Abstract
The roles of an aspartate and an arginine, which are completely conserved in the active sites of β-class carbonic anhydrases, were investigated by steady-state kinetic analyses of replacement variants of the β-class enzyme (Cab) from the archaeon Methanobacterium thermoautotrophicum. Previous kinetic analyses of wild-type Cab indicated a two-step zinc-hydroxide mechanism of catalysis in which the kcat/Km value depends only on the rate constants for the CO2 hydration step, whereas kcat also depends on rate constants from the proton transfer step (K. S. Smith, N. J. Cosper, C. Stalhandske, R. A. Scott, and J. G. Ferry, J. Bacteriol. 182:6605-6613, 2000). The recently solved crystal structure of Cab shows the presence of a buffer molecule within hydrogen bonding distance of Asp-34, implying a role for this residue in the proton transport step (P. Strop, K. S. Smith, T. M. Iverson, J. G. Ferry, and D. C. Rees, J. Biol. Chem. 276:10299-10305, 2001). The kcat/Km values of Asp-34 variants were decreased relative to those of the wild type, although not to an extent which supports an essential role for this residue in the CO2 hydration step. Parallel decreases in kcat and kcat/Km values for the variants precluded any conclusions regarding a role for Asp-34 in the proton transfer step; however, the kcat of the D34A variant was chemically rescued by replacement of 2-(N-morpholino)propanesulfonic acid buffer with imidazole at pH 7.2, supporting a role for the conserved aspartate in the proton transfer step. The crystal structure of Cab also shows Arg-36 with two hydrogen bonds to Asp-34. Arg-36 variants had both kcat and kcat/Km values that were decreased at least 250-fold relative to those of the wild type, establishing an essential function for this residue. Imidazole was unable to rescue the kcat of the R36A variant; however, partial rescue of the kinetic parameter was obtained with guanidine-HCl indicating that the guanido group of this residue is important.
Carbonic anhydrase is a zinc-containing enzyme that catalyzes the reversible hydration of carbon dioxide:
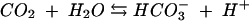 |
(1) |
This enzyme, which is present in species from all three domains of life, plays a critical role in many diverse physiological processes such as respiration, photosynthesis, and CO2 fixation.
Based on amino acid sequence comparisons, carbonic anhydrases belong to four genetically distinct classes (α, β, γ, and δ) of independent origins (38). The crystal structures for representatives of the α, β, and γ classes have now been determined (3, 7, 9-13, 16, 18, 19, 23, 26, 35, 36). Although the structure of the recently identified δ-class prototype from the diatom Thalassiosira weissflogii has not yet been solved, extended X-ray absorption fine-structure analyses suggest that the active-site zinc is coordinated by three histidines and one water molecule, as found in the α and γ classes (6). The structures of the β-class carbonic anhydrases (7, 18, 26, 36) indicate striking differences between this class and the others. For example, the active-site zinc in the β-class enzymes is coordinated by two cysteines, one histidine, and one water molecule.
The kinetic properties of the human α-class isozymes have been comprehensively investigated and follow a common zinc-hydroxide mechanism for catalysis (24, 30). The catalytically active group in this mechanistic model is the zinc-bound water, which ionizes to a metal-bound hydroxyl which attacks CO2. Despite considerable structural differences in the active sites of these enzymes, the catalytic mechanisms of both γ- and β-class enzymes resemble those of human α-class HCA II (1, 14, 15, 28, 31). The overall enzyme-catalyzed reaction occurs in two mechanistically distinct steps (where E = enzyme and B = buffer):
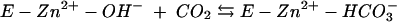 |
(2a) |
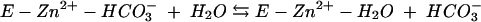 |
(2b) |
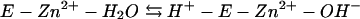 |
(2c) |
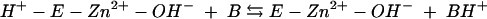 |
(2d) |
The first step is the interconversion between carbon dioxide and bicarbonate (equations 2a and 2b) and involves the nucleophilic attack of the zinc-bound hydroxyl on the CO2 molecule. In the α-class enzyme, the zinc-bound oxygen forms a hydrogen bond with Thr-199 (22, 42, 43). The role of this conserved gatekeeper residue is to prevent nonprotonated atoms from binding effectively. In addition, Thr-199 has been proposed to electrophilically activate the CO2 molecule by forming a hydrogen bond with CO2 through its backbone amide. The second step of the proposed mechanism is the regeneration of the zinc hydroxide at the active site of the enzyme (equations 2c and 2d), involving proton transfer events. The kcat/Km value depends only on the rate constants for the CO2 hydration steps (equations 2a and 2b), whereas kcat also depends on rate constants from the proton transfer steps (equations 2c and 2d). Carbonic anhydrases with slower rates (kcat of less than 104 s−1) transfer the proton directly to buffer or water molecules in solution, as this is the fastest rate at which protons can transfer from an acidic group with a pKa of 7 to water (25). Faster carbonic anhydrases with turnover numbers in the range of 104 to 106 s−1 transfer the proton from the metal-bound water molecule to an intermediate proton shuttle residue and then to an external buffer molecule. In the α-class HCAII, the proton first undergoes intramolecular transfer to the proton shuttle residue His-64 (40). In the γ-class enzyme Cam, this proton transfer is to Glu-84 (37). The proton is subsequently transferred to an accepting buffer molecule in the surrounding media during an intermolecular proton transfer step. Imidazole is able to rescue the kcat of variant carbonic anhydrases in which the proton shuttle residue is replaced with alanine, presumably by entering the active site and replacing the function of the proton shuttle residue.
Previously, the lack of a structure for a β-class carbonic anhydrase hindered work on this class relative to that of the α class and even the recently discovered γ class. However, the structures of four β-class enzymes have been reported in the last 2 years (7, 18, 26, 36). Comparison of these structures, in addition to phylogenetic analysis, indicates that the β class is composed of two subclasses (34, 38). The plant-type subclass is represented by the Pisum sativum (pea) enzyme and is composed of enzymes from the Eucarya domain and gram-negative species in the Bacteria domain. The other subclass, the cab-type, is represented by the enzyme Cab from the thermophilic methane-producing archaeon Methanobacterium thermoautotrophicum. The cab-type subclass includes carbonic anhydrases from the Archaea domain and gram-positive species from the Bacteria domain.
Little is known concerning what residues play a critical role in the zinc hydroxide mechanism for the β-class carbonic anhydrases compared with the α and γ classes. Only five residues are completely conserved in the diverse β class (33, 34). Three of these residues (two cysteines and one histidine) chelate the active-site zinc. Roles for the other two conserved residues, an aspartate and an arginine, have not been determined. A variant of the Spinacia oleracea enzyme in which the conserved aspartate was replaced (D152N) retained approximately 1% of the specific activity of the wild-type enzyme; however, no kinetic analyses were reported (4). Therefore, the role of the conserved aspartate in the β-class enzymes remains unclear. Here, we report the first kinetic characterization of Cab variants with replacements at the conserved aspartate (Asp-34) and arginine (Arg-36).
MATERIALS AND METHODS
Enzyme purification.
The wild-type and variant carbonic anhydrases were heterologously produced in Escherichia coli BL21(DE3) (Novagen, Inc.) and purified as previously described (31, 32). Enzyme activity during purification was measured at room temperature by using a modification of the electrometric method of Wilbur and Anderson (41). Protein concentrations were determined by the Bradford method with Bio-Rad dye reagent and bovine serum albumin (Sigma) as the standard (5).
Site-directed mutagenesis.
Mutagenesis was performed by oligonucleotide-directed in vitro mutagenesis by using the QuikChange kit (Stratagene) according to the manufacturer's instructions. Plasmid pMBTCA13 (32), a derivative of the plasmid pET15a (Novagen, Inc.) containing the M. thermoautotrophicum cab gene, was the target plasmid for site-directed mutagenesis. The mutations were confirmed by dye termination cycle sequencing with an ABI PRISM 377 DNA sequencer (Perkin Elmer Life Sciences) at the Nucleic Acid Facility at Pennsylvania State University.
Steady-state kinetics.
Initial rates of CO2 hydration were determined by stopped-flow spectroscopy (KinTek stopped-flow apparatus; KinTek, State College, Pa.) at 25°C by using the changing pH indicator method (17). Saturated solutions of CO2 (32.9 mM) were prepared by bubbling CO2 into distilled deionized water at 25°C, and the final CO2 concentrations (6 to 24 mM) were obtained by dilution with N2-saturated buffer. The following buffer and pH indicator pairs (and wavelengths) were used: at pH 6.4 and 6.8, MES [2-(N-morpolino)ethanesulfonic acid] (pKa = 6.1) and chlorophenol red (574 nm); at pH 7.2, MOPS [2-(N-morpholino)propanesulfonic acid] (pKa = 7.2) and p-nitrophenol (400 nm); at pH 7.2, imidazole (pKa = 6.9) and p-nitrophenol (400 nm); at pH 7.6, HEPES (pKa = 7.5) and phenol red (557 nm); at pH 8.0 to 9.0, TAPS [N-tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid] (pKa = 8.4) and m-cresol purple (578 nm). The observed initial rates were corrected for the uncatalyzed rate of the reaction, which was at least five times lower than the catalyzed rate. The steady-state parameters kcat and kcat/Km and their standard errors were then determined by fitting the observed initial rates to the Michaelis-Menten equation. All fits described were performed by using KaleidaGraph (Synergy Software, Reading, Pa.).
Materials.
Chemicals were purchased from Sigma, VWR Scientific, or Fisher. Oligonucleotide primers for site-directed mutagenesis were obtained from Integrated DNA Technologies, Inc.
RESULTS
Initial characterization of Asp-34 and Arg-36 replacement variants of Cab.
The roles of the two completely conserved residues in the β class (excluding zinc ligands) were examined in Cab to begin to understand the catalytic mechanism of β-class carbonic anhydrases. Thus, Asp-34 and Arg-36 of Cab were replaced by site-directed mutagenesis (Table 1), and the variant enzymes were kinetically characterized. Gel filtration chromatography indicated that the variants are all tetramers, in accordance with the wild-type enzyme. In addition, all the variants exhibited patterns of temperature stability similar to those of the wild-type (data not shown). These results indicate that the variants experienced no gross structural perturbations.
TABLE 1.
Steady-state kinetic parameters for wild-type Cab and Cab variants with TAPS buffer (pH 8.5)a
| Variant | kcat (104 s−1) | Km in CO2 (mM) | kcat/Km (106 s−1 M−1) |
|---|---|---|---|
| Wild type | 2.4 ± 0.2 | 2.9 ± 0.3 | 8.3 ± 0.7 |
| D34A | 0.42 ± 0.01 | 3.6 ± 0.4 | 1.2 ± 0.3 |
| D34E | 0.36 ± 0.1 | 1.8 ± 0.2 | 2.0 ± 0.5 |
| R36A | 0.0046 ± 0.0008 | 36.2 ± 1.5 | 0.0013 ± 0.0005 |
| R36K | 0.0092 ± 0.0007 | 7.2 ± 0.2 | 0.013 ± 0.004 |
| D34A-R36A | 0.052 ± 0.0006 | 9.2 ± 0.8 | 0.56 ± 0.03 |
Values are means ± standard errors.
Kinetic analysis of Asp-34 and Arg-36 replacement variants of Cab.
The variants were analyzed by stopped-flow spectroscopy in the direction of CO2 hydration. As previously shown for wild-type Cab (31), the assay progress curves for all the variants were consistent with Michaelis-Menten kinetics (data not shown). The kcat or kcat/Km values for both Asp-34 replacement variants decreased no more than 10-fold relative to those of the wild type when assayed with either MOPS or TAPS buffer (Tables 1 and 2); however, both of the Arg-36 replacement variants had kcat or kcat/Km values that were reduced between 250- and 6,000-fold compared to those of the wild type. The D36K variant had kcat and kcat/Km values greater than those of the D36A variant. Analysis of the D34A-R36A double variant revealed steady-state parameters that were intermediate to those of the D34A and R36A variants.
TABLE 2.
Steady-state kinetic parameters for wild-type Cab and Cab variants with MOPS and imidazole buffers (pH 7.2)
| Variant | Buffer (50 mM) | kcat (104 s−1) | Km in CO2 (mM) | kcat/Km (106 s−1 M−1) |
|---|---|---|---|---|
| Wild type | MOPS | 1.1 ± 0.2 | 2.7 ± 0.2 | 4.1 ± 0.3 |
| Imidazole | 1.3 ± 0.3 | 4.2 ± 0.3 | 3.2 ± 0.4 | |
| D34A | MOPS | 0.11 ± 0.1 | 3.5 ± 0.2 | 2.9 ± 0.2 |
| Imidazole | 0.91 ± 0.05 | 15.3 ± 1.5 | 0.6 ± 0.3 | |
| R36A | MOPS | 0.0024 ± 0.0004 | 35.3 ± 1.7 | 0.00067 ± 0.00003 |
| Imidazole | 0.0030 ± 0.0003 | 47.1 ± 2.2 | 0.00064 ± 0.00006 | |
| D34A-R36A | MOPS | 0.026 ± 0.03 | 9.2 ± 0.3 | 0.029 ± 0.05 |
| Imidazole | 0.15 ± 0.02 | 5.8 ± 0.2 | 0.26 ± 0.08 |
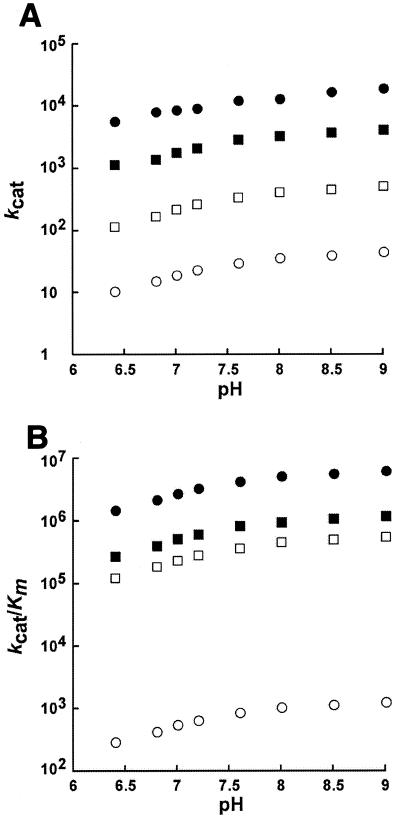
The kcat and kcat/Km values for wild-type Cab and all of the variants were pH dependent between pH 6.4 and 9.0 and increased with increasing pH (Fig. 1). These results indicate that the fundamental zinc hydroxide mechanism is operable for the variants, as determined previously for wild-type Cab (31). As previously reported for wild-type Cab (31), the pH profiles of either steady-state parameter for the variants could not be fit to a theoretical titration curve with one, two, or three ionizations.
FIG. 1.
pH-dependent steady-state kinetic parameters for wild-type Cab and Cab variants. CO2 hydration was measured at 25°C in 50 mM buffer with an ionic strength of 0.1 M. The observed steady-state parameters were determined over a pH range of 6.4 to 9.0. (A) kcat values for wild-type (filled circles), D34A (filled squares), R36A (open circles), and D34A-R36A (open squares) enzymes are shown. (B) kcat/Km values for wild-type (filled circles), D34A (filled squares), R36A (open circles), and D34A-R36A (open squares) enzymes are shown.
Imidazole rescue of Cab variants.
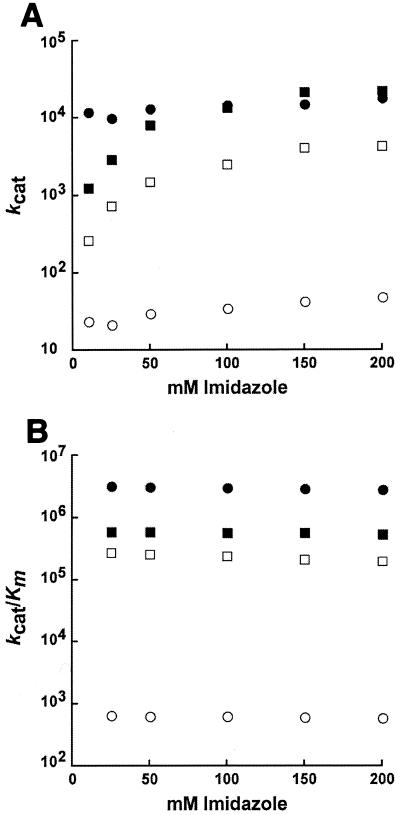
The ability of imidazole to rescue kcat for both α- and γ-class carbonic anhydrase variants is considered strong experimental evidence for involvement of the replaced residue in the intramolecular proton transfer step of catalysis. Thus, the kcat values for wild-type Cab and the variants were determined in the presence of either imidazole or MOPS buffer at pH 7.2 (Table 2). The kcat values for the wild type and the R36A variant were not significantly altered with either buffer. However, the kcat values for the D34A and D34A-R36A variants were eight- and sixfold greater in the presence of imidazole than in the presence of MOPS. The kcat value for the D34A variant was also dependent on the imidazole concentration, with kcat approaching a maximum value at approximately 150 mM (Fig. 2A). The kcat values for the wild type and the R36A variant were independent of the concentration (0 to 200 mM) of imidazole (Fig. 2A). Replacing MOPS with imidazole did not significantly change the kcat/Km value for the wild type or the R36A variant (Table 2). However, the kcat/Km value for the D34A variant decreased nearly 5-fold, whereas the kinetic parameter for the D34A-R36A variant increased nearly 10-fold with imidazole compared to MOPS. The kcat/Km value for the wild type or the variants was independent of the concentration (25 to 200 mM) of imidazole (Fig. 2B).
FIG. 2.
Dependence on imidazole concentration of steady-state kinetic parameters for wild-type Cab and Cab variants. CO2 hydration was measured at 25°C with an ionic strength of 0.1 M and buffer concentrations of 20 to 200 mM imidazole, pH 7.2. (A) kcat values for wild-type (filled circles), D34A (filled squares), R36A (open circles), and D34A-R36A (open squares) enzymes are shown. (B) kcat/Km values for wild-type (filled circles), D34A (filled squares), R36A (open circles), and D34A-R36A (open squares) enzymes are shown.
Guanidine-HCl rescue of Cab variants.
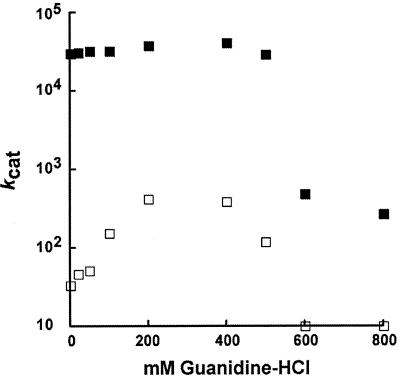
Replacement of the conserved Arg-36 had a substantial effect on the kcat and kcat/Km values of Cab, suggesting that the guanido side chain of Arg-36 may be important for catalysis. The steady-state parameters of the R36A variant were examined in the presence of increasing concentrations of guanidine-HCl (Fig. 3) to address the function of this residue. The kcat values of the wild-type Cab remained constant between 0 and 400 mM guanidine-HCl, whereas the kcat of the R36A variant increased between 0 and 200 mM. However, guanidine-HCl concentrations greater than 400 mM decreased the kcat values for both the wild type and the variant. The effect of guanidine-HCl on the kcat value of the R36K variant was similar to that of the wild-type Cab (data not shown). Concentrations of 100 to 1,000 mM urea or methylamine had no significant effect on the kcat of either Arg-36 replacement variant (data not shown).
FIG. 3.
Effect of guanidine-HCl on the steady-state parameters of the wild type and the R36A variant. CO2 hydration was measured at 25°C in 50 mM TAPS (pH 8.5) with an ionic strength of 0.1 M in the presence of various concentrations of guanidine-HCl. kcat values for wild-type (filled squares) and R36A (open squares) enzymes are shown.
DISCUSSION
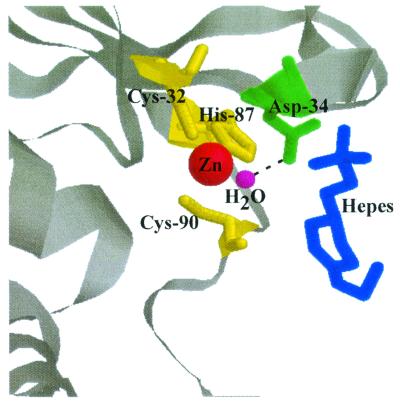
Although the β-class carbonic anhydrases appear to be the most widely distributed in nature and the only class to have been documented in all three domains (33, 38), less is known about the biochemistry of this class than about either of the other two classes. Of the β-class carbonic anhydrases, only two plant-type β-class carbonic anhydrases and one cab-type β-class carbonic anhydrase have been kinetically characterized (14, 15, 28, 31). Even before the structure of any β-class enzyme had been determined, Asp-34 and Arg-36 of Cab were obvious targets for site-specific replacement to determine their roles in catalysis. They represent the only residues that are completely conserved in all enzymes of the β class, aside from the two cysteines and one histidine which chelate zinc (33). A variant of the S. oleracea enzyme, in which the conserved aspartate (Asp-152) was replaced with asparagine (4), had 1% of the activity of the wild type when assayed in 25 mM EPPS buffer (4) at pH 8.0 and 9% of the activity of the wild type when assayed in 25 mM imidazole buffer at pH 8.0. These results are consistent with a role for Asp-152 as a proton shuttle residue; however, a kinetic analysis of the D152N variant has not been reported to further investigate this proposed role. No studies are reported which address the role of the conserved aspartate in any of the β-class carbonic anhydrases for which the crystal structure is known. Thus, the role of the conserved aspartate in the catalytic mechanism for any of the β-class carbonic anhydrases has not been resolved. Based on the crystal structure, it was proposed that one possible function for the Asp-34 of Cab is to orient CO2 for attack by the zinc-bound hydroxyl (36). The replacement of Asp-34 in Cab with either alanine or glutamate produced variants with modest decreases in kcat/Km values relative to those of the wild-type enzyme, suggesting that Asp-34 has no essential role in the CO2 hydration step of catalysis. The crystal structure of Cab complexed with a HEPES buffer molecule is also consistent with a role for Asp-34 as a proton shuttle residue (36). Asp-34 forms a hydrogen bond to the zinc-bound water molecule, and a HEPES molecule is found approximately 8 Å from the catalytic zinc, within hydrogen bonding distance of Asp-34 (Fig. 4). Therefore, one possible pathway for proton transfer in the cab-type β-class enzymes is from the zinc-bound water molecule to Asp-34 and then to HEPES. The solvent hydrogen isotope effect on kcat (2.1 ± 0.1) for wild-type Cab suggests that an intramolecular proton transfer step is at least partially rate determining (31); however, comparable decreases in both kcat and kcat/Km for the Asp-34 replacement variants preclude any conclusions from the kinetic analyses regarding a proton transfer role for this residue in catalysis. Nonetheless, imidazole rescue of kcat for the D34A variant and the D34A-R36A double variant suggests a role for Asp-34 in proton transfer. A route for proton transfer could not be discerned from the crystal structures of the Porphyridium purpureum, P. sativum, or E. coli β-class enzymes (7, 18, 26).
FIG. 4.
View of the Cab active site complexed with HEPES. The active site is shown with the three conserved zinc ligands Cys-32, His-87, and Cys-90 in yellow, the catalytic zinc in red, the zinc-bound water in magenta, the conserved Asp-34 in green, and the putative proton-accepting HEPES buffer molecule in blue. Hydrogen bonds are indicated by broken lines. The image was prepared with Web Lab Viewer.
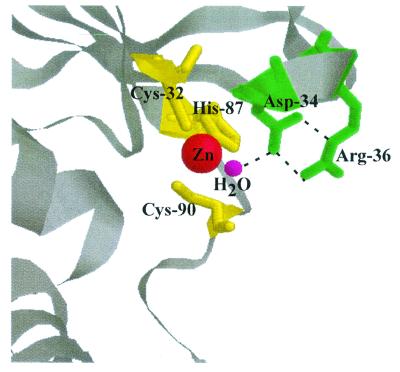
In Cab, the conserved aspartate (Asp-34) is held in place by a pair of hydrogen bonds from Arg-36, the only other residue completely conserved among the β-class sequences (36) (Fig. 5). Replacements of the conserved arginine in Cab produced variants with substantial decreases in both kcat and kcat/Km values relative to those of the wild type. Replacement of Arg-36 with lysine had a similar effect as replacement with alanine, suggesting that, in addition to the positive charge at this position, the two-point interaction of the guanido group with Asp-34 is important. Chemical rescue of kinetic parameters with guanido derivatives has been reported for arginine replacement variants of several enzymes (2, 8, 20, 21, 27, 29), including Cam, the prototype of the γ-class carbonic anhydrase (39). These derivatives are thought to occupy the cavity vacated by the arginine side chain in a productive conformation which mimics the guanido group of arginine. Guanidine-HCl rescued the kcat for the Arg-36 replacement variants of Cab. The inability of urea to rescue the variants argues against a role for guanidine-HCl in rescuing the variants simply by restoring the conformation of the active site. The inability of methylamine to rescue the variants indicates that a positive charge alone is not sufficient for rescue. The results suggest that guanidine-HCl diffuses into the active site of the variants to mimic the guanido group of Arg-36. Higher guanidine-HCl concentrations (above 400 mM) resulted in a large decrease in kcat and kcat/Km values for both the R36A variant and the wild-type enzyme consistent with denaturation.
FIG. 5.
View of the Cab active site showing the hydrogen bonding between Arg-36 and Asp-34. The active site is shown with the three conserved zinc ligands Cys-32, His-87, and Cys-90 in yellow, the catalytic zinc in red, the zinc-bound water in magenta, and the conserved Asp-34 and Arg-36 in green. Hydrogen bonds are indicated by broken lines. The image was prepared with Web Lab Viewer.
An explanation for why replacement of Arg-36 decreased the kinetic parameters of Cab may come from inspection of the duplicated active sites in the crystal structure of the P. purpureum enzyme (26). The conserved arginine does not form hydrogen bonds to the conserved aspartate in either of the duplicated active sites; instead, the aspartates coordinate to zinc while the arginine side chains are flipped away from the active sites. This structure led the authors to propose a modified zinc hydroxide mechanism in which the aspartate ligand to zinc is exchanged with a hydroxyl during catalysis. Aspartate ligation to zinc may be occurring in the Arg-36 replacement variants of Cab where the hydrogen bonds to Asp-34 would be disrupted. In the R36A variant, Asp-34 may swing toward the active site and ligate zinc in a dead-end inactive conformation. If this hypothesis is correct, a variant in which both Asp-34 and Arg-36 are replaced with alanine would be expected to have kcat and kcat/Km values greater than those of the R36A variant since Asp-34 could no longer ligate zinc in the double variant. Indeed, the kcat and kcat/Km values for the D34A-R36A variant in TAPS buffer were 11- and 430-fold greater than those for the R36A variant. These results are consistent with the function of Arg-36 to hold Asp-34 in the active site through two hydrogen bonds; however, an exchange of ligands to Asp-34 during catalysis cannot be ruled out. Indeed, an exchange of ligands may be necessary if the pKa of Asp-34 were decreased when hydrogen bonded to Arg-36 such that Asp-34 is unable to abstract a proton from the zinc-bound water. Another potential function for Arg-36 may be to bind bicarbonate after release from zinc, thereby facilitating product removal, as proposed for arginine in the active site of Cam (39).
Acknowledgments
We thank Matthew Kimber for invaluable discussions regarding the active sites of β-class carbonic anhydrases.
This work was supported by grants from the National Institutes of Health to J.G.F. (no. GM44661) and from the NASA-Ames Cooperative Agreement NCC2-1057 to The Pennsylvania State University Astrobiology Research Center.
REFERENCES
- 1.Alber, B. E., C. M. Colangelo, J. Dong, C. M. V. Stalhandske, T. T. Baird, C. Tu, C. A. Fierke, D. N. Silverman, R. A. Scott, and J. G. Ferry. 1999. Kinetic and spectroscopic characterization of the gamma carbonic anhydrase from the methanoarchaeon Methanosarcina thermophila. Biochemistry 38:13119-13128. [DOI] [PubMed]
- 2.Boehlein, S. K., E. S. Walworth, N. G. Richards, and S. M. Schuster. 1997. Mutagenesis and chemical rescue indicate residues involved in beta-aspartyl-AMP formation by Escherichia coli asparagine synthetase B. J. Biol. Chem. 272:12384-12392. [DOI] [PubMed] [Google Scholar]
- 3.Boriack-Sjodin, P. A., R. W. Heck, P. J. Laipis, D. N. Silverman, and D. W. Christianson. 1995. Structure determination of murine mitochondrial carbonic anhydrase V at 2.45 A resolution: implications for catalytic proton transfer and inhibitor design. Proc. Natl. Acad. Sci. USA 92:10949-10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bracey, M. H., J. Christiansen, P. Tovar, S. P. Cramer, and S. G. Bartlett. 1994. Spinach carbonic anhydrase: investigation of the zinc-binding ligands by site-directed mutagenesis, elemental analysis, and EXAFS. Biochemistry 33:13126-13131. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Cox, E. H., G. L. McLendon, F. M. Morel, T. W. Lane, R. C. Prince, I. J. Pickering, and G. N. George. 2000. The active site structure of Thalassiosira weissflogii carbonic anhydrase 1. Biochemistry 39:12128-12130. [DOI] [PubMed] [Google Scholar]
- 7.Cronk, J. D., J. A. Endrizzi, M. R. Cronk, J. W. O'Neill, and K. Y. J. Zhang. 2001. Crystal structure of E. coli β-carbonic anhydrase, an enzyme with an unusual pH-dependent activity. Protein Sci. 10:911-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhalla, A. M., B. Li, M. F. Alibhai, K. J. Yost, J. M. Hemmingsen, W. M. Atkins, J. Schineller, and J. J. Villafranca. 1994. Regeneration of catalytic activity of glutamine synthetase mutants by chemical activation: exploration of the role of arginines 339 and 359 in activity. Protein Sci. 3:476-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eriksson, A. E., P. M. Kylsten, T. A. Jones, and A. Liljas. 1988. Crystallographic studies of inhibitor binding sites in human carbonic anhydrase II: a pentacoordinated binding of the SCN− ion to the zinc at high pH. Proteins 4:283-293. [DOI] [PubMed] [Google Scholar]
- 10.Eriksson, A. E., and A. Liljas. 1993. Refined structure of bovine carbonic anhydrase-III at 2.0 angstrom resolution. Proteins 16:29-42. [DOI] [PubMed] [Google Scholar]
- 11.Hakansson, K., and A. Wehnert. 1992. Structure of cobalt carbonic anhydrase complexed with bicarbonate. J. Mol. Biol. 228:1212-1218. [DOI] [PubMed] [Google Scholar]
- 12.Huang, S., Y. Xue, E. Sauer-Eriksson, L. Chirica, S. Lindskog, and B. H. Jonsson. 1998. Crystal structure of carbonic anhydrase from Neisseria gonorrhoeae and its complex with the inhibitor acetazolamide. J. Mol. Biol. 283:301-310. [DOI] [PubMed] [Google Scholar]
- 13.Iverson, T. M., B. E. Alber, C. Kisker, J. G. Ferry, and D. C. Rees. 2000. A closer look at the active site of γ-class carbonic anhydrase: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila. Biochemistry 39:9222-9231. [DOI] [PubMed] [Google Scholar]
- 14.Johansson, I. M., and C. Forsman. 1993. Kinetic studies of pea carbonic anhydrase. Eur. J. Biochem. 218:439-446. [DOI] [PubMed] [Google Scholar]
- 15.Johansson, I. M., and C. Forsman. 1994. Solvent hydrogen isotope effects and anion inhibition of CO2 hydration catalysed by carbonic anhydrase from Pisum sativum. Eur. J. Biochem. 224:901-907. [DOI] [PubMed] [Google Scholar]
- 16.Kannan, K. K., B. Notstrand, K. Fridborg, S. Lovgren, A. Ohlsson, and M. Petef. 1975. Crystal structure of human erythrocyte carbonic anhydrase B. Three-dimensional structure at a nominal 2.2 A resolution. Proc. Natl. Acad. Sci. USA 72:51-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khalifah, R. G. 1971. The carbon hydroxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isozymes B and C. J. Biol. Chem. 246:2561-2573. [PubMed] [Google Scholar]
- 18.Kimber, M. S., and E. F. Pai. 2000. The active site architecture of Pisum sativum β-carbonic anhydrase is a mirror image of that of α-carbonic anhydrases. EMBO J. 19:1407-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kisker, C., H. Schindelin, B. E. Alber, J. G. Ferry, and D. C. Rees. 1996. A left-hand beta-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila. EMBO J. 15:2323-2330. [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, H. J., M. D. Lloyd, I. J. Clifton, K. Harlos, A. Dubus, J. E. Baldwin, J. M. Frere, and C. J. Schofield. 2001. Alteration of the co-substrate selectivity of deacetoxycephalosporin C synthase. The role of arginine 258. J. Biol. Chem. 276:18290-18295. [DOI] [PubMed] [Google Scholar]
- 21.Lehoux, I. E., and B. Mitra. 2000. Role of arginine 277 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate binding and transition state stabilization. Biochemistry 39:10055-10065. [DOI] [PubMed] [Google Scholar]
- 22.Liang, Z., Y. Xue, G. Behravan, B. H. Jonsson, and S. Lindskog. 1993. Importance of the conserved active-site residues Tyr7, Glu106 and Thr199 for the catalytic function of human carbonic anhydrase II. Eur. J. Biochem. 211:821-827. [DOI] [PubMed] [Google Scholar]
- 23.Liljas, A., K. K. Kannan, P. C. Bergsten, I. Waara, K. Fridborg, B. Strandberg, U. Carlbom, L. Jarup, S. Lovgren, and M. Petef. 1972. Crystal structure of human carbonic anhydrase C. Nat. New Biol. 235:131-137. [DOI] [PubMed] [Google Scholar]
- 24.Lindskog, S. 1997. Structure and mechanism of carbonic anhydrase. Pharmacol. Ther. 74:1-20. [DOI] [PubMed] [Google Scholar]
- 25.Lindskog, S., and J. E. Coleman. 1973. The catalytic mechanism of carbonic anhydrase. Proc. Natl. Acad. Sci. USA 70:2505-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitsuhashi, S., T. Mizushima, E. Yamashita, M. Yamamoto, T. Kumasaka, H. Moriyama, T. Ueki, S. Miyachi, and T. Tsukihara. 2000. X-ray structure of β-carbonic anhydrase from the red alga, Porphridium purpureum, reveals a novel catalytic site for CO2 hydration. J. Biol. Chem. 275:5521-5526. [DOI] [PubMed] [Google Scholar]
- 27.Phillips, M. A., L. Hedstrom, and W. J. Rutter. 1992. Guanidine derivatives restore activity to carboxypeptidase lacking arginine-127. Protein Sci. 1:517-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowlett, R. S., M. R. Chance, M. D. Wirt, D. E. Sidelinger, J. R. Royal, M. Woodroffe, Y. F. Wang, R. P. Saha, and M. G. Lam. 1994. Kinetic and structural characterization of spinach carbonic anhydrase. Biochemistry 33:13967-13976. [DOI] [PubMed] [Google Scholar]
- 29.Rynkiewicz, M. J., and B. A. Seaton. 1996. Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ornithine transcarbamylase. Biochemistry 35:16174-16179. [DOI] [PubMed] [Google Scholar]
- 30.Silverman, D. N., and S. Lindskog. 1988. The catalytic mechanism of carbonic anhydrase: implications of a rate-limiting proteolysis of water. Acc. Chem. Res. 21:30-36. [Google Scholar]
- 31.Smith, K. S., N. J. Cosper, C. Stalhandske, R. A. Scott, and J. G. Ferry. 2000. Structural and kinetic characterization of an archaeal β-class carbonic anhydrase. J. Bacteriol. 182:6605-6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, K. S., and J. G. Ferry. 1999. A plant-type (β-class) carbonic anhydrase from the thermophilic methanoarchaeon Methanobacterium thermoautotrophicum. J. Bacteriol. 181:6247-6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, K. S., and J. G. Ferry. 2000. Prokaryotic carbonic anhydrases. FEMS Microbiol. Rev. 24:335-366. [DOI] [PubMed] [Google Scholar]
- 34.Smith, K. S., C. Jakubzick, T. S. Whittam, and J. G. Ferry. 1999. Carbonic anhydrase is an ancient enzyme widespread in prokaryotes. Proc. Natl. Acad. Sci. USA 96:15184-15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stams, T., S. K. Nair, T. Okuyama, A. Waheed, W. S. Sly, and D. W. Christianson. 1996. Crystal structure of the secretory form of membrane-associated human carbonic anhydrase IV at 2.8 A resolution. Proc. Natl. Acad. Sci. USA 93:13589-13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strop, P., K. S. Smith, T. M. Iverson, J. G. Ferry, and D. C. Rees. 2001. Crystal structure of the “cab”-type beta class carbonic anhydrase from the archaeon Methanobacterium thermoautotrophicum. J. Biol. Chem. 276:10299-10305. [DOI] [PubMed] [Google Scholar]
- 37.Tripp, B. C., and J. G. Ferry. 2000. A structure-function study of a proton transport pathway in the γ-class carbonic anhydrase from Methanosarcina thermophila. Biochemistry 39:9232-9240. [DOI] [PubMed] [Google Scholar]
- 38.Tripp, B. C., K. S. Smith, and J. G. Ferry. 2001. Carbonic anhydrase: new insights for an ancient enzyme. J. Biol. Chem. 276:48615-48618. [DOI] [PubMed] [Google Scholar]
- 39.Tripp, B. C., C. Tu, and J. G. Ferry. 2002. Role of arginine 59 in the γ-class carbonic anhydrases. Biochemistry 41:669-678. [DOI] [PubMed] [Google Scholar]
- 40.Tu, C. K., D. N. Silverman, C. Forsman, B. H. Jonsson, and S. Lindskog. 1989. Role of histidine 64 in the catalytic mechanism of human carbonic anhydrase II studied with a site-specific mutant. Biochemistry 28:7913-7918. [DOI] [PubMed] [Google Scholar]
- 41.Wilbur, K. M., and N. G. Anderson. 1948. Electrometric and colorimetric determination of carbonic anhydrase. J. Biol. Chem. 176:147-154. [PubMed] [Google Scholar]
- 42.Xue, Y., A. Liljas, B. H. Jonsson, and S. Lindskog. 1993. Structural analysis of the zinc hydroxide-Thr-199-Glu-106 hydrogen-bond network in human carbonic anhydrase II. Proteins 17:93-106. [DOI] [PubMed] [Google Scholar]
- 43.Xue, Y., J. Vidgren, L. A. Svensson, A. Liljas, B. H. Jonsson, and S. Lindskog. 1993. Crystallographic analysis of Thr-200→His human carbonic anhydrase II and its complex with the substrate, HCO3. Proteins 15:80-87. [DOI] [PubMed] [Google Scholar]