual avrPtoB homologs were subcloned into the broad-host-range vector pCPP45 to create pAvrPtoBT1, pavrPtoBPT23, pavrPtoBB728a, and phopPmaL, and this was followed by introduction into ΔavrPtoΔavrPtoB using triparental mating to obtain ΔavrPtoΔavrPtoB expressing avrPtoB homologs under the control of an hrp promoter. ΔavrPtoΔavrPtoB strains containing an empty vector [referred to below as ΔavrPtoΔavrPtoB(ev)] and avrPtoB [referred to below as ΔavrPtoΔavrPtoB (avrPtoB)] were constructed in a previous study (26).
Plant assays.
Tomato (Solanum lycopersicon) Rio-Grande (RG) lines were grown as previously described (23) and included four genotypes: RG-PtoR (Pto/Pto, Prf/Prf), RG-pto11 (pto11/pto11, Prf/Prf), RG-prf3 (Pto/Pto, prf3/prf3), and RG-PtoS, which is nearly isogenic with RG-PtoR (35). For pathogenicity assays, bacterial suspensions containing 1 × 104 CFU ml−1 in 10 mM MgCl2 and 0.002% Silwet L-77 (Cromptom Co., Middlebury, CT) were vacuum infiltrated into leaves of 4-week-old plants. Disease development and bacterial populations were monitored every other day after inoculation. Bacteria recovered from plants were plated on KB medium containing rifampin. Numbers of specks were determined for nine independent 1-cm2 areas. Fully expanded leaflets were used for recovery of bacteria and counting of lesions.
Construction of strains for Agrobacterium-mediated transient assay.
KpnI/XbaI fragments of the pCR2.1 constructs, which were generated previously for yeast two-hybrid interactions, were subcloned into a KpnI/XbaI-digested pBTEX binary vector. The pBTEX constructs were electroporated into A. tumefaciens GV2260, and the strains were used to syringe infiltrate leaves of 4-week-old tomato plants and 6-week-old Nicotiana benthamiana plants at a final OD600 of 0.1. A. tumefaciens strains carrying transgenes of avrPto, avrPtoB, or Pto were constructed in previous studies (1, 39). Agrobacterium-mediated transient assays were performed as described previously (37).
Ion leakage assays.
Leaf disks excised from the areas infiltrated with Agrobacterium were incubated in water at room temperature for 3 h. Each solution was used to measure sample conductivity with an Acorn series CON5 m (OAKTON Instruments, Vernon Hills, IL). Leaf disks taken from areas where there was no infiltration with Agrobacterium were used as blank controls to normalize sample conductivity. Ion leakage was expressed as the difference in conductivity between the blank control and samples.
RESULTS
Sequence comparisons of avrPtoB homologs.
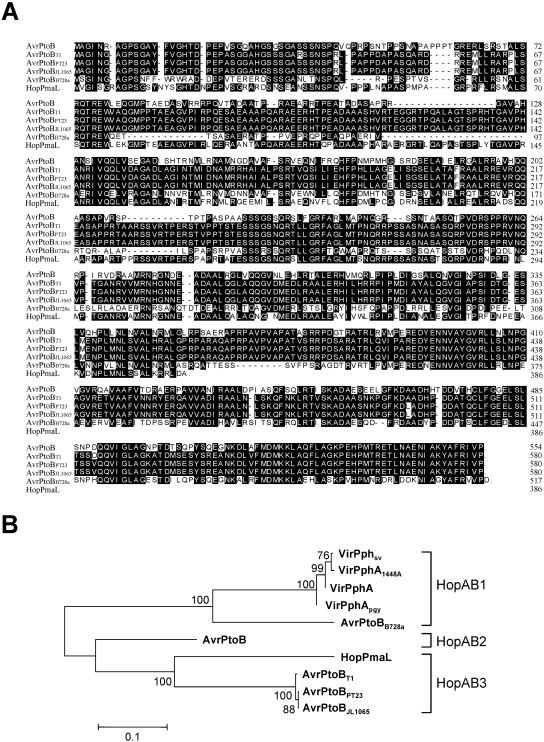
Southern blot analysis revealed that many Pseudomonas spp. strains contain avrPtoB sequences (20, 24; data not shown). Among these, P. syringae pv. tomato strains T1, PT23, and JL1065, P. syringae pv. syringae B728a and P. syringae pv. maculicola ES4326 were chosen for cloning and characterization of their avrPtoB homologs. Previously, an AvrPtoB domain involved in recognition by Pto was mapped to amino acids 1 to 308 of AvrPtoB, and an anti-PCD domain was identified in the C terminus (1). Alignments obtained with ClustalW (40) revealed that these two domains are for the most part conserved among AvrPtoB and its homologs, although many amino acid substitutions and numerous insertions and deletions distinguish them, particularly in the region from amino acid 128 to amino acid 202 (Fig. 1A).
FIG. 1.
Sequence comparisons for AvrPtoB and AvrPtoB homologs. (A) Alignment of the amino acid sequences of AvrPtoB and five AvrPtoB homologs. Alignment was performed using ClustalW (40). Conserved residues are highlighted. The domain required for Pto interaction in a yeast two-hybrid system encompasses AvrPtoB residues 1 to 308. The domain required for the anti-PCD function consists of AvrPtoB residues 309 to 553 (1). The GenBank accession numbers are DQ133533 (avrPtoBT1), DQ133534 (avrPtoBPT23), and DQ133535 (avrPtoBJL1065). (B) Neighbor-joining tree for AvrPtoB and eight other members of the AvrPtoB/VirPphA (HopAB) family, which was constructed using MEGA, version 2.1 (25). The horizontal branch lengths are proportional to the estimated numbers of amino acid substitutions, and bootstrap scores (expressed as percentages) are indicated at the nodes.
As reported previously, AvrPtoB exhibits 52% sequence similarity with VirPphA. Homologs of VirPphA were identified in P. syringae pv. phaseolicola 1448A, P. syringae pv. glycinea (VirPphAPgy; accession no. AJ439728), and P. syringae pv. savastanoi (VirPphAPsv; accession no. AJ439729) (19). We performed phylogenetic and molecular evolutionary analyses of the AvrPtoB/VirPphA family (also known as the HopAB family) using MEGA, version 2.1 (25). These analyses showed that AvrPtoB homologs from P. syringae pv. tomato strains T1, PT23, and JL1065 are closely related to HopPmaL from P. syringae pv. maculicola ES4326 (Fig. 1B). The AvrPtoB homolog from P. syringae pv. syringae B728a is more similar to VirPphA from P. syringae pv. phaseolicola than to AvrPtoB. Based on a recently introduced nomenclature (27), the homologs that we cloned were derived from the HopAB1 and HopAB3 families (although note that HopPmaL is a truncated version of the other members of the HopAB3 family [Fig. 1A]). At present, AvrPtoB is the only member of the HopAB2 family. In order to highlight the many functional similarities of the new homologs to AvrPtoB (see below), we refer to them below as avrPtoBT1, avrPtoBPT23, avrPtoBJL1065, and avrPtoBB728a.
avrPtoB is 77% identical at the nucleotide level (66% identical at the amino acid level) to avrPtoBT1, avrPtoBPT23, and avrPtoBJL1065. The nucleotide sequences of avrPtoBPT23 and avrPtoBJL1065 are identical, and each exhibits 99.7% identity to avrPtoBT1. Because AvrPtoBPT23 and AvrPtoBJL1065 are identical, only AvrPtoBPT23 was characterized further. All three homologs encode 579-amino-acid predicted proteins with molecular masses of 61.9 kDa. AvrPtoBT1 differs from AvrPtoBPT23 and AvrPtoBJL1065 by only three amino acids, located at residues 35, 493, and 515. avrPtoBB728a exhibits 69% nucleotide identity (52% amino acid identity) with avrPtoB and encodes a 516-amino-acid predicted protein with a molecular mass of 57.6 kDa. hopPmaL encodes a shorter protein (385 amino acids) with a molecular mass of 42 kDa, and the HopPmaL protein is therefore similar to the C-terminal truncated form of AvrPtoB, AvrPtoBΔ6 (1, 11). In the region that overlaps with avrPtoB, hopPmaL exhibits 66.5% nucleotide identity (58% amino acid identity).
Expression of endogenous avrPtoB homologs in P. syringae strains.
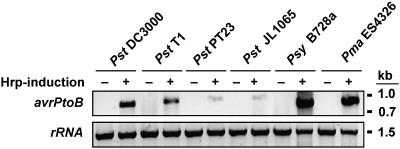
Based on previous observations, P. syringae pv. tomato strains containing an avrPtoB homolog would be expected to be avirulent on tomato plants expressing Pto. However, only P. syringae pv. tomato PT23 and P. syringae pv. tomato JL1065 are avirulent on Pto-expressing tomatoes, whereas P. syringae pv. tomato T1 and P. syringae pv. maculicola ES4326 are virulent (data not shown for P. syringae pv. maculicola ES4326). RT-PCR using gene-specific primers (Table 2) was performed to test whether the differences are due to lack of expression of the avrPtoB-related genes in T1 and ES4326. Abundant hrp-dependent accumulation of endogenous avrPtoB, avrPtoBT1, avrPtoBB728a, and hopPmaL transcripts was observed, whereas endogenous avrPtoBPT23 and avrPtoBJL1065 transcripts were present at much lower levels (Fig. 2). The lower transcript abundance in P. syringae pv. tomato PT23 and P. syringae pv. tomato JL1065 was not due to primer problems because the primer sets used for avrPtoBT1, avrPtoBPT23, and avrPtoBJL1065 amplified these genes with similar efficiencies when genomic DNA was used as a template (data not shown). Therefore, the faint bands detected for P. syringae pv. tomato PT23 and P. syringae pv. tomato JL1065 likely indicate that their avrPtoB homologs are not expressed at high levels when these two strains are grown in hrp-inducing minimal medium (HrpMM).
FIG. 2.
RT-PCR analysis of avrPtoB homolog transcripts. Total RNA was isolated from each Pseudomonas strain after growth without (lanes −) or with (lanes +) Hrp induction. RT-PCR was performed as described in Materials and Methods using gene-specific primers; primers for 16S rRNA were used as an internal control. The PCR products were separated on an agarose gel and stained with ethidium bromide. Transcripts for each of the AvrPtoB homologs could be detected in the strain from which the corresponding gene was isolated.
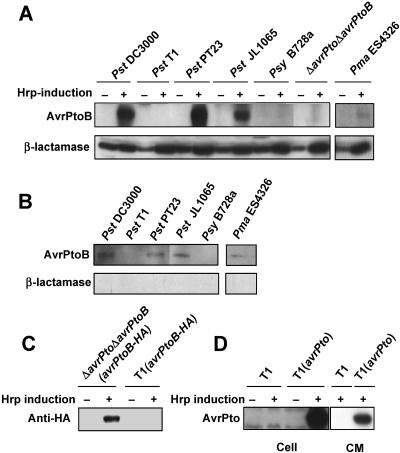
All AvrPtoB proteins tested except AvrPtoBT1 are produced and secreted in an hrp-dependent manner.
The accumulation of a protein corresponding to AvrPtoB and its homologs was examined using anti-AvrPtoB antibodies. Production of endogenous AvrPtoB, AvrPtoBPT23, AvrPtoBJL1065, and HopPmaL proteins was detected in both bacterial cells and culture medium after growth in HrpMM, indicating that these four proteins accumulate and are secreted in an hrp-dependent manner apparently not correlated with transcript levels (Fig. 3A and B).
FIG. 3.
Analysis of protein accumulation and secretion of AvrPtoB and AvrPtoB homologs. (A) AvrPtoB and AvrPtoB homolog proteins were detected in cells of P. syringae strains by Western blotting using anti-AvrPtoB antibodies (see Materials and Methods). β-Lactamase was included as a loading control. Strains were grown without (lanes −) or with (lanes +) Hrp induction. AvrPtoB or AvrPtoB homolog proteins were detected in P. syringae pv. tomato DC3000, P. syringae pv. tomato PT23, P. syringae pv. tomato JL1065, and P. syringae pv. maculicola ES4326. (B) Secretion of AvrPtoB and AvrPtoB homologs into the culture medium in an Hrp-dependent manner was analyzed by Western blotting using anti-AvrPtoB antibodies. Secretion of AvrPtoB proteins was observed for all strains except T1 and B728a (see Results). Strains were grown in Hrp-inducing conditions. β-Lactamase antibodies were used as a control, and, as expected, no β-lactamase was detected in the culture medium. (C) Analysis of AvrPtoB-HA protein accumulation in strains DC3000ΔavrPtoΔavrPtoB and T1 by Western blotting using anti-HA antibodies. An AvrPtoB-HA-expressing plasmid was transformed into the two strains, and the strains were grown without (lanes −) or with (lanes +) Hrp induction. AvrPtoB was observed to accumulate in DC3000ΔavrPtoΔavrPtoB but not in T1. (D) Analysis of accumulation and secretion of AvrPto in strain T1. T1 and a derivative expressing AvrPto from a plasmid were grown without (lanes −) or with (lanes +) Hrp induction. Extracts were prepared from cells or from the culture medium (CM). Western blotting with anti-AvrPto antibodies detected AvrPto in both total cells and the culture medium, indicating that, as expected, this protein is expressed by and secreted from strain T1.
Surprisingly, no AvrPtoB-related proteins were detected in P. syringae pv. tomato T1 or P. syringae pv. syringae B728a. Although AvrPtoB and AvrPtoBB728a exhibit 52% amino acid sequence identity to each other, regions of similarity are dispersed throughout the proteins. Therefore, it is possible that our anti-AvrPtoB antibodies did not cross-react with AvrPtoBB728a. Indeed, AvrPtoBB728a protein could be detected by using anti-HA antibody but not by using anti-AvrPtoB antibody when an HA-tagged version of AvrPtoBB728a was expressed in P. syringae pv. syringae B728a (data not shown). However, AvrPtoBT1 differs by only three amino acids from AvrPtoBPT23 and AvrPtoBJL1065, and we detected both of the latter proteins using our anti-AvrPtoB antibodies. Thus, it is unlikely that the anti-AvrPtoB antibodies fail to cross-react with AvrPtoBT1. To examine this observation further, a broad-host-range vector expressing an HA-tagged version of AvrPtoB was introduced into ΔavrPtoΔavrPtoB and P. syringae pv. tomato T1, and AvrPtoB protein expression was examined using anti-HA antibodies. hrp-dependent expression of the AvrPtoB::HA protein was detected in ΔavrPtoΔavrPtoB(avrPtoB) but not in P. syringae pv. tomato T1 expressing avrPtoB::HA (Fig. 3C). To test whether the type III secretion system (TTSS) is functional in P. syringae pv. tomato T1, P. syringae pv. tomato T1 carrying avrPto under control of its native promoter [referred to below as P. syringae pv. tomato T1(avrPto)] was used to perform the same translation/secretion assay used with the AvrPtoB proteins. In P. syringae pv. tomato T1(avrPto), AvrPto was detected in both bacterial cells and culture medium after induction of bacterial gene expression in HrpMM, indicating that the TTSS is functional in P. syringae pv. tomato T1 (Fig. 3D). Thus, we concluded that strain T1 appears to selectively interfere with accumulation of both AvrPtoBT1 and AvrPtoB by an unknown mechanism.
AvrPtoB homologs interact with Pto in a yeast two-hybrid system and confer avirulence activity to ΔavrPtoΔavrPtoB in tomato leaves expressing Pto.
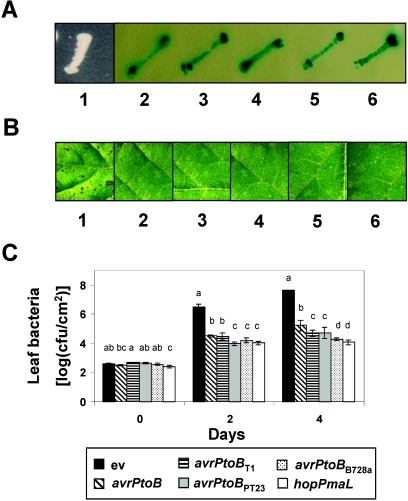
AvrPtoB was originally isolated as a Pto-interacting protein from a yeast two-hybrid screening analysis. We found that like AvrPtoB, all of the AvrPtoB homologs interact with Pto in a yeast two-hybrid system (Fig. 4A). To examine the ability of each AvrPtoB protein to elicit Pto-mediated resistance, we relied on the avrPto/avrPtoB deletion mutant of P. syringae pv. tomato DC3000 (Table 1) (29). This mutant causes disease in tomato lines expressing Pto and has a reduced ability to produce specks on susceptible tomato plants (26). Introduction of AvrPtoB on a broad-host-range vector into the avrPto/avrPtoB mutant restored its ability to elicit Pto-mediated resistance and to produce specks similar to those produced by wild-type P. syringae pv. tomato DC3000. This mutant therefore was a useful tool to study the avirulence and virulence activities of avrPtoB homologs from different Pseudomonas strains.
FIG. 4.
AvrPtoB homologs interact with Pto and elicit Pto-dependent resistance. (A) Yeast two-hybrid interactions of Pto with AvrPto or the AvrPtoB homologs. Image 1, empty vector; image 2, AvrPtoB; image 3, AvrPtoBT1; image 4, AvrPtoBPT23; image 5, AvrPtoBB728a; image 6, HopPmaL. (B) Elicitation of Pto-dependent resistance by infiltration of RG-PtoR leaves with 104 CFU ml−1 ΔavrPtoΔavrPtoB expressing the following genes: none (empty vector) (leaf 1), avrPtoB (leaf 2), avrPtoBT1 (leaf 3), avrPtoBPT23 (leaf 4), avrPtoBB728a (leaf 5), and hopPmaL (leaf 6). Photographs were taken 6 days after inoculation. Disease developed 3 days after inoculation of RG-PtoR leaves with only ΔavrPtoΔavrPtoB(ev) (leaf 1). (C) Bacterial populations in RG-PtoR tomato leaves at 0, 2, and 4 days after inoculation with DC3000ΔavrPtoΔavrPtoB expressing the genes indicated. Leaves were vacuum infiltrated with 104 CFU ml−1 of bacteria. The error bars indicate the standard deviations for three replicates. Data analysis was performed using Duncan's multiple-range test. Means with the same letter above the bars are not different at a significance level of 5%. The experiments were performed twice, and similar results were obtained.
To determine whether the interaction with Pto in the yeast two-hybrid system correlated with the avirulence activity of avrPtoB homologs in Pseudomonas spp., each gene was introduced on a broad-host-range plasmid into ΔavrPtoΔavrPtoB and tested for its ability to elicit resistance in tomato lines expressing Pto. Pto-dependent resistance was observed on leaves that were vacuum infiltrated with ΔavrPtoΔavrPtoB expressing avrPtoB, avrPtoBT1, avrPtoBPT23, avrPtoBB728a, or hopPmaL, whereas disease developed on tomato leaves inoculated with control strain ΔavrPtoΔavrPtoB(ev) (Fig. 4B). The sizes of the bacterial populations were restricted to 104 to 105 CFU cm−2 in leaves infiltrated with ΔavrPtoΔavrPtoB expressing avrPtoB and each of the avrPtoB homologs (Fig. 4C). In contrast, bacteria grew to a level of 5 × 107 CFU cm−2 in leaves infiltrated with the control ΔavrPtoΔavrPtoB(ev) strain. Therefore, like avrPtoB, all of the avrPtoB homologs confer avirulence activity to ΔavrPtoΔavrPtoB.
avrPtoB homologs enhance lesion-forming ability and growth of ΔavrPtoΔavrPtoB in susceptible tomato leaves.
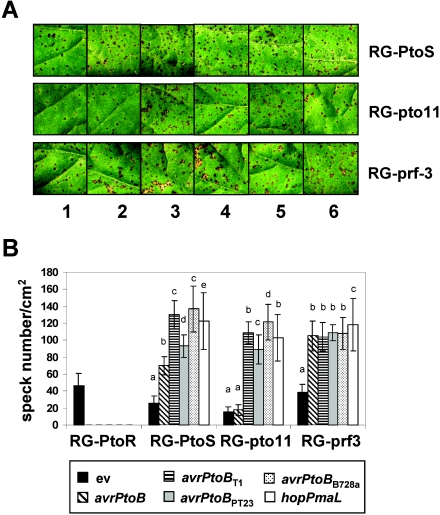
Susceptible tomato lines RG-PtoS, RG-pto11, and RG-prf3 (see Materials and Methods) were used for pathogenicity assays. Although some variability was associated with certain tomato lines and avrPtoB homologs, compared to leaves infiltrated with ΔavrPtoΔavrPtoB(ev), more specks were observed on tomato leaves inoculated with ΔavrPtoΔavrPtoB expressing avrPtoB or one of the homologs (Fig. 5). The only exception was the finding that avrPtoB did not restore the lesion-forming ability of ΔavrPtoΔavrPtoB when it was inoculated onto leaves of tomato line RG-pto11. These results indicate that each of the avrPtoB homologs promotes bacterial virulence, in part, by enhancing the development of disease symptoms.
FIG. 5.
Lesion-forming ability on susceptible tomato leaves of DC3000ΔavrPtoΔavrPtoB strains expressing AvrPtoB or the AvrPtoB homologs. (A) Host defense response or disease symptoms in RG-PtoS, RG-pto11, and RG-prf3 plants 6 days after inoculation with DC3000ΔavrPtoΔavrPtoB strains expressing the following genes: none (empty vector) (leaf 1), avrPtoB (leaf 2), avrPtoBT1 (leaf 3), avrPtoBPT23 (leaf 4), avrPtoBB728a (leaf 5), and hopPmaL (leaf 6). Photographs were taken under a dissecting microscope 6 days after inoculation, and each area shown is 1 cm2. (B) Numbers of specks on tomato leaves inoculated with DC3000ΔavrPtoΔavrPtoB expressing the genes indicated. The numbers of specks per cm2 were counted for nine independent areas of the leaflet, and the error bars indicate standard deviations. Leaflets at the same position of each plant were chosen for counting numbers of specks and photography. Means with the same letter above the bars are not different at a significance level of 5%. The experiments were performed twice, and similar results were obtained.
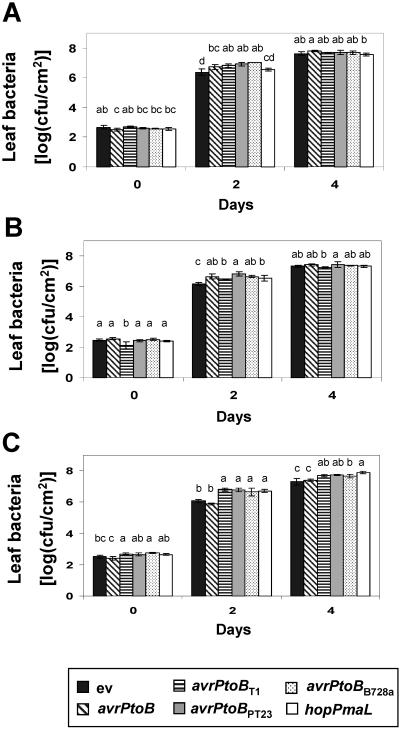
To determine if the increased disease symptoms that we observed correlated with increased bacterial growth, we next measured populations of ΔavrPtoΔavrPtoB expressing avrPtoB or the avrPtoB homologs in leaves of each tomato genotype. At day 2, in tomato lines RG-PtoS and RG-prf3 ΔavrPtoΔavrPtoB expressing avrPtoB or avrPtoB homologs showed between 2- and 10-fold enhancement of bacterial growth compared to ΔavrPtoΔavrPtoB(ev) (Fig. 6A and B). However, the sizes of the populations of all strains were similar 4 days after inoculation. We reported similar observations previously for AvrPtoB (26) and hypothesized that the enhanced disease symptoms seen with the AvrPtoB-expressing strain, compared to the empty-vector control strain, were probably due to the faster growth of the former strain at early times.
FIG. 6.
Growth of DC3000ΔavrPtoΔavrPtoB expressing AvrPtoB or the AvrPtoB homologs. (A) RG-PtoS. (B) RG-prf3. (C) RG-pto11. The bacterial strains were vacuum infiltrated into leaves of Rio-Grande tomato lines, and the bacterial populations were measured 0, 2, and 4 days after inoculation. The error bars indicate the standard deviations for three 1-cm-diameter leaf disks. Data analysis was performed using Duncan's multiple-range test. Means with the same letter above the bars are not different at a significance level of 5%. ev, empty vector pCPP45. The experiments were performed twice, and similar results were obtained.
In leaves of tomato line RG-pto11 2 days after inoculation, expression of each avrPtoB homolog promoted 5- to 10-fold more bacterial growth in ΔavrPtoΔavrPtoB than in either ΔavrPtoΔavrPtoB(ev) or ΔavrPtoΔavrPtoB(avrPtoB) (Fig. 6C). At day 4, the populations of ΔavrPtoΔavrPtoB(ev) and ΔavrPtoΔavrPtoB(avrPtoB) were still smaller than the populations of the strains expressing the avrPtoB homologs (Fig. 6C). The smaller populations of ΔavrPtoΔavrPtoB(ev) and ΔavrPtoΔavrPtoB(avrPtoB) correlated with the lower numbers of specks produced by these two strains on RG-pto11 (Fig. 5B).
AvrPtoBT1, AvrPtoBPT23, and AvrPtoBB728a inhibit PCD.
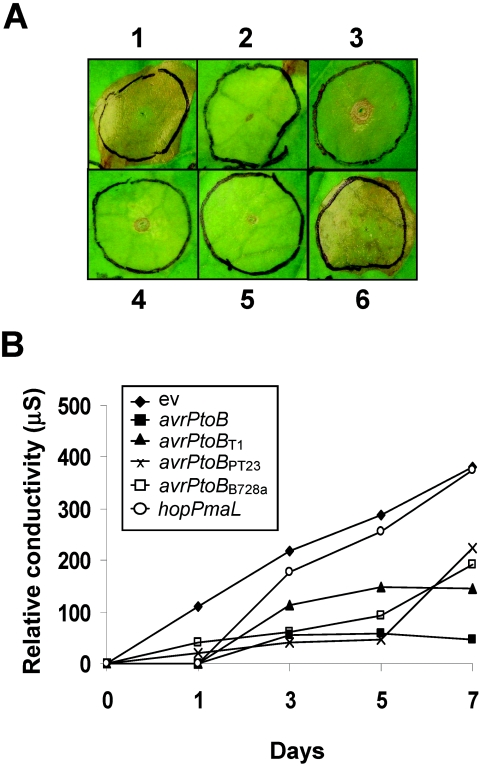
AvrPtoB has the ability to suppress plant immunity by inhibiting PCD (1). The anti-PCD domain has been mapped to the C terminus of AvrPtoB, which exhibits 73% sequence identity with AvrPtoBT1 and AvrPtoBPT23 and 59% sequence identity with AvrPtoBB728a. To determine if these three avrPtoB homologs have anti-PCD activity, avrPto and Pto were coexpressed with avrPtoBT1, avrPtoBPT23, or avrPtoBB728a using Agrobacterium-mediated transient expression in leaves of N. benthamiana. Three days after agroinfiltration, the cell death normally initiated by AvrPto and Pto was inhibited by coexpression of AvrPtoB. AvrPtoBT1, AvrPtoBPT23, or AvrPtoBB728a each inhibited cell death triggered by AvrPto/Pto (Fig. 7A). Ion leakage in the area infiltrated with A. tumefaciens was measured over a 7-day period to quantify host cell death. Each of the AvrPtoB homologs suppressed AvrPto/Pto-mediated ion leakage compared to the empty-vector control, and AvrPtoB showed the most stable suppression activity (Fig. 7B). Like AvrPtoBΔ6, a C terminus truncated form of AvrPtoB, HopPmaL triggered cell death in N. benthamiana when it was overexpressed in an Agrobacterium-mediated transient assay (data not shown). Lacking the anti-PCD domain, HopPmaL cannot inhibit cell death triggered by AvrPto/Pto (Fig. 7B).
FIG. 7.
Inhibition of programmed cell death by AvrPtoB in AvrPtoB homologs in N. benthamiana leaves. (A) The following proteins were coexpressed with AvrPto and Pto in mature leaves of N. benthamiana using Agrobacterium-mediated transient expression: none (empty vector) (leaf 1), AvrPtoB (leaf 2), AvrPtoBT1 (leaf 3), AvrPtoBPT23 (leaf 4), AvrPtoBB728a (leaf 5), and HopPmaL (leaf 6). Photographs were taken 7 days after Agrobacterium infiltration. (B) Ion leakage in the area infiltrated with A. tumefaciens expressing AvrPto, Pto, AvrPtoB, or AvrPtoB homologs 1, 3, 5, and 7 days after infiltration. Conductivity is indicated on the y axis.
DISCUSSION
The initial observation that AvrPtoB-related sequences are widely distributed among phytopathogenic bacteria suggested that members of this type III effector family might play an important role in bacterial virulence (23). Indeed, additional molecular studies have shown that AvrPtoB is a chimeric protein with distinct virulence activities in its N- and C-terminal domains (1). The C-terminal domain (amino acids 309 to 553) is sufficient for suppressing host programmed cell death elicited by Pto/AvrPto and a variety of other cell death inducers. Recent data suggest that the N-terminal domain (amino acids 1 to 308), which interacts with Pto, plays a role in promoting disease symptoms on susceptible plants (3; F. Xiao and G. B. Martin, unpublished data). Here we characterized representatives from each branch of the AvrPtoB/VirPphA (HopAB) family. These homologs have sequences that are divergent (52 to 65%) compared with the AvrPtoB sequence. Despite these differences, each of the AvrPtoB homologs interacts with Pto, elicits Pto-dependent resistance when it is expressed in ΔavrPtoΔavrPtoB, and restores lesion-forming ability and enhances growth on susceptible tomato leaves when it is expressed in the ΔavrPtoΔavrPtoB mutant. Except for HopPmaL, which lacks the C-terminal domain, each of the AvrPtoB homologs suppresses PCD associated with immunity. These observations indicate that despite the fact that AvrPtoB homologs have undergone considerable evolution of their DNA sequences, key residues involved in virulence have been conserved among diverse AvrPtoB homologs. These homologs therefore should be useful tools in future studies to determine which residues play key roles in avirulence and virulence activities.
Our studies demonstrated that the avrPtoB homolog in P. syringae pv. maculicola ES4326, hopPmaL, is transcribed and translated and that the protein is secreted from bacterial cells in an hrp-dependent manner in vitro. Furthermore, HopPmaL both interacts with Pto in the yeast two-hybrid system and elicits Pto-specific resistance when it is delivered into tomato cells via ΔavrPtoΔavrPtoB. Nevertheless, P. syringae pv. maculicola ES4326 is fully virulent on Pto-expressing tomato leaves. We have not investigated the basis of this observation further, but one possible explanation for this discrepancy is that P. syringae pv. maculicola ES4326, like P. syringae pv. phaseolicola 1448a, from which VirPphA was first identified (18), might carry another effector which masks the avirulence activity of the AvrPtoB homolog.
We also observed that P. syringae pv. tomato T1, despite being virulent for Pto-expressing lines, also has an AvrPtoB homolog with an intact open reading frame. The AvrPtoBT1 protein was functionally identical to AvrPtoB in terms of both its avirulence and virulence activities in our various assays (except for its activity on RG-pto11 [see below]). However, in contrast to P. syringae pv. maculicola ES4326, we were unable to detect the AvrPtoBT1 protein in P. syringae pv. tomato T1, although the avrPtoBT1 gene was transcribed. Notably, we found that AvrPtoB also did not accumulate in P. syringae pv. tomato T1 and that AvrPtoBT1 is expressed normally in ΔavrPtoΔavrPtoB. Because transcription and translation are coupled in bacteria, this observation suggests that P. syringae pv. tomato T1 employs an unknown posttranslational mechanism to control the accumulation of AvrPtoB-like proteins. This mechanism appears to be specific because the AvrPto protein accumulated at normal levels in P. syringae pv. tomato T1 and was secreted in an hrp-dependent fashion.
Why are AvrPtoB-like proteins unable to accumulate in P. syringae pv. tomato T1? One possibility is that a helper protein of AvrPtoB (e.g., a chaperone), which is required for stabilization of the AvrPtoB protein or regulation of its translation or secretion, is missing in P. syringae pv. tomato T1. In certain bacterial pathogens of animals, storage of some virulence proteins and flagellar subunits before secretion involves association with specific chaperones (16). TTSS chaperones may function as (i) antiaggregation and stabilizing factors, (ii) signals for secretion and hierarchy-determining factors, (iii) antifolding factors, or iv) regulators of expression (9, 32). An additional role for chaperones as molecules that participate in coupling of translation to secretion was revealed for the anti-σ28 factor, FlgM, and its cognate chaperone, FlgN, in the flagellar system (22). Several chaperones in plant pathogens have been shown to interact with their cognate substrates and are required for secretion of their substrates into culture medium and translocation into plant cells. For example, ShcFPto is known to stabilize its substrate (HopFPto) when it is expressed in P. syringae pv. phaseolicola (4, 21, 38, 42). Chaperone genes are typically located next to the gene encoding the chaperone substrate. However, no candidate chaperone genes have been found near avrPtoB coding sequences in DC3000. In addition, no obvious candidate helper proteins were identified in a yeast two-hybrid screening analysis for AvrPtoB-interacting proteins from DC3000 (A. S. Iyer and G. B. Martin, unpublished data). A second possibility to explain why AvrPtoB-like proteins do not accumulate in P. syringae pv. tomato T1 is that a specific negative regulator may function in P. syringae pv. tomato T1 to degrade AvrPtoBT1 as soon as the protein is made. It has been demonstrated that a Lon protease in P. syringae pv. syringae 61, mediating degradation of HrpR, functions as a negative regulator of type III effectors. The Lon protein also mediates the proteolytic degradation of type III effectors to regulate the secretion of several effectors (5, 29). We are currently using different approaches to further dissect the mechanism underlying the regulation of AvrPtoB in P. syringae pv. tomato T1.
Although AvrPtoB and AvrPtoB homologs have similar avirulence and virulence activities, AvrPtoB had weaker effects on restricting bacterial growth on RG-PtoR, on promoting disease symptoms in susceptible tomato lines RG-PtoS and RG-pto11, and on enhancing bacterial growth in tomato line RG-pto11. Most notable was the inability of AvrPtoB, when expressed on a broad-host-range vector in ΔavrPtoΔavrPtoB, to enhance disease symptoms on leaves of RG-pto11 (Fig. 5B). avrPtoB, however, conferred increased virulence to ΔavrPtoΔavrPtoB in RG-PtoS and RG-prf3, indicating that avrPtoB is expressed and translocated at sufficient levels to alter pathogen growth in these plants. This observation suggests that the lack of enhanced growth in RG-pto11 is not the result of poor AvrPtoB translocation into the host. Rather, it might be due to recognition of AvrPtoB by the Rsb gene present in RG-pto11 (1).
The fact that many Pseudomonas pathovars encode AvrPtoB-like proteins with diverse C-terminal domains that nevertheless suppress host PCD highlights the importance of this virulence strategy. Inhibition of resistance gene-mediated responses may be one of the most common virulence functions of type III effectors. This phenomenon was demonstrated in P. syringae pv. phaseolicola 1448a race 6, in which both VirPphA and AvrPphC were found to suppress gene-for-gene resistance elicited by other effector proteins (18, 41). Later, AvrPtoB was shown to act generally as a cell death suppressor (1). More recent studies of type III effectors of DC3000 revealed that at least six effectors can suppress host PCD, strengthening the hypothesis that suppression of plant immunity is a general virulence mechanism utilized by type III effectors (3, 6, 8, 20, 28). In the future, it will be important to understand the molecular mechanisms underlying AvrPtoB suppression of PCD to gain further insight into its role in disease development.
The results reported here and in the previous study of virPphA and its homologs provide compelling evidence that broadly distributed type III effectors play important roles in bacterial pathogenesis. Further study of AvrPtoB homologs in their source bacterial strains should lead to insights into the molecular basis of this virulence activity and why this activity is so important to pathogen fitness.
Acknowledgments
We thank Alan Collmer for the kind gifts of pCPP2318 and anti-β-lactamase antibody. We acknowledge Hye-sook Oh for her assistance with the secretion assay and Olga del Pozo for her help with the ion leakage assay.
This work was supported by NSF grant DBI-0077622 (to G.B.M.) and by a government scholarship from the Ministry of Education, Taiwan, Republic of China (to N.-C.L.).
REFERENCES
- 1.Abramovitch, R. B., Y.-J. Kim, S. Chen, M. B. Dickman, and G. B. Martin. 2003. Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J. 22:60-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abramovitch, R. B., and G. B. Martin. 2005. AvrPtoB: a bacterial type III effector that both elicits and suppresses programmed cell death associated with plant immunity. FEMS Microbiol. Lett. 245:1-8. [DOI] [PubMed] [Google Scholar]
- 3.Abramovitch, R. B., and G. B. Martin. 2004. Strategies used by bacterial pathogens to suppress plant defense. Curr. Opin. Plant Biol. 7:356-364. [DOI] [PubMed] [Google Scholar]
- 4.Badel, J. L., K. Nomura, S. Bandyopadhyay, R. Shimizu, A. Collmer, and S. Y. He. 2003. Pseudomonas syringae pv. tomato DC3000 HopPtoM (CEL ORF3) is important for lesion formation but not growth in tomato and is secreted and translocated by the Hrp type III secretion system in a chaperone-dependent manner. Mol. Microbiol. 49:1239-1251. [DOI] [PubMed] [Google Scholar]
- 5.Bretz, J., L. Losada, K. Lisboa, and S. W. Hutcheson. 2002. Lon protease functions as a negative regulator of type III protein secretion in Pseudomonas syringae. Mol. Microbiol. 45:397-409. [DOI] [PubMed] [Google Scholar]
- 6.Bretz, J. R., N. M. Mock, J. C. Charity, S. Zeyad, C. J. Baker, and S. W. Hutcheson. 2003. A translocated protein tyrosine phosphatase of Pseudomonas syringae pv. tomato DC3000 modulates plant defence response to infection. Mol. Microbiol. 49:389-400. [DOI] [PubMed] [Google Scholar]
- 6a.Charlowski, A. O., H.-C. Huang, and A. Collmer. 1997. Altered localization of HrpZ in Pseudomonas syringae pv. syringae hrp mutants suggests that different components of the type III secretion pathway control protein translocation across the inner and outer membranes of gram-negative bacteria. J. Bacteriol. 179:3866-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohn, J. R., and G. B. Martin. 2005. Pseudomonas syringae pv. tomato type III effectors AvrPto and AvrPtoB promote ethylene-dependent cell death in tomato. Plant J. 44:139-154. [DOI] [PubMed] [Google Scholar]
- 8.Espinosa, A., M. Guo, V. C. Tam, Z. Q. Fu, and J. R. Alfano. 2003. The Pseudomonas syringae type III-secreted protein HopPtoD2 possesses protein tyrosine phosphatase activity and suppresses programmed cell death in plants. Mol. Microbiol. 49:377-387. [DOI] [PubMed] [Google Scholar]
- 9.Feldman, M. F., and G. R. Cornelis. 2003. The multitalented type III chaperones: all you can do with 15 kDa. FEMS Microbiol. Lett. 219:151-158. [DOI] [PubMed] [Google Scholar]
- 10.Golemis, E., J. Gyuris, and R. Brent. 1996. Interaction trap/two-hybrid system to identify interacting protein, p. 20.1.1-20.1.28. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons Inc., New York, N.Y.
- 11.Guttman, D. S., B. A. Vinatzer, S. F. Sarkar, M. V. Ranall, G. Kettler, and J. T. Greenberg. 2002. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295:1722-1726. [DOI] [PubMed] [Google Scholar]
- 12.Ham, J. H., D. W. Bauer, D. E. Fouts, and A. Collmer. 1998. A cloned Erwinia chrysanthemi Hrp (type III protein secretion) system functions in Escherichia coli to deliver Pseudomonas syringae Avr signals to plant cells and to secrete Avr proteins in culture. Proc. Natl. Acad. Sci. USA 95:10206-10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan, D. 1985. Techniques for transformation of E. coli, p. 109-135. In D. M. Glover (ed.), DNA cloning: a practical approach. IRL Press, Oxford, United Kingdom.
- 14.Harper, S., and D. W. Speicher. 1997. Expression and purification of GST fusion proteins, p. 6.6.1-6.6.21. In J. E. Coligan, B. M. Dunn, D. W. Speicher, P. T. Wingfield, and H. L. Ploegh (ed.), Current protocols in protein science. John Wiley & Sons, Inc., New York, N.Y.
- 15.Hauck, P., R. Thilmony, and S. Y. He. 2003. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA 100:8577-8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He, S. Y., K. Nomura, and T. S. Whittam. 2004. Type III protein secretion mechanism in mammalian and plant pathogens. Biochim. Biophys. Acta 1694:181-206. [DOI] [PubMed] [Google Scholar]
- 17.Huynh, T. V., D. Dahlbeck, and B. J. Staskawicz. 1989. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245:1374-1377. [DOI] [PubMed] [Google Scholar]
- 18.Jackson, R. W., E. Athanassopoulos, G. Tsiamis, J. W. Mansfield, A. Sesma, D. L. Arnold, M. J. Gibbon, J. Murillo, J. D. Taylor, and A. Vivian. 1999. Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola. Proc. Natl. Acad. Sci. USA 96:10875-10880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson, R. W., J. W. Mansfield, H. Ammouneh, L. C. Dutton, B. Wharton, A. Ortiz-Barredo, D. L. Arnold, G. Tsiamis, A. Sesma, D. Butcher, J. Boch, Y.-J. Kim, G. B. Martin, S. Tegli, J. Murillo, and A. Vivian. 2002. Location and activity of members of a family of virPphA homologues in pathovars of Pseudomonas syringae and P. savastanoi. Mol. Plant Pathol. 3:205-216. [DOI] [PubMed] [Google Scholar]
- 20.Jamir, Y., M. Guo, H. S. Oh, T. Petnicki-Ocwieja, S. Chen, X. Tang, M. B. Dickman, A. Collmer, and, J. R. Alfano 2004. Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. Plant J. 37:554-565. [DOI] [PubMed] [Google Scholar]
- 21.Kabisch, U., A. Landgraf, J. Krause, U. Bonas, and J. Boch. 2005. Type III secretion chaperones ShcS1 and ShcO1 from Pseudomonas syringae pv. tomato DC3000 bind more than one effector. Microbiology 151:269-280. [DOI] [PubMed] [Google Scholar]
- 22.Karlinsey, J. E., J. Lonner, K. L. Brown, and K. T. Hughes. 2000. Translation/secretion coupling by type III secretion systems. Cell 102:487-497. [DOI] [PubMed] [Google Scholar]
- 23.Kim, Y.-J., N.-C. Lin, and G. B. Martin. 2002. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109:589-598. [DOI] [PubMed] [Google Scholar]
- 24.King, E., M. Ward, and D. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 25.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 26.Lin, N.-C., and G. B. Martin. 2005. An avrPto/avrPtoB mutant of Pseudomonas syringae pv. tomato DC3000 does not elicit Pto-mediated resistance and is less virulent on tomato. Mol. Plant-Microbe Interact. 18:43-51. [DOI] [PubMed] [Google Scholar]
- 27.Lindeberg, M., J. Stavrinides, J. H. Chang, J. R. Alfano, A. Collmer, J. L. Dangl, J. T. Greenberg, J. W. Mansfield, and D. S. Guttman. 2005. Proposed guidelines for a unified nomenclature and phylogenetic analysis of type III Hop effector proteins in the plant pathogen Pseudomonas syringae. Mol. Plant-Microbe Interact. 18:275-282. [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Solanilla, E., P. A. Bronstein, A. R. Schneider, and A. Collmer. 2004. HopPtoN is a Pseudomonas syringae Hrp (type III secretion system) cysteine protease effector that suppresses pathogen-induced necrosis associated with both compatible and incompatible plant interactions. Mol. Microbiol. 54:353-365. [DOI] [PubMed] [Google Scholar]
- 29.Losada, L. C., and S. W. Hutcheson. 2005. Type III secretion chaperones of Pseudomonas syringae protect effectors from Lon-associated degradation. Mol. Microbiol. 55:941-953. [DOI] [PubMed] [Google Scholar]
- 30.McBride, K. E., and K. R. Summerfelt. 1990. Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 14:269-276. [DOI] [PubMed] [Google Scholar]
- 31.Olmsted, J. B. 1981. Affinity purification of antibodies from diazotized paper blots of heterogeneous protein samples. J. Biol. Chem. 256:11955-11957. [PubMed] [Google Scholar]
- 32.Page, A. L., and C. Parsot. 2002. Chaperones of the type III secretion pathway: jacks of all trades. Mol. Microbiol. 46:1-11. [DOI] [PubMed] [Google Scholar]
- 33.Pedley, K. F., and G. B. Martin. 2003. Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu. Rev. Phytopathol. 41:215-243. [DOI] [PubMed] [Google Scholar]
- 34.Ronald, P. C., J. M. Salmerson, F. Carland, M., and B. J. Staskawicz. 1992. The cloned avirulence gene avrPto induces disease resistance in tomato cultivars containing the Pto resistance gene. J. Bacteriol. 174:1604-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salmeron, J. M., S. J. Barker, F. M. Carland, A. Y. Mehta, and B. J. Staskawicz. 1994. Tomato mutants altered in bacterial disease resistance provide evidence for a new locus controlling pathogen recognition. Plant Cell 6:511-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Sessa, G., M. D'Ascenzo, and G. B. Martin. 2000. Thr38 and Ser198 are Pto autophosphorylation sites required for the AvrPto-Pto-mediated hypersensitive response. EMBO J. 19:2257-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shan, L., H. S. Oh, J. Chen, M. Guo, J. Zhou, J. R. Alfano, A. Collmer, X. Jia, and X. Tang. 2004. The HopPtoF locus of Pseudomonas syringae pv. tomato DC3000 encodes a type III chaperone and a cognate effector. Mol. Plant-Microbe Interact. 17:447-455. [DOI] [PubMed] [Google Scholar]
- 39.Tang, X., R. D. Frederick, J. Zhou, D. A. Halterman, Y. Jia, and G. B. Martin. 1996. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274:2060-2063. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsiamis, G., J. W. Mansfield, R. Hockenhull, R. W. Jackson, A. Sesma, E. Athanassopoulos, M. A. Bennett, C. Stevens, A. Vivian, J. D. Taylor, and J. Murillo. 2000. Cultivar-specific avirulence and virulence functions assigned to avrPphF in Pseudomonas syringae pv. phaseolicola, the cause of bean halo-blight disease. EMBO J. 19:3204-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Dijk, K., V. C. Tam, A. R. Records, T. Petnicki-Ocwieja, and J. R. Alfano. 2002. The ShcA protein is a molecular chaperone that assists in the secretion of the HopPsyA effector from the type III (Hrp) protein secretion system of Pseudomonas syringae. Mol. Microbiol. 44:1469-1481. [DOI] [PubMed] [Google Scholar]