Abstract
The genus Pseudomonas (sensu stricto) represents a group of microorganisms directly involved in functions conferring plant health. We performed a study in the DOK long-term agricultural field experiment on the basis of previously published Pseudomonas-selective PCR primers in order to investigate the community structure of the microbial groups defined by the target range of these primers. Three different agricultural management systems, i.e., conventional, biodynamic, and bio-organic, along with mineral and unfertilized controls were investigated, with each system planted with either winter wheat or a grass-clover ley. Amplified small-subunit rRNA gene fragments were analyzed using the genetic profiling techniques restriction fragment length polymorphism (RFLP) and denaturing gradient gel electrophoresis (DGGE), revealing distinct differences between soils planted with winter wheat and grass clover but only minor differences between the management systems. Phylogenetic analyses of 59 clone sequences retrieved from bio-organic and unfertilized systems identified sequences related to Pseudomonas fluorescens and a novel cluster termed Cellvibrio-related Pseudomonadaceae (CRP). The CRP clones were exclusively isolated from winter wheat soil samples and were responsible for the crop-specific differences observed in RFLP and DGGE profiles. New primers were designed for the amplification of CRP targets directly from soil DNA, yielding strong signals exclusively for winter wheat soils. We concluded that crop-associated CRP exist in agricultural soils and that genetic profiling followed by specific probe design represents a valuable approach for identification as well as sensitive and rapid monitoring of novel microbial groups in the environment.
In order to maintain the fertility of agricultural soils, it is important to learn more about the structures and functions of the indigenous microbial communities. A promising strategy may be to gain detailed information about microbial community structures in relation to various agricultural practices. For bacteria and fungi, a number of studies have been performed to investigate the relative influences of factors such as soil properties, agricultural practices, or plant types on their community structures. Soil type has consistently been reported as one of the main determinants of microbial community structures (15, 47). Furthermore, agricultural practices, including types of fertilizer, the addition of lime, and pesticide application, have been shown to significantly affect microbial community structures as well (14, 19, 48). The influence of plants on soil microbial communities has been assessed intensively in rhizosphere soil (7, 20, 41) as well as in bulk soil (15, 19, 48). Whereas clear differences between bulk and rhizosphere soil and also between different plant species have been detected, the influence of plants on microbial communities in bulk soil has often been reported as less significant (15, 19, 48).
In the DOK field experiment, located in northwestern Switzerland, three different farming systems, i.e., biodynamic (BIODYN), bio-organic (BIOORG), and conventional (CONFYM) management, and two control treatments with mineral fertilization (CONMIN) or no fertilization (NOFERT) have been established since 1978 (25). A study of soils from winter wheat plots from the DOK field experiment based on Biolog profiles revealed differences between the farming systems, but variation among field replications was relatively high (10). Recently, these differences have been confirmed with Biolog and bacterial terminal restriction fragment length polymorphism (T-RFLP) analyses performed on soils from winter wheat and grass-clover ley plots (48). These studies have shown that long-term farmyard manure (FYM) application has a highly significant effect on soil bacterial community structures, while short-term crop effects were weaker but still significant. Differences among BIODYN, BIOORG, and CONFYM were not significant. However, analyses of soil organic matter in the DOK system have indicated that distinct differences exist in the differently managed systems (9).
While the detection of entire bacterial or fungal communities has proven to be a successful approach for assessing changes in dominant community components, the resolution provided by current community-level profiling methods may not be sufficient to obtain information on the structure of specific microbial subgroups. This limitation can be overcome by directly targeting specific subgroups of particular interest.
The bacterial genus Pseudomonas represents one such subgroup that plays important roles in agricultural ecosystems. Specifically, it is involved in disease suppression in many crops (23, 46), exhibits plant growth-promoting properties (16, 40), and is able to degrade a variety of pesticides used in agriculture for crop protection purposes (30, 39). Besides these beneficial traits, some members of the genus Pseudomonas also represent plant pathogens (35, 37, 50) and cause reductions in crop yields. Limited information is available on environmental or anthropogenic factors that determine or influence the occurrence, distribution, and activity of indigenous Pseudomonas populations in soil (1, 3, 21). The majority of studies on soil Pseudomonas communities have relied on cultivation-dependent techniques (1, 21, 33). Although selective cultivation of Pseudomonas spp. has been successfully applied, only a small fraction of the entire soil Pseudomonas community can be retrieved by cultivation (24, 34). On the other hand, few studies have been performed to analyze soil Pseudomonas communities by using less biased molecular biological techniques (12, 28, 32, 34, 49). The agronomic importance and the saprophytic nature of Pseudomonas spp. make them an important microbial group for investigation in agricultural management systems such as the long-term DOK field experiment.
Our hypothesis was that differences in soil organic matter quality among BIODYN, BIOORG, and CONFYM derived from system-specific (FYM) qualities influence saprophytic Pseudomonas populations in soil. In addition, different crops may also have an influence on different components of soil Pseudomonas communities. In order to address these issues, we first performed a theoretical reevaluation of the Pseudomonas-selective PCR primers published by Widmer et al. (49). Second, the structure of the bacterial group defined by the Pseudomonas-selective PCR primers was examined in soils of the different farming systems of the DOK field experiment planted with either winter wheat or a grass-clover ley.
MATERIALS AND METHODS
Site description and sample collection.
The DOK field experiment was established in 1978 near Basel, in northwestern Switzerland, to compare three different agricultural management systems regarding soil fertility and productivity, nutrient dynamics, and impact on the environment (26). The soil at the site is classified as haplic luvisol (sandy loam) on deep deposits of alluvial loess. The experiment consists of BIODYN, BIOORG, and CONFYM farming systems, which all receive FYM as fertilizer. Two controls include a system with exclusive mineral fertilization (CONMIN) and an unfertilized system (NOFERT). The 7-year crop rotation includes potato, winter wheat 1, soy, maize, and winter wheat 2 followed by 2 years of grass-clover ley and is implemented in all treatments in three temporally shifted parallels. Each combination of management system and crop is replicated four times in the field (25). Altogether, 96 single plots with a size of 5 by 20 m are arranged in a split-split plot design on an area of 1.4 ha.
Soil samples (40 total) were obtained in March 2000 from each replicate plot for all five treatments planted with either winter wheat 1 (preceding crop, potato) or the grass-clover ley in its second year (for details, see references 15, 19, and 48). Briefly, 16 soil cores per plot were sampled to a depth of 20 cm, sieved to 2 mm, and stored on ice until further processing. DNA was extracted from the soil of each replicate plot, but for efficient screening of treatment-dependent population changes, equal amounts of DNA from the four corresponding replicates were pooled. This strategy allows for efficient screening and, if necessary, reanalysis of individual replicates.
DNA extraction and quantification.
DNA was extracted following the protocol of Bürgmann et al. (4), using a bead-beating procedure with slight modifications. Briefly, 0.5 g of soil (dry weight) was subjected to three repeated extractions using a bead beater (FP 120; Savant Instruments Inc., NY). Supernatants from all three extractions were pooled and subsequently purified with one chloroform-isoamyl alcohol (24/1) extraction. DNA concentrations were determined using a fluorometric assay with PicoGreen (Molecular Probes, Eugene, OR) as described previously (4).
Reevaluation of primers Ps-for and Ps-rev.
Theoretical reevaluation of the Ps-for and Ps-rev primer sequences (49) was performed by calculating the coverage (percentage of Pseudomonas small-subunit [SSU] rRNA gene sequences that contain the primer sequence) and specificity (ratio of Pseudomonas SSU rRNA gene sequences that contain the primer sequence to the number of bacterial SSU rRNA gene sequences [including Pseudomonas] that contain the primer sequence) of the primers. For these analyses, exclusively bacterial SSU rRNA sequences longer than 1,200 bp and deposited in RDP-II release 9.24 (January 2005; 44,890 sequences) were used. The Primer_Match algorithm in RDP-II does not distinguish between sequences that lack one primer site due to missing sequence information and those that display mismatches with primer sequences.
PCR amplification, RFLP, and denaturing gradient gel electrophoresis (DGGE) analyses.
Pseudomonas-selective PCR was performed with primers Ps-for (5′-GGTCTGAGAGGATGATCAGT-3′) and Ps-rev (5′-TTAGCTCCACCTCGCGGC-3′) (49). Reaction cocktails (50 μl) contained 2 mM MgCl2, 0.2 μM each primer, 0.4 mM each deoxyribonucleoside, 0.6 mg ml−1 bovine serum albumin, 2 U of HotStar Taq polymerase (QIAGEN GmbH, Hilden, Germany), 1× PCR buffer (QIAGEN), and 40 ng soil DNA. The cycling conditions consisted of an initial denaturation step for 15 min at 95°C to activate the HotStar polymerase, followed by 40 cycles of denaturation for 45 s at 94°C, primer annealing for 45 s at 65°C, and DNA synthesis for 45 s at 72°C. A final synthesis step for 5 min at 72°C was performed to complete the reaction.
Prior to RFLP analyses, PCR products were mixed with concentration conversion buffer (18) to convert the PCR mixture into a solution suitable for restriction digestion. For digestion, 3 U of the restriction endonuclease HaeIII (Promega, Madison, WI) was added and incubated overnight at 37°C. Digestion products were cleaned using a Montage SEQ96 sequencing reaction cleanup kit (Millipore Corporation, Billerica, MA) and resuspended in 30 μl Tris-EDTA buffer. Restriction fragments were electrophoresed in 12% polyacrylamide gels (bisacrylamide/acrylamide, 1/37.5) for 6 h at 150 V and 35°C (Dcode system; Bio-Rad Laboratories Ltd., Hercules, CA). Gels were stained for 30 min with SYBR green I nucleic acid gel stain (1:5,000 in 1× Tris-acetate-EDTA buffer; Molecular Probes, Eugene, OR) and were subsequently analyzed using the GelDoc EQ system and Quantity One software (Bio-Rad).
For preparation of PCR products suitable for DGGE analyses, a nested PCR approach for Ps-for/Ps-rev PCR products was developed. Two hundred picograms of Ps-for/Ps-rev PCR product or 1 ng of plasmid DNA was used as the template for PCR with the primers UNI-b-for (5′-TGCCAGCMGCCGCGGTA-3′ [modified from reference 13]; positions 516 to 532 according to Escherichia coli numbering) and 520-rev (5′-CGTGGACTACCAGGGTATC-3′ [49]; positions 791 to 809, including a 40-bp GC clamp at the 5′ end [29]). Nested PCR yielded products of approximately 300 bp. Reaction cocktails (50 μl) and cycling conditions were the same as those described above, except for the annealing temperature, which was set to 55°C. DGGE analyses were performed in 16- by 16-cm polyacrylamide gels (10%) containing 1× Tris-acetate-EDTA buffer, 2% glycerol, with a denaturing gradient of formamide and urea of 35 to 65% (29). Electrophoresis was performed with the Dcode system (Bio-Rad) for 14 h at 70 V and 60°C. Gel staining and analysis were the same as those described above for RFLP analysis. A mixture of SSU rRNA gene fragments from the following organisms was used as a DGGE migration standard: (i) Pseudomonas fluorescens ATCC 26663, (ii) Bacillus subtilis ATCC 14893, (iii) Rhizobium meliloti DSM 1981, (iv) Flavobacterium capsulatum DSM 30196, and (v) Arthrobacter globiformis DSM 20124.
DNA cloning and sequencing.
Ps-for/Ps-rev PCR products were cloned using a pGEM-T Easy cloning kit (Promega, Madison, WI). The approximately 990-bp PCR products were sequenced on both strands according to the method of Widmer et al. (49), using an ABI3100 genetic analyzer (Applied Biosystems, Foster City, CA).
DNA sequence analyses.
Sequences were subjected to theoretical RFLP analyses using the online program RestrictionMapper (http://www.restrictionmapper.org/). Clones were classified into restriction types labeled A, B, C, etc., according to the calculated RFLP patterns.
The 59 clone sequences from this study and 26 control sequences retrieved from RDP-II (6) were aligned using BioEdit, version 7.01 (17). Pairwise Jukes-Cantor distance determinations and cluster analysis by the unweighted-pair group method with mathematical averages (UPGMA) with 100 bootstrap resamplings were performed with Treecon, version 1.3b (44). Maximum likelihood analysis was performed with the fastDNAml routine in BioEdit.
Primer design, testing, and application.
Primer sequences for the detection of Cellvibrio-related Pseudomonadaceae (CRP) were deduced from the sequence alignment established for phylogenetic analyses. The determined primer sequences were evaluated using the Probe_Match tool from RDP-II (5) as well as the BLASTn tool from GenBank (2). The sequence of the forward primer, CRP 149-for (positions 440 to 460), was 5′-CAGTGGGGAGAAARRTCTG-3′, and the sequence of the reverse primer, CRP 565-rev (positions 837 to 856), was 5′-CCACTAAAGCCTCAAGGACT-3′. The PCR conditions for these primers were optimized using clones and soil DNA extracts from this study. Reaction cocktails and cycling conditions were the same as those described above, except for the annealing temperature, which was set to 57°C.
Nucleotide sequence accession numbers.
The nucleotide sequences of the 59 clones retrieved in this study are deposited in GenBank under accession numbers AY854436 to AY854494.
RESULTS
Reevaluation of primers Ps-for and Ps-rev.
Primer Ps-for covered 1,370 (96.8%) and primer Ps-rev covered 1,338 (94.6%) of 1,415 Pseudomonas (sensu stricto) SSU rRNA sequences deposited in RDP-II. The combination of both primers detected 1,299 of 1,415 Pseudomonas sequences, corresponding to 91.8% coverage. Primer Ps-for revealed an exact match with 364 non-Pseudomonas bacterial SSU rRNA sequences in RDP-II (79.0% specificity), while primer Ps-rev matched 588 non-Pseudomonas sequences (69.5% specificity). For the combination of both primers, only 12 non-Pseudomonas sequences were identified, leading to a specificity of 99.1%. Among these nontarget sequences, five were associated with the genus Cellvibrio (sequence accession numbers AF006505, AJ289162, AF452103, AY212574, and AY038046), which is the closest known relative to Pseudomonas. The remaining seven sequences were classified as Oceanospirillales (two Zooshikella sp. sequences [AB021682 and AF195410] and one unclassified sequence [AB020600]), unclassified γ-Proteobacteria (three sequences [AY676463, AF006502, and AF006506]), and unclassified β-Proteobacteria (one sequence [AF006507]).
Pseudomonas-selective RFLP analysis.
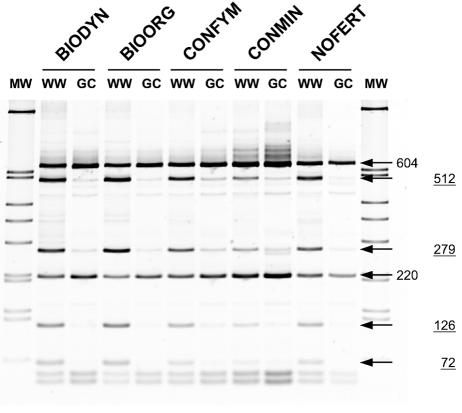
Distinct changes in HaeIII RFLP patterns were observed between winter wheat and grass-clover bulk soils across all five treatments in the DOK field experiment (Fig. 1). Prominent bands with calculated fragment sizes of 512, 279, 126, and 72 bp were detected in bulk soil from winter wheat plots but were absent or only faint for soil samples from grass-clover plots. Band intensities and fragment sizes observed in RFLP patterns from winter wheat or grass-clover bulk soil appeared highly similar across the five management systems.
FIG. 1.
HaeIII RFLP analysis of PCR-amplified SSU rRNA gene fragments from bulk soil DNA extracts with Pseudomonas-selective primers. Samples represent five different treatments of the DOK field experiment (lanes BIODYN, BIOORG, CONFYM, CONMIN, and NOFERT) planted with winter wheat (lanes WW) or grass-clover (lanes GC). Bands referred to in the text are identified with arrows and DNA fragment sizes (bp). Underlined fragment size values indicate bands that revealed clear intensity changes across different soils and crops.
Pseudomonas-selective DGGE analyses.
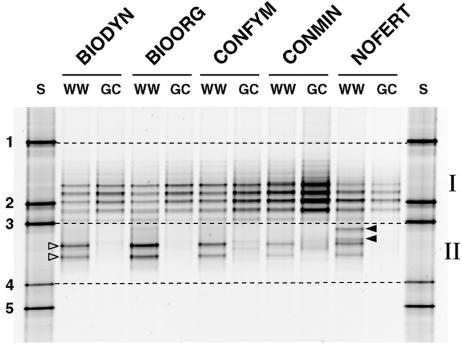
The DGGE banding patterns for winter wheat plot soils of all five treatments revealed two distinct additional bands in the higher denaturing region of the gel compared to the corresponding soils from grass-clover plots (Fig. 2, region II, open arrowheads). For soils from grass-clover plots of the farming systems BIODYN, BIOORG, and NOFERT, these bands were not detectable, and for soils from the systems CONFYM and CONMIN, they were only weakly detectable. DGGE patterns derived from soils of winter wheat plots showed high similarity among BIODYN, BIOORG, CONFYM, and CONMIN, while NOFERT soils planted with winter wheat displayed two additional bands that were not detected in the other soils (Fig. 2, closed arrowheads).
FIG. 2.
DGGE analysis of PCR-amplified SSU rRNA gene fragments based on Pseudomonas-selective PCR products from bulk soil DNA extracts. Samples represent five different treatments of the DOK field experiment (BIODYN, BIOORG, CONFYM, CONMIN, and NOFERT) planted with winter wheat (WW) or grass-clover (GC). Migration standard (S) bands 1 and 3 define the migration region for sequences associated with Pseudomonas (sensu stricto) (lower denaturing region [I]), and bands 3 and 4 define the migration region for sequences of CRP (higher denaturing region [II]). Open arrowheads, dominant bands detected in all winter wheat soils; closed arrowheads, bands exclusively detected in NOFERT system planted with winter wheat.
Cloning of Pseudomonas-selective PCR products.
Four gene libraries of cloned Ps-for/Ps-rev PCR products were constructed from the BIOORG and NOFERT systems for both crops. The libraries were screened by HaeIII RFLP analysis, and a total of 59 randomly selected clones were isolated and sequenced (Table 1). Theoretical restriction analysis of individual clone sequences with the restriction endonuclease HaeIII revealed eight different RFLP patterns (Table 1). Pattern A, which yielded the dominant fragment sizes observed in the whole community patterns for grass-clover soils (Fig. 1, size labels without underlining), was also dominant (100% and 81.3%) in the two libraries derived from grass-clover soils (Table 1). Accordingly, pattern B, which yielded the additional bands in the community patterns for winter wheat soils (Fig. 1, underlined size labels), was most frequently (23.1 and 48%) found in the libraries from winter wheat soils.
TABLE 1.
HaeIII RFLP types and frequencies among Ps-for/Ps-rev PCR product clone libraries from BIOORG and NOFERT plot soils planted with either winter wheat or grass-clover
| RFLP type | DGGE groupb | No. of clones (%) in four SSU rRNA gene librariesa
|
|||
|---|---|---|---|---|---|
| BIOORG
|
NOFERT
|
||||
| Grass-clover (DOK_O_GC) | Winter wheat (DOK_O_WW) | Grass-clover (DOK_N_GC) | Winter wheat (DOK_N_WW) | ||
| A | Low | 5 (100) | 5 (38.5) | 13 (81.3) | 11 (44) |
| B | Highc | 0 | 3 (23.1) | 2 (12.5) | 12 (48) |
| C | Low | 0 | 0 | 1 (6.3) | 2 (8) |
| D | High | 0 | 1 (7.7) | 0 | 0 |
| E | Low | 0 | 1 (7.7) | 0 | 0 |
| F | High | 0 | 1 (7.7) | 0 | 0 |
| G | Low | 0 | 1 (7.7) | 0 | 0 |
| H | Low | 0 | 1 (7.7) | 0 | 0 |
| Total | 5 (100) | 13 (100) | 16 (100) | 25 (100) | |
Clone libraries labeled according to their sources, i.e., BIOORG grass-clover, DOK_O_GC; BIOORG winter wheat, DOK_O_WW; NORFERT grass-clover, DOK_N_GC; and NOFERT winter wheat, DOK_N_WW.
Clones were divided into a lower and a higher denaturing group according to their migration positions in DGGE gels relative to the standard (Fig. 2).
One clone from the DOK_N_GC library migrated with the lower DGGE group.
Of the 59 isolated clones, 45 differing within the 300 bp amplified by nested PCR were used for DGGE classification into lower and higher denaturing groups (Fig. 2). Clones characterized by HaeIII RFLP types A, C, E, G, and H were exclusively assigned to the lower denaturing group (Table 1). Clones with RFLP types B, D, and F were assigned to the higher denaturing group, except for clone DOK_N_GC 60 (RFLP type B), which migrated in the lower denaturing group. Five clones (DOK_N_WW 07, 45, 21, 13, and 52) from the higher denaturing group migrated at exactly the positions of the two additional bands observed for the NOFERT system planted with winter wheat (data not shown).
Phylogenetic analysis.
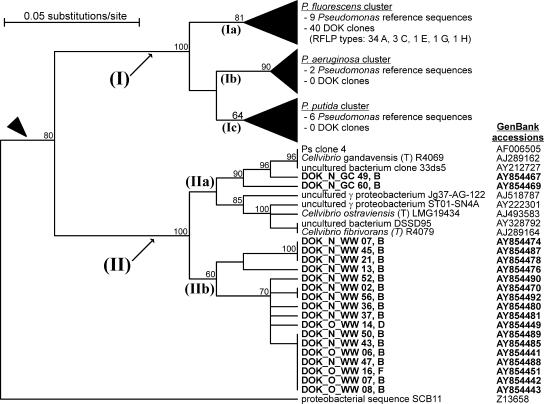
Cluster analysis revealed that all 59 sequences matched the target range of the Pseudomonas-selective primers and were associated with three branches within the Pseudomonadaceae (Fig. 3). Forty sequences (68%) grouped within the P. fluorescens branch (Fig. 3, branch Ia) of the Pseudomonas (sensu stricto) cluster (Fig. 3, cluster I). In DGGE profiles, these sequences corresponded to the lower denaturing group, i.e., region I in Fig. 2. Two sequences (3%), one in the lower (DOK_N_GC 60) and the other in the higher (DOK_N_GC 49) denaturing group, were associated with the genus Cellvibrio (Fig. 3, branch IIa). Seventeen sequences (29%) formed a novel cluster (Fig. 3, branch IIb) located close to Cellvibrio and therefore termed Cellvibrio-related Pseudomonadaceae (CRP). In DGGE analysis, these sequences migrated in the higher denaturing group, i.e., region II in Fig. 2. Clones DOK_N_WW 07, 13, 21, 45, and 52 associated with the CRP cluster and accounted for the two unique bands in the soil from winter wheat plots in the NOFERT system (Fig. 2, closed arrowheads).
FIG. 3.
Phylogenetic tree of SSU rRNA gene sequences amplified with Pseudomonas-selective PCR primers. Sequences from this study are printed in bold and are annotated with their RFLP types (A to H; see Table 1). The arrowhead marks the branch including the family Pseudomonadaceae. Cluster I includes the genus Pseudomonas (sensu stricto); collapsed subclusters Ia, Ib, and Ic are defined by the marker species P. fluorescens, P. aeruginosa, and P. putida, respectively (49). Cluster II includes the genus Cellvibrio (IIa) and CRP (IIb). The proteobacterial sequence SCB11 represents the outgroup, as originally defined for the design of the Ps-for/Ps-rev primers (49). Maximum likelihood calculation supported the definition of these clusters and the locations of all retrieved clones within these clusters.
The sequences, which grouped in the P. fluorescens cluster (Fig. 3, branch Ia), originated from all four clone libraries. The sequences associated with the Cellvibrio cluster (Fig. 3, branch IIa) derived from NOFERT soils planted with grass-clover. The sequences from the CRP cluster (Fig. 3, branch IIb) derived exclusively from soils planted with winter wheat. No sequences were retrieved which clustered within the Pseudomonas aeruginosa (Fig. 3, branch Ib) and the Pseudomonas putida (Fig. 3, branch Ic) branches. Phylogenetic analyses using maximum likelihood revealed an identical basic tree topology to that by UPGMA cluster analysis (data not shown).
Selective detection of winter wheat-associated CRP.
Theoretical analyses in RDP-II revealed that in combination, the newly designed primers CRP 149-for and CRP 565-rev discriminated all published nontarget sequences by at least 3 bp, except for one uncultured γ-proteobacterium (GenBank accession number AJ518787), which contained only two mismatches within the forward and none within the reverse primer. Experimental testing of the primers was performed, with the 45 different clones as templates. Positive PCR signals were exclusively observed for clones associated with the CRP cluster (data not shown). Clones belonging to the Pseudomonas (sensu stricto) and Cellvibrio clusters (Fig. 3) yielded no PCR products (data not shown).
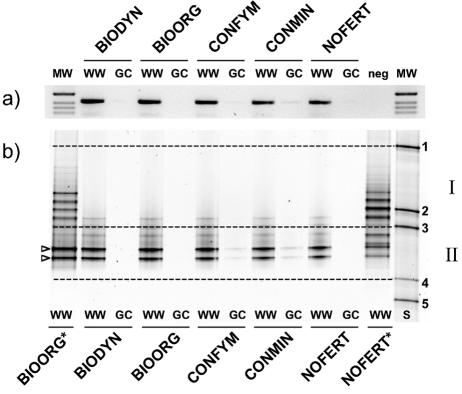
PCR analyses of soil DNA extracts from all farming systems, controls, and crops resulted in strong products of the expected size from soils planted with winter wheat, while no or faint signals were detected in soil samples from grass-clover plots (Fig. 4a). With DGGE analysis, two dominant bands were resolved (Fig. 4b). These bands corresponded to those detected by Pseudomonas-selective PCR in soils planted with winter wheat (Fig. 2 and 4b, open arrowheads). For soils from grass-clover plots, only very weak bands were detected in DGGE patterns from the CONFYM and CONMIN systems, whereas no signals were detected for the BIODYN, BIOORG, and NOFERT systems. This finding was in perfect agreement with the Pseudomonas-selective DGGE profiles (Fig. 2).
FIG. 4.
PCR (a) and DGGE (b) analyses of CRP SSU rRNA gene fragments amplified from bulk soil DNA extracts. Analyzed samples included five different soils from the DOK field experiment (lanes BIODYN, BIOORG, CONFYM, CONMIN, and NOFERT) planted with winter wheat (lanes WW) or grass-clover (lanes GC). For PCR analysis, a 1-kb marker (lane MW) was used as a DNA size standard, and a negative control (neg) was included. The size of the CRP PCR products was 417 bp. The migration standard (S) and migration regions (I and II) for DGGE analysis are described in the legend to Fig. 2. Asterisks designate community DGGE patterns from soils and were included for direct comparison to the data shown in Fig. 2. Open arrowheads, dominant bands detected in all winter wheat soils.
DISCUSSION
Data from RFLP and DGGE profiling were directly comparable because the analyses were based on identical PCR products generated with the primer pair Ps-for and Ps-rev (Fig. 1 and 2) (49). Both techniques produced consistent results by clearly discriminating between target populations in soils planted with winter wheat and grass-clover. DGGE analysis yielded higher resolution within the Pseudomonas (sensu stricto) and CRP clusters but required a nested PCR step, potentially leading to increased bias by preferential amplification of certain sequence types (42). On the other hand, the limited resolution power of the applied RFLP approach for analysis of Pseudomonas spp. could be overcome by using multiple enzymes for the digestion of PCR products (32). Furthermore, both profiling techniques yielded objectively analyzable information for detection of and discrimination between Pseudomonas spp. and CRP. In RFLP analysis and DGGE analysis, fragment sizes of particular bands and lower and higher denaturing regions, respectively, represented the diagnostic criteria (Fig. 1 and 2). Although performed on pooled DNA samples, our analyses allowed us to determine that both fingerprinting techniques do not have the sensitivity to resolve possible differences within the soil Pseudomonas (sensu stricto) communities. However, we could clearly demonstrate a pronounced crop effect on the CRP community.
The majority of SSU rRNA gene sequences retrieved, i.e., 55% for winter wheat plots and 90% for grass-clover plots, were derived from Pseudomonas (sensu stricto) (Table 1; Fig. 3). These sequences exclusively associated with the P. fluorescens branch, although our theoretical analyses and previous experimental testing (49) revealed that the Ps primers also amplify SSU rRNA genes from the P. aeruginosa and P. putida clusters (49). The results from the present study underscore the importance of repeated theoretical and experimental reevaluations of primer specificities as new sequences are continuously added to databases. In silico reevaluation of primers Ps-for and Ps-rev (49) in RDP-II revealed high coverage (91.8%) and specificity (99.1%) for the target group Pseudomonas (sensu stricto) and fully justified the use of these primers in our analysis. Subsequent phylogenetic analysis of 59 clone sequences demonstrated that the primers were highly selective for the originally defined target range characterized by the outgroup sequence SCB11 (Fig. 3) (49).
A group of sequences isolated from soils planted with winter wheat clustered apart from Pseudomonas (sensu stricto) and formed the novel CRP cluster (Fig. 3, branch IIb). It is premature to define whether CRP constitute a new species of the genus Cellvibrio or a new genus of the Pseudomonadaceae. The isolation of CRP and detailed analyses will be required for this definition. It was surprising that close relatives of Pseudomonas (sensu stricto) have only rarely been detected in soil and that the genus Cellvibrio has only been recently isolated by selective cultivation from bulk soil (22, 45). These isolates were reported to display agarolytic or cellulolytic properties. A few Cellvibrio SSU rRNA gene sequences have also recently been retrieved from soil and plants, including bulk agricultural soil (27, 49), maize rhizospheres (36), and potato endospheres (38). The detection of Cellvibrio spp. in plant rhizo- and endospheres suggested that they may be preferentially associated with specific plants. This would be in agreement with our finding that the presence of CRP was highly correlated with winter wheat.
The novel CRP primers, i.e., CRP 149-for and CRP 565-rev, allowed us to confirm the results obtained with RFLP and DGGE analyses based on Pseudomonas-selective PCR. Forty PCR cycles were used for the detection of Pseudomonas spp. and CRP, suggesting a comparable abundance of CRP and Pseudomonas spp. in soils planted with winter wheat. The same conclusion could be drawn from the comparable band intensities observed for Pseudomonas spp. and CRP in the RFLP and DGGE analyses (Fig. 1 and 2). Pseudomonas spp. have been reported to account for approximately 1% of the bacterial communities in soil (24), which may also represent the approximate abundance of CRP in the winter wheat plots. If more detailed quantitative analyses of these microbial groups are required, specific quantitative PCR approaches will have to be developed (43). The independent analysis of CRP also provided strong evidence that the observed differences in RFLP and DGGE profiles (Fig. 1 and 2) reflected actual differences in bacterial community structures and were not related to a PCR bias. The identification of changing components in genetic profiles by cloning and sequencing has been frequently performed in microbial community analysis (7, 41). However, the gained information has rarely been confirmed by designing specific primers for independent analysis of the changing community components (31). It has previously been shown that such a strategy can also provide tools with increased detection sensitivities (31). We strongly believe that, if consistently applied, this approach will provide an increasing set of tools for indicator detection of high value for research and soil quality analyses.
The main agricultural factor determining CRP community structure in the DOK field soils appeared to be the plant type. Currently, it is unknown whether CRP are promoted in the presence of winter wheat or whether they are excluded by the grass-clover ley. Applying the developed detection tools to soil samples from different crops at different time points during growth and at different locations will provide further information regarding the mechanisms involved in enriching CRP in soils and their possible functions. Previous analyses of the same DOK field samples have focused on the bacterial domain by using T-RFLP analyses (48). These data revealed that the type of fertilizer, i.e., organic versus mineral or no fertilization, dominated plant effects (48), suggesting that bacterial subgroups which responded to the fertilizer regimen were present. In agreement with these results, Kennedy et al. (19) reported that bacterial community structures were influenced by lime and nitrogen additions, and to a much smaller extent, by the plant species. This apparent difference compared to our findings may be explained by the fact that a bacterial community profile represents a composite of various bacterial subgroups, which may display different responses. Relatively small groups may either not be detected in bacterial community profiles (8, 29) or be outweighed by other components in statistical analysis (11, 18), thus emphasizing the necessity of genus-level analyses in microbial ecology investigations.
Conclusions.
With RFLP and DGGE analyses, no clear differences were detected in the Pseudomonas soil communities in the DOK field experiment. Therefore, we had to reject our initial hypothesis that Pseudomonas communities respond to the different treatments in the DOK long-term field experiment. However, the novel group of soil CRP identified in the present study appeared to be highly responsive to the crop type. The isolation and detailed characterization of CRP will allow us to taxonomically and functionally define this interesting novel soil bacterial group.
Acknowledgments
We thank Anne Grundschober for technical assistance with cloning and sequencing analysis. We acknowledge Andreas Fliessbach and Frank Rasche for help with soil sampling and DNA extraction and Michel Aragno for providing the DGGE migration standard. Martin Hartmann and Jürg Enkerli are acknowledged for their valuable comments on the manuscript.
REFERENCES
- 1.Achouak, W., J. M. Thiery, P. Roubaud, and T. Heulin. 2000. Impact of crop management on intraspecific diversity of Pseudomonas corrugata in bulk soil. FEMS Microbiol. Ecol. 31:11-19. [DOI] [PubMed] [Google Scholar]
- 2.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2005. GenBank. Nucleic Acids Res. 33:D34-D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergsma-Vlami, M., M. E. Prins, and J. M. Raaijmakers. 2005. Influence of plant species on population dynamics, genotypic diversity and antibiotic production in the rhizosphere by indigenous Pseudomonas spp. FEMS Microbiol. Ecol. 52:59-69. [DOI] [PubMed] [Google Scholar]
- 4.Bürgmann, H., M. Pesaro, F. Widmer, and J. Zeyer. 2001. A strategy for optimizing quality and quantity of DNA extracted from soil. J. Microbiol. Methods 45:7-20. [DOI] [PubMed] [Google Scholar]
- 5.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duineveld, B. M., G. A. Kowalchuk, A. Keijzer, J. D. van Elsas, and J. A. van Veen. 2001. Analysis of bacterial communities in the rhizosphere of chrysanthemum via denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA as well as DNA fragments coding for 16S rRNA. Appl. Environ. Microbiol. 67:172-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2000. Assessment of microbial diversity in four southwestern U.S. soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 66:2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fliessbach, A., and P. Mäder. 2000. Microbial biomass and size-density fractions differ between soils of organic and conventional agricultural systems. Soil Biol. Biochem. 32:757-768. [Google Scholar]
- 10.Fliessbach, A., and P. Mäder. 1997. Carbon source utilization by microbial communities in soils under organic and conventional farming practices, p. 109-120. In H. Insam and A. Rangger (ed.), Microbial communities—functional versus structural approaches. Springer-Verlag, Berlin, Germany.
- 11.Fromin, N., J. Hamelin, S. Tarnawski, D. Roesti, K. Jourdain-Miserez, N. Forestier, S. Teyssier-Cuvelle, F. Gillet, M. Aragno, and P. Rossi. 2002. Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns. Environ. Microbiol. 4:634-643. [DOI] [PubMed] [Google Scholar]
- 12.Garbeva, P., J. A. van Veen, and J. D. van Elsas. 2004. Assessment of the diversity, and antagonism towards Rhizoctonia solani AG3, of Pseudomonas species in soil from different agricultural regimes. FEMS Microbiol. Ecol. 47:51-64. [DOI] [PubMed] [Google Scholar]
- 13.Giovannoni, S. J., E. F. Delong, G. J. Olsen, and N. R. Pace. 1988. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J. Bacteriol. 170:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girvan, M. S., J. Bullimore, A. S. Ball, J. N. Pretty, and A. M. Osborn. 2004. Responses of active bacterial and fungal communities in soils under winter wheat to different fertilizer and pesticide regimens. Appl. Environ. Microbiol. 70:2692-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girvan, M. S., J. Bullimore, J. N. Pretty, A. M. Osborn, and A. S. Ball. 2003. Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl. Environ. Microbiol. 69:1800-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall, J. A., D. Peirson, S. Ghosh, and B. R. Glick. 1996. Root elongation in various agronomic crops by the plant growth promoting rhizobacterium Pseudomonas putida GR12-2. Isr. J. Plant Sci. 44:37-42. [Google Scholar]
- 17.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 18.Hartmann, M., B. Frey, R. Kölliker, and F. Widmer. 2005. Semi-automated genetic analyses of soil microbial communities: comparison of T-RFLP and RISA based on descriptive and discriminative statistical approaches. J. Microbiol. Methods 61:349-360. [DOI] [PubMed] [Google Scholar]
- 19.Kennedy, N., E. Brodie, J. Connolly, and N. Clipson. 2004. Impact of lime, nitrogen and plant species on bacterial community structure in grassland microcosms. Environ. Microbiol. 6:1070-1080. [DOI] [PubMed] [Google Scholar]
- 20.Kuske, C. R., L. O. Ticknor, M. E. Miller, J. M. Dunbar, J. A. Davis, S. M. Barns, and J. Belnap. 2002. Comparison of soil bacterial communities in rhizospheres of three plant species and the interspaces in an arid grassland. Appl. Environ. Microbiol. 68:1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latour, X., T. S. Corberand, G. Laguerre, F. Allard, and P. Lemanceau. 1996. The composition of fluorescent pseudomonad populations associated with roots is influenced by plant and soil type. Appl. Environ. Microbiol. 62:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lednicka, D., J. Mergaert, M. C. Cnockaert, and J. Swings. 2000. Isolation and identification of cellulolytic bacteria involved in the degradation of natural cellulosic fibres. Syst. Appl. Microbiol. 23:292-299. [DOI] [PubMed] [Google Scholar]
- 23.Lemanceau, P., and C. Alabouvette. 1991. Biological control of fusarium diseases by fluorescent Pseudomonas and nonpathogenic Fusarium. Crop Prot. 10:279-286. [Google Scholar]
- 24.Lloyd-Jones, G., A. D. Laurie, and A. C. Tizzard. 2005. Quantification of the Pseudomonas population in New Zealand soils by fluorogenic PCR assay and culturing techniques. J. Microbiol. Methods 60:217-224. [DOI] [PubMed] [Google Scholar]
- 25.Mäder, P., S. Edenhofer, T. Boller, A. Wiemken, and U. Niggli. 2000. Arbuscular mycorrhizae in a long-term field trial comparing low-input (organic, biological) and high-input (conventional) farming systems in a crop rotation. Biol. Fertil. Soils 31:150-156. [Google Scholar]
- 26.Mäder, P., A. Fliessbach, D. Dubois, L. Gunst, P. Fried, and U. Niggli. 2002. Soil fertility and biodiversity in organic farming. Science 296:1694-1697. [DOI] [PubMed] [Google Scholar]
- 27.Marilley, L., and M. Aragno. 1999. Phylogenetic diversity of bacterial communities differing in degree of proximity of Lolium perenne and Trifolium repens roots. Appl. Soil Ecol. 13:127-136. [Google Scholar]
- 28.Milling, A., K. Smalla, F. X. Maidl, M. Schloter, and J. C. Munch. 2005. Effects of transgenic potatoes with an altered starch composition on the diversity of soil and rhizosphere bacteria and fungi. Plant Soil 266:23-39. [Google Scholar]
- 29.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S ribosomal RNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penalozavazquez, A., G. L. Mena, L. Herreraestrella, and A. M. Bailey. 1995. Cloning and sequencing of the genes involved in glyphosate utilization by Pseudomonas pseudomallei. Appl. Environ. Microbiol. 61:538-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pesaro, M., F. Widmer, G. Nicollier, and J. Zeyer. 2003. Effects of freeze-thaw stress during soil storage on microbial communities and methidathion degradation. Soil Biol. Biochem. 35:1049-1061. [Google Scholar]
- 32.Porteous, L. A., F. Widmer, and R. J. Seidler. 2002. Multiple enzyme restriction fragment length polymorphism analysis for high resolution distinction of Pseudomonas (sensu stricto) 16S rRNA genes. J. Microbiol. Methods 51:337-348. [DOI] [PubMed] [Google Scholar]
- 33.Rangarajan, S., P. Loganathan, L. M. Saleena, and S. Nair. 2001. Diversity of pseudomonads isolated from three different plant rhizospheres. J. Appl. Microbiol. 91:742-749. [DOI] [PubMed] [Google Scholar]
- 34.Reiter, B., N. Wermbter, S. Gyamfi, H. Schwab, and A. Sessitsch. 2003. Endophytic Pseudomonas spp. populations of pathogen-infected potato plants analysed by 16S rDNA- and 16S rRNA-based denaturating gradient gel electrophoresis. Plant Soil 257:397-405. [Google Scholar]
- 35.Sands, D. C., M. N. Schroth, and D. C. Hildebrand. 1970. Taxonomy of phytopathogenic pseudomonads. J. Bacteriol. 101:9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmalenberger, A., and C. C. Tebbe. 2002. Bacterial community composition in the rhizosphere of a transgenic, herbicide-resistant maize (Zea mays) and comparison to its non-transgenic cultivar Bosphore. FEMS Microbiol. Ecol. 40:29-37. [DOI] [PubMed] [Google Scholar]
- 37.Schroth, M. N., V. B. Vitanza, and D. C. Hildebrand. 1971. Pathogenic and nutritional variation in halo blight group of fluorescent pseudomonads of bean. Phytopathology 61:852-857. [Google Scholar]
- 38.Sessitsch, A., B. Reiter, U. Pfeifer, and E. Wilhelm. 2002. Cultivation-independent population analysis of bacterial endophytes in three potato varieties based on eubacterial and Actinomycetes-specific PCR of 16S rRNA genes. FEMS Microbiol. Ecol. 39:23-32. [DOI] [PubMed] [Google Scholar]
- 39.Shapir, N., and R. T. Mandelbaum. 1997. Atrazine degradation in subsurface soil by indigenous and introduced microorganisms. J. Agric. Food Chem. 45:4481-4486. [Google Scholar]
- 40.Sharma, A., B. N. Johri, A. K. Sharma, and B. R. Glick. 2003. Plant growth-promoting bacterium Pseudomonas sp. strain GRP(3) influences iron acquisition in mung bean (Vigna radiata L. Wilzeck). Soil Biol. Biochem. 35:887-894. [Google Scholar]
- 41.Smalla, K., G. Wieland, A. Buchner, A. Zock, J. Parzy, S. Kaiser, N. Roskot, H. Heuer, and G. Berg. 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67:4742-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van de Peer, Y., and R. De Wachter. 1994. Treecon for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 45.Voget, S., C. Leggewie, A. Uesbeck, C. Raasch, K. E. Jaeger, and W. R. Streit. 2003. Prospecting for novel biocatalysts in a soil metagenome. Appl. Environ. Microbiol. 69:6235-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weller, D. M., J. M. Raaijmakers, B. B. M. Gardener, and L. S. Thomashow. 2002. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40:309-348. [DOI] [PubMed] [Google Scholar]
- 47.Widmer, F., A. Fliessbach, E. Laczko, J. Schulze-Aurich, and J. Zeyer. 2001. Assessing soil biological characteristics: a comparison of bulk soil community DNA-, PLFA-, and Biolog™-analyses. Soil Biol. Biochem. 33:1029-1036. [Google Scholar]
- 48.Widmer, F., F. Rasche, M. Hartmann, and A. Fliessbach. Community structures and substrate utilization of bacteria in soils from organic and conventional farming systems of the DOK long-term field experiment. Appl. Soil Ecol., in press.
- 49.Widmer, F., R. J. Seidler, P. M. Gillevet, L. S. Watrud, and G. D. Di Giovanni. 1998. A highly selective PCR protocol for detecting 16S rRNA genes of the genus Pseudomonas (sensu stricto) in environmental samples. Appl. Environ. Microbiol. 64:2545-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young, J. M. 1991. Pathogenicity and identification of the lilac pathogen, Pseudomonas syringae pv. syringae van Hall 1902. Ann. Appl. Biol. 118:283-298. [Google Scholar]