Abstract
Cell-free supernatants from growing Bacillus cereus strain ATCC 10987 induced luminescence in a Photorhabdus luminescens ΔluxS mutant, indicating the production of functional autoinducer 2 (AI-2). The exogenous addition of in vitro synthesized AI-2 had an inhibitory effect on biofilm formation by B. cereus and promoted release of the cells from a preformed biofilm.
Studies on Vibrio harveyi have uncovered a signaling molecule called autoinducer 2 (AI-2) (17). In this species, AI-2 acts in conjunction with AI-1, an acyl-homoserine lactone signal, to regulate the luminescence in response to cell density. AI-2 synthesis is linked to the metabolism of S-adenosylmethionine. Indeed, reactions of methylation frequently use S-adenosylmethionine as the methyl donor and generate S-adenosylhomocysteine (SAH). In most bacteria, SAH is converted into homocysteine, adenine and 4,5-dihydroxy-2,3-pentanedione (DPD) by the sequential action of the Pfs and LuxS enzymes (14, 15). The by-product DPD can spontaneously cyclize and/or interact with borate to form at least two different interconvertible molecules described as AI-2 (3, 10). A broad range of gram-positive and gram-negative bacteria produce AI-2 (1, 16, 20). In every case, an AI-2 synthase highly similar to the V. harveyi LuxS protein is required for its synthesis (17). The genes encoding Pfs- and LuxS-like enzymes are also present in the recently sequenced genomes of Bacillus cereus, Bacillus thuringiensis, and Bacillus anthracis. In B. anthracis, the LuxS protein is essential for AI-2 synthesis (6). AI-2 has been shown to control a variety of cellular processes, such as production of pathogenicity factors in Streptococcus pyogenes (8), toxin production in Clostridium perfringens (11), and formation of mixed biofilm between the two oral bacteria Streptococcus gordonii and Porphyromonas gingivalis (9). AI-2 could therefore be a universal signaling factor for intra- and interspecies communication in response to cell density. Until now, the mechanism of AI-2 detection and the signal transduction pathway have been established only for V. harveyi, Vibrio cholerae, Salmonella enterica serovar Typhimurium, and Escherichia coli (2, 19, 21). In V. harveyi, two proteins, LuxP and LuxQ, function together as the AI-2 sensor (2). LuxP is a periplasmic binding protein, and LuxQ is a hybrid two-component protein that contains sensor kinase and response regulator domains. In S. enterica serovar Typhimurium, AI-2 is imported into the bacteria via the Lsr ABC transporter (19).
B. cereus is a gram-positive, spore-forming bacterium closely related to the lethal pathogen B. anthracis. B. cereus is frequently identified as the causative agent of food-borne diseases. As such, the interest in this bacterium is growing. This ubiquitous organism can easily contaminate food production or processing systems (7) and forms biofilms that are highly resistant to cleaning procedures (12). In the present work, we show that AI-2 is produced by the biofilm-forming strain B. cereus ATCC 10987 and that this molecule inhibits biofilm formation.
Formation of biofilms by B. cereus.
The ability of the B. cereus sequenced strains ATCC 14579 and ATCC 10987 to form biofilms was tested. Precultures in the exponential phase of growth were inoculated at an optical density at 600 nm (OD600) of 0.01 into fresh LB medium (10 g/liter bactopeptone, 5 g/liter yeast extract, 5 g/liter NaCl) in 96-well polyvinylchloride microtiter plates (Falcon 35911). After 72 h of incubation at 30°C, the biofilm density was measured as follows: the microtiter plate wells were washed once with phosphate-buffered saline, and bound cells were stained with a 1% (wt/vol) crystal violet solution at room temperature for 20 min (5). The wells were then washed with phosphate-buffered saline three times, and the dye was solubilized with a 20%/80% acetone/ethanol mixture. The absorbance at 595 nm of the solubilized dye was subsequently determined. ATCC 10987 made biofilms in polyvinylchloride plates (Fig. 1A), whereas no biofilm was observed for ATCC 14579 under the same conditions (data not shown). Within the Bacillus cereus group, these two strains are genetically distant (13) and might be different in their cell surface properties and/or exopolysaccharide production, both of these being important for biofilm formation.
FIG. 1.
Biofilm formation by B. cereus ATCC 10987. (A) Photograph of a B. cereus biofilm stained with crystal violet. Panels: 1, LB medium inoculated with strain ATCC 10987; 2, LB medium alone. (B) OD595 of solubilized crystal violet from microtiter plate assay (filled circles) and CFU/ml of attached cells (open circles) in biofilms over time. After various times of incubation, biofilm density was measured as described in the text. The data represent the means of three independent experiments. The error bars represent standard deviations.
To determine the kinetics of biofilm formation, a microtiter plate was inoculated with the ATCC 10987 strain as described above. A measurable amount of biofilm was detected after 16 h of inoculation (Fig. 1B). The number of viable cells in the biofilm rings was determined as follows. The biofilm was manually scraped from the sides of the wells using a pipette tip and resuspended in LB medium. After serial dilutions, cells were plated onto LB medium. The increase in crystal violet staining with time of incubation was proportional to the increase in the number of viable cells in the biofilm (Fig. 1B).
Synthesis of biologically active AI-2 by B. cereus.
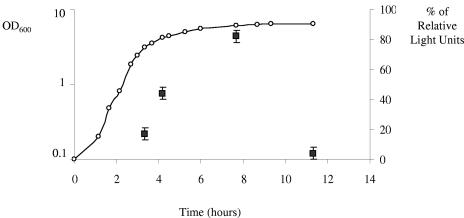
After a Blastp search of the complete B. cereus ATCC 10987 genome sequence (http://www.ncbi.nlm.nih.gov), we detected two genes, bce4456 and bce4946, encoding a Pfs-like protein and a LuxS-like protein, respectively. The Bce4456 protein shares 67% identity with the SAH nucleosidase from Bacillus subtilis strain 168, while the predicted Bce4946 polypeptide is 100% identical to LuxS from B. anthracis strain Ames and 82% identical to LuxS from B. subtilis. The ability of B. cereus ATCC 10987 to synthesize active AI-2 was determined using a Photorhabdus luminescens AI-2 reporter assay. In this system, addition of supernatants from cultures of AI-2-producing bacteria restores the luminescence of a P. luminescens ΔluxS mutant (4). Addition of LB medium alone to P. luminescens wild-type strain TT01 and to the ΔluxS mutant P12012 was used to define the reference levels of luminescence. Cell-free supernatants (CFS) were collected from a culture of B. cereus ATCC 10987 at various time points (Fig. 2). The culture was grown at 37°C with vigorous shaking at 200 rpm. CFS were prepared by centrifugation at 13,000 rpm for 5 min and filtration of the supernatant (0.2-μm-pore-size Millipore filter). The P. luminescens ΔluxS strain was grown overnight at 30°C in Schneider medium, cultures were diluted to an OD600 of 0.1 in fresh medium, and the CFS to be tested was added at a final concentration of 10%. Bioluminescence was measured on 10-μl aliquots and expressed as relative light units (RLU) by using a luminometer. Figure 2 shows that the CFS of B. cereus ATCC 10987 led to a significant increase in luminescence. The level of light induction exhibited a growth-phase dependence with a maximum corresponding to the late-exponential culture as previously observed in B. anthracis and S. gordonii (6, 9). Our results indicated that B. cereus ATCC 10987 synthesizes active AI-2 recognized by the lux quorum-sensing system.
FIG. 2.
Growth-dependent AI-2 production by B. cereus ATCC 10987. B. cereus ATCC 10987 was grown in LB medium (open circles). At various time points, the amount of AI-2 in CFS (filled squares) was measured by using the P. luminescens bioassay. Addition of LB medium alone to P. luminescens wild-type strain TT01 (100% RLU) and to the ΔluxS mutant P12102 (0% RLU) was used to define the reference levels of luminescence. The data indicated by squares represent the means (± standard deviations) of three independent preparations.
Effect of in vitro synthesized AI-2 on biofilm formation by B. cereus.
We further studied the direct effect of in vitro synthesized AI-2 on biofilm formation. For this purpose, the pfs and luxS genes from P. luminescens were amplified from genomic DNA, and the PCR products were inserted into the NdeI and XhoI sites of the pET22b expression vector (Novagen). The resulting plasmids pET22bpfs and pET22bluxS were introduced into the E. coli BL21(DE3) strain. To overproduce the recombinant N-terminally His-tagged proteins, the transformed BL21(DE3) strain was grown in Hyper BrothTM (Athena Enzyme Systems) and induced at an OD600 of 3.0 with 3 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 h. The His-tagged proteins were subsequently purified using NiSO4 chelation columns. DPD was synthesized as previously described (15) using equimolar concentrations of Pfs and LuxS proteins and 1 mM SAH. The DPD concentration was quantified by measuring homocysteine concentration with Ellman's reagent [5,5′-dithiobis-(2-nitrobenzoic acid)] (15). The concentration of DPD synthetized was about 170 μM in the 1-ml reaction mixture. The presence of 8 nM DPD in P. luminescens ΔluxS cell culture increased the level of luminescence 1.4-fold, thus indicating the functionality of AI-2 synthesized in vitro.
The effect of AI-2 on biofilm formation was then tested. An overnight culture of B. cereus ATCC 10987 was inoculated into microtiter plates (OD600, 0.01) in the presence of 1 μM to 6.8 μM of AI-2. After 24 h of incubation, increasing the amount of AI-2 supplied in the medium resulted in a decrease in the biofilm density (Fig. 3A). As controls, the presence of SAH, the substrate of the reaction mixture, and of adenine or homocysteine, the secondary products of the in vitro reaction, did not affect the biofilm formation at 1, 5, or 10 μM (data not shown). We also verified that AI-2 had no effect on planktonic cell growth. The growth rates of the strain ATCC 10987 cultured in flasks at 30°C in the presence of 0, 1, or 6.8 μM of AI-2 were identical (data not shown). These results showed that AI-2 has an inhibitory effect on the formation of biofilms by B. cereus. We then assessed the time course of biofilm formation in the presence of 1 μM AI-2 (Fig. 3B). After 24 h and 30 h of incubation, the biofilm was 2.3-fold less dense when AI-2 was present, whereas after 48 h of incubation, biofilm reduction in the presence of AI-2 was only 1.2-fold. This could be due to reduction of the amount of AI-2 in the medium used in the experiment.
FIG. 3.
Effect of AI-2 on biofilm formation by B. cereus ATCC 10987. (A) Different concentrations of in vitro synthetized AI-2 were added to microtiter wells inoculated with strain ATCC 10987 in LB medium. After 24 h of incubation, the biofilm density was measured. The data represent the means (± standard deviations) of triplicate experiments. (B) Time course of biofilm formation in the presence (gray bars) or absence (white bars) of 1 μM AI-2. Experiments were run in triplicate.
Effect of AI-2 on preformed biofilms.
To determine at which steps AI-2 can inhibit the formation of biofilms, we tested whether AI-2 also had an effect on a preformed biofilm. For this purpose, AI-2 was added only after 24 h of culture incubation in microtiter plates. As measured 16 h and 24 h after AI-2 addition, no reduction in biofilm formation was observed (data not shown). However, we could not exclude the possibility that AI-2 was titrated from the medium by the planktonic cells present in the wells after 24 h of incubation, since bacteria can eliminate AI-2 from the medium by internalization (17, 19).
To test this hypothesis, the medium and thus the planktonic cells were removed after 24 h of incubation and replaced by fresh medium containing different concentrations of AI-2. Incubation was continued for another 24 h. As shown in Fig. 4, increasing the level of AI-2 in the fresh medium resulted in a decrease in the biofilm density. This result indicates that the presence of AI-2 can also elicit the release of a large proportion of the cells from the biofilm.
FIG. 4.
Effect of AI-2 on mature biofilms. After 24 h of incubation in microtiter plates, free cells were removed and replaced by fresh medium alone or fresh medium containing 3, 4.5, or 6 μM AI-2. Incubation was continued for 24 h, and biofilm density was measured. The means of three independent experiments are indicated.
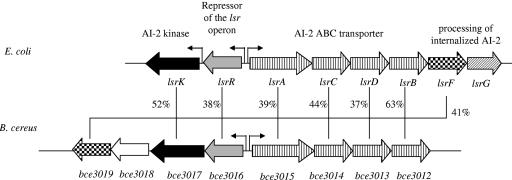
An Lsr-like system is present in B. cereus.
To inhibit B. cereus biofilm formation, AI-2 must be sensed by the bacterial cells. A Blastp search of sequence databases (http://www.ncbi.nlm.nih.gov) revealed that the genome of B. cereus does not encode homologs of V. harveyi LuxP and LuxQ proteins, whereas lsrACDB-like genes are present (Fig. 5). It also contains a lsrR-like gene, which encodes the regulator of the lsr operon, and lsrK and lsrF-like genes, whose products are necessary for the processing of internalized AI-2 (18). Therefore, the Lsr-like system could be responsible for the AI-2 uptake and processing. It is worth noting that the Lsr-like system was not found in any other sequenced gram-positive bacterium, including B. subtilis, Bacillus halodurans or Listeria spp. Elucidation of the role of the Lsr-like system in the transport and processing of AI-2 in B. cereus would certainly be of a great interest.
FIG. 5.
Genetic organization and sequence analysis of the lsr region in B. cereus ATCC 10987 and E. coli K-12. Putative transcription start sites are indicated by broken arrows. For each gene product, the similarity between the B. cereus and E. coli proteins is indicated as a percentage of identity. LsrB, periplasmic AI-2 binding protein; LsrC and LsrD, channel proteins; LsrA, ATPase; LsrF, a protein similar to aldolases; LsrG and Bce3018, proteins with no significant similarity with known proteins; LsrR, repressor of the lsr operon; LsrK, AI-2 kinase. The genes encoding the ABC transporters are represented by striped boxes, the LsrR-like regulators are represented by gray boxes, and the processing enzymes are represented by checkered boxes (lsrF-like) or a diagonally striped box (lsrG).
We have reported that B. cereus synthetizes and recognizes AI-2 as an extracellular signal. Most particularly, we have shown that AI-2 inhibits biofilm formation in a concentration-dependent manner. The genome of B. cereus contains genes encoding an Lsr-like system that could be involved in the internalization and processing of AI-2.
Acknowledgments
We thank M. F. Hullo for her technical help during this work. We are grateful to A. Sorokin, I. Martin-Verstraete, and C. Tinsley for critical reading of the manuscript.
REFERENCES
- 1.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 3.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 4.Derzelle, S., E. Duchaud, F. Kunst, A. Danchin, and P. Bertin. 2002. Identification, characterization, and regulation of a cluster of genes involved in carbapenem biosynthesis in Photorhabdus luminescens. Appl. Environ. Microbiol. 68:3780-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamon, M. A., and B. A. Lazazzera. 2001. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 42:1199-1209. [DOI] [PubMed] [Google Scholar]
- 6.Jones, M. B., and M. J. Blaser. 2003. Detection of a luxS-signaling molecule in Bacillus anthracis. Infect. Immun. 71:3914-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotiranta, A., K. Lounatmaa, and M. Haapasalo. 2000. Epidemiology and pathogenesis of Bacillus cereus infections. Microbes Infect. 2:189-198. [DOI] [PubMed] [Google Scholar]
- 8.Lyon, W. R., J. C. Madden, J. C. Levin, J. L. Stein, and M. G. Caparon. 2001. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol. Microbiol. 42:145-157. [DOI] [PubMed] [Google Scholar]
- 9.McNab, R., S. K. Ford, A. El-Sabaeny, B. Barbieri, G. S. Cook, and R. J. Lamont. 2003. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller, S. T., K. B. Xavier, S. R. Campagna, M. E. Taga, M. F. Semmelhack, B. L. Bassler, and F. M. Hughson. 2004. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 15:677-687. [DOI] [PubMed] [Google Scholar]
- 11.Ohtani, K., H. Hayashi, and T. Shimizu. 2002. The luxS gene is involved in cell-cell signalling for toxin production in Clostridium perfringens. Mol. Microbiol. 44:171-179. [DOI] [PubMed] [Google Scholar]
- 12.Peng, J. S., W. C. Tsai, and C. C. Chou. 2002. Inactivation and removal of Bacillus cereus by sanitizer and detergent. Int. J. Food Microbiol. 77:11-18. [DOI] [PubMed] [Google Scholar]
- 13.Rasko, D. A., J. Ravel, O. A. Okstad, E. Helgason, R. Z. Cer, L. Jiang, K. A. Shores, D. E. Fouts, N. J. Tourasse, S. V. Angiuoli, J. Kolonay, W. C. Nelson, A. B. Kolsto, C. M. Fraser, and T. D. Read. 2004. The genome sequence of Bacillus cereus ATCC 10987 reveals metabolic adaptations and a large plasmid related to Bacillus anthracis pXO1. Nucleic Acids Res. 32:977-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schauder, S., and B. L. Bassler. 2001. The languages of bacteria. Genes Dev. 15:1468-1480. [DOI] [PubMed] [Google Scholar]
- 15.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 16.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taga, M. E., S. T. Miller, and B. L. Bassler. 2003. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol. 50:1411-1427. [DOI] [PubMed] [Google Scholar]
- 19.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 20.Winzer, K., K. R. Hardie, and P. Williams. 2003. LuxS and autoinducer-2: their contribution to quorum sensing and metabolism in bacteria. Adv. Appl. Microbiol. 53:291-396. [DOI] [PubMed] [Google Scholar]
- 21.Xavier, K. B., and B. L. Bassler. 2005. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187:238-248. [DOI] [PMC free article] [PubMed] [Google Scholar]