Abstract
Field and laboratory studies were performed to determine whether slugs could act as novel vectors for pathogen (e.g., Escherichia coli O157) transfer from animal feces to salad vegetables. Escherichia coli O157 was isolated from 0.21% of field slugs from an Aberdeenshire sheep farm. These isolates carried the verocytotoxin genes (vt1 and vt2) and the attaching and effacing gene (eae), suggesting that they are potentially pathogenic to humans. Strain typing using multilocus variable number tandem repeats analysis showed that slug and sheep isolates were indistinguishable. Laboratory experiments using an E. coli mutant resistant to nalidixic acid showed that the ubiquitous slug species Deroceras reticulatum could carry viable E. coli on its external surface for up to 14 days. Slugs that had been fed E. coli shed viable bacteria in their feces with numbers showing a short but statistically significant linear log decline. Further, it was found that E. coli persisted for up to 3 weeks in excreted slug feces, and hence, we conclude that slugs have the potential to act as novel vectors of E. coli O157.
Infection caused by Escherichia coli O157 is an emerging zoonoses in many countries including the United Kingdom and the United States (39, 47). The incidence of human disease is relatively low in industrialized countries compared to other enteric pathogens such as Campylobacter and Salmonella spp. (4, 25, 35). However, the sequelae can be severe, with approximately 5 to 10% of patients exhibiting hemolytic-uremic syndrome with a mortality rate of 3 to 5% (28, 48). Scotland has the highest ratio of human infections in the United Kingdom with 4.1 cases per 100,000 in 2004 (25).
Farm ruminants are the major reservoirs of E. coli O157, which is carried asymptomatically and shed transiently in their feces (21, 38, 52). Both cattle and sheep can excrete high levels of this pathogen (>104 CFU/g), leading to gross contamination of the farm environment and pasture (34, 45, 49). Escherichia coli O157 has been reported to have long-term survival (in manure, soil, and pasture), and manure application can lead to the contamination of vegetable crops and the surrounding environment (11, 20, 22, 30, 32, 52).
Escherichia coli O157 has caused a number of outbreaks associated with meat and dairy products of bovine origin, and in recent years, other foodstuffs, including fruit and vegetables (cabbage, alfalfa sprouts, celery, coriander, cress sprouts, and lettuce) (6), have been implicated as vehicles for human infection. Such outbreaks are frequently traced back to growers where a potential local E. coli source is identified, e.g., cattle feedlots, deer feces, or other ruminant fecal sources (2, 7, 8, 15). One of the largest outbreaks worldwide occurred in Japan with >6,000 cases (29) where the vehicle was identified as radish sprouts.
Although ruminants are recognized as the major reservoir of E. coli O157, it has also been isolated from a number of other animals including wild birds and rodents (31). Rabbits have been implicated as the vector of E. coli O157 when visitors to a petting farm were infected (37). This demonstrates the potential of alternate sources of E. coli O157 where ruminants are not directly present. In terms of invertebrates carrying this pathogen, pathogenic serotypes of E. coli have been isolated from the darkling beetle in both adult and larva forms and from the lesser mealworm (Alphitobus diaperinus) in poultry farms where Salmonella and Campylobacter spp. were also present (1, 14, 18). Laboratory experiments have further confirmed lesser mealworms as potential vectors of E. coli, Salmonella, and Campylobacter (27, 46). Escherichia coli O157 has also been isolated from 2.1% of houseflies sampled at a cattle farm in Japan (17) and 2.2% of houseflies collected from a cattle farm in the United States. Laboratory research has shown that flies can transmit E. coli from an environmental source to food products, e.g., the transfer of E. coli to apples by fruit flies (Drosophila melanogaster) (19). These studies demonstrate the potential of invertebrates to act as vectors in the transmission of E. coli O157 (12).
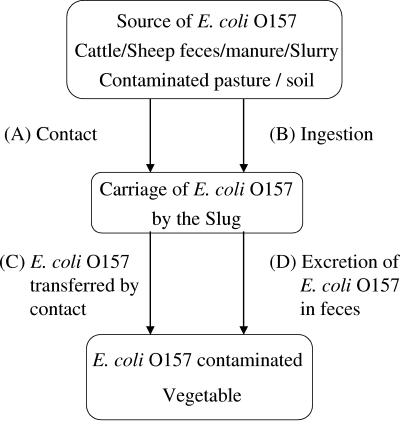
Slugs are widespread agricultural pests and frequently contaminate leafy vegetables, some of which are grown for human consumption without prior cooking (36). Slugs continuously ingest bacteria from the soil and their environment (51) and therefore have the potential to become contaminated with E. coli O157, both internally and externally. In previous work on the yellow slug (Limax flavus) and great gray slug (Limax maximus), both species were shown to carry coliforms both on the surface and in their gastrointestinal tract. A significant proportion of these were E. coli (10, 55), which suggests that slugs could be potential carriers of E. coli O157. However, neither of these large slug species are usually associated with leafy salad crops (10, 55). In this study, we have focused our laboratory studies on the gray field slug (Deroceras reticulatum), which is a widely distributed agricultural and horticultural pest, and as such, it frequently contaminates lettuce and other salad vegetables (36). This invertebrate may therefore play a role in the transfer of E. coli O157 to fruit and vegetables, either by direct contact or contamination with feces (Fig. 1).
FIG. 1.
Proposed transfer pathways of E. coli O157 from an environmental source to vegetable crops with slugs as the vector.
The aims of this study are to demonstrate that E. coli O157 and commensal E. coli can be found in slugs within the farming environment and that carriage and transfer of this pathogen to vegetable crops by slugs is feasible.
MATERIALS AND METHODS
Field survey.
Slugs were collected during an 8-day period in July and a 3-day period in August 2004 from a farm in Aberdeenshire, Scotland, confirmed previously as having sheep shedding E. coli O157. During the study period, sheep feces were also collected and analyzed for continuing presence of E. coli O157 (50). The average number of slugs collected each day was 43 (23 to 98). The mean number of slugs pooled for enrichment was 15, with a variation of 2 to 26 depending on size and numbers available. On each sampling day, the slugs were pooled into groups of similar mass (mean ± standard deviation, 11.6 g ± 4.8 g) for the detection of E. coli O157. A total of 474 slugs were collected and pooled into 33 groups throughout the study period.
Isolation of E. coli O157.
The slugs were homogenized in a sterile blender and enriched in a 10× volume of buffered peptone water (Oxoid CM509) supplemented with vancomycin (8 mg liter−1) for 6 h at 42°C. This was followed by immunomagnetic separation (IMS) as described by Omisakin et al. (34). IMS beads were resuspended in 0.1 ml wash buffer and spread equally onto sorbitol MacConkey agar (SMAC, Oxoid CM813) supplemented with cefixime (0.05 mg liter−1) and potassium tellurite (2.5 mg liter−1) (CT-SMAC; Mast Diagnostics, Merseyside, United Kingdom) and on Harlequin SMAC BCIG agar (Lab M, IDG, Bury, United Kingdom) supplemented with cefixime and tellurite (Harlequin CT-BCIG). Both plates were incubated for 18 to 24 h at 37°C. Presumptive E. coli O157 colonies (non-sorbitol fermenting) were confirmed by latex agglutination (Oxoid DR620). Positive isolates were stored in a 13% glycerol solution and frozen at −20°C for further analysis.
Commensal E. coli carriage.
A total of 40 slugs were separated into 2 equal groups and homogenized in a sterile blender, and 1 g from each was serially diluted in phosphate-buffered saline (PBS) and plated onto MacConkey agar. The plates were incubated for 24 h at 37°C, and presumptive target colonies were counted and confirmed by the detection of indole at 44°C from tryptone water (26).
Identification of E. coli O157 virulence genes.
The E. coli O157 determinants of virulence (vt1, vt2, and eae) from isolates of both slug and sheep feces were identified using PCR (23). The PCR protocol was modified as follows: denaturing at 95°C for 2 min, followed by 30 cycles of 95°C for 30 s, 65°C for 30 s, and 72°C for 30 s, and then finally 4 min at 72°C for the VT1 primer. The PCR temperature sequence for VT2 and eae was the same except that the annealing temperature of 65°C was reduced to 55°C. PCR products were separated on a 1.4% agarose gel and visualized under UV.
Multilocus VNTR typing.
The E. coli O157 strains from sheep feces with identical virulence profiles to those from slugs were subtyped by variable number tandem repeat (VNTR) typing (24). Here, PCR was used to amplify 7 loci that had previously identified nucleotide sequences which are repeated in tandem. The PCR temperature profile was 95°C for 4 min, followed by 30 cycles of 95°C for 30 s, an annealing temperature of 60°C for 60 s, 72°C for 60 s, and extension of 4 min at 72°C for all the primers except for primer 2, which required a lower annealing temperature of 55°C. Products from the seven primers were purified (QIAquick) and sequenced using dye-labeled dideoxy-labeled nucleotides. The number of times each set of base pairs is repeated was identified and used to fingerprint individual strains.
Nalidixic acid-resistant strains of commensal E. coli.
Five strains (EC61, EC378, NCTC9001, ECB, and EC63) of non-O157 E. coli (all isolated previously from food except the NCTC strain, which was isolated from urine) were screened to produce nalidixic acid-resistant mutants (N+) (5). Suspensions were prepared by adding 10-μl aliquots of each N+ strain to 100 ml tryptic soy broth and incubated at 37°C for 24 h prior to centrifugation (5,000 × g), washing three times, and resuspension in 0.1 M PBS. All assays used MacConkey agar supplemented with 50 μl ml−1 nalidixic acid (MacN+). Experimental samples were kept at 16°C, the average spring/autumn temperature when slugs are most active.
Carriage and transfer of E. coli from the external surface of the slug via direct contact.
Two tests were performed where slugs were inoculated by placement into 90-mm petri dishes containing 5 ml of suspension (see above) of N+ E. coli. The slugs were removed after 2 h and transferred to sterile dishes for 1 h to eliminate excess N+ solution. Control batches of slugs were treated with sterile PBS.
In the first experiment, the inoculating suspension contained 6.0 × 109 CFU ml−1, and 5 slugs were tested by wiping each slug directly over two MacN+ plates. In the second experiment (5.8 × 109 CFU ml−1), 5 slugs were swabbed with a cotton wool bud, previously dampened with PBS, and the cotton bud was spread over the MacN+ plate. All plates were incubated for 24 h at 37°C, and colony numbers were recorded. This was repeated at 0 and 6 h and 1, 2, 3, 5, 7, 14, and 21 days. Each slug was transferred to a new sterile petri dish every 24 h to prevent recontamination.
Excretion of E. coli in slug feces.
Slug food consisting of Ready Brek (an oat-based breakfast cereal), nonfat milk powder, and calcium carbonate (2:1:1) was hydrated with E. coli in PBS. In each of three experiments, five slugs were left exposed to food inoculated with the nalidixic acid mutant strain in separate 90-mm petri dishes containing filter paper moistened with 1.5 ml sterile PBS. One slug consumed no food and was excluded from further testing. After 24 h, feces were collected, weighed, serially diluted, and plated onto MacConkey agar, and the slugs were transferred to new sterile dishes with uninoculated food. This was repeated daily for 14 days. Controls were performed with uninoculated food.
Survival of E. coli in slug feces.
Fresh feces were collected from slugs fed slug food (described above) for 48 h. The feces were divided into four equal portions of 0.41 g and each placed separately into 1.5-ml microcentrifuge tubes. Three of the tubes contained 65 μl N+ E. coli in PBS (control was sterile PBS), and all tubes were mixed thoroughly. The three tubes of inoculated feces had an average initial E. coli concentration of 8.7 log10 CFU/g. At each sampling time (0 and 6 h and 1, 2, 3, 5, 7, 14, and 21 days), 0.04 g of slug feces was mixed with 360 μl PBS, serially diluted, and plated in duplicate onto MacN+ agar.
RESULTS
Carriage of E. coli by slugs in the field.
From the field sampling, 1/33 pooled samples (total of 474 slugs) tested positive for E. coli O157. Two colonies were confirmed by the latex agglutination test, and due to the low number of positive samples, it is plausible that both originated from the same slug. This equates to a field slug prevalence of 0.21%. The two pooled samples, tested for commensal E. coli carriage from field slugs, were positive with values of 4.92 and 6.04 log10 CFU g−1, respectively. Sheep feces collected from the same field on the same day had E. coli counts of 5.90 and 6.50 log10 CFU g−1.
Molecular analyses (PCR and VNTR).
Analysis of virulence factors by PCR showed that the E. coli O157 strain isolated from the slugs was positive for both verocytotoxin genes (vt1 and vt2) and the attaching and effacing gene (eae). Slug and sheep isolates with the same virulence profiles (vt1, vt2, and eae) were analyzed by VNTR, and results are shown in Table 1. The slug and sheep isolates showed identical VNTR fingerprints. The time period between isolation of E. coli O157 from sheep feces and slug was 14 days. Interestingly, both sheep and slug isolates did not produce PCR products for VNTR primers 1 and 4, unlike three other E. coli O157 isolates from sheep (data not presented), including the reference EDL 933.
TABLE 1.
Number of base pair tandem repeats at the loci isolated by each primer used from E. coli O157 isolates from slugs and sheep and a control (EDL 933)
| Isolate | No. of repeats at locus no.:
|
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| Slug 1 | NDb | 1 | 5 | ND | 1 | 0 | 11 |
| Slug 2 | ND | 1 | 5 | ND | 1 | 0 | 11 |
| Sheep Feces 1 | ND | 1 | 5 | ND | 1 | 0 | 11 |
| Sheep Feces 2 | ND | 1 | 5 | ND | 1 | 0 | 11 |
| EDL 933a | 3 | 9 | 1 | 0 | 11 | ||
Reference strain.
ND, no PCR product for this locus.
Carriage and transfer of E. coli from the external surface of the slug via contact.
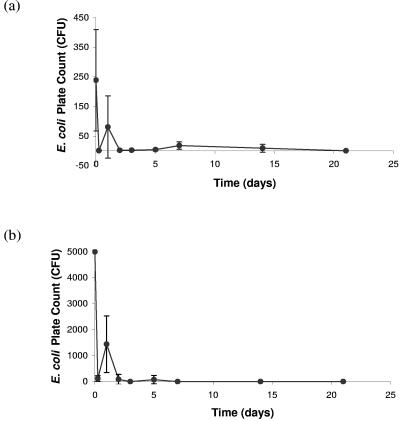
The transfer of E. coli from the external surface of the slug varied greatly between individuals and sampling times, with no obvious pattern except a general overall decline (Fig. 2). However, on comparison of the two different methods used (Fig. 2), a similar pattern is evident with a reduction in mean counts at 6 h followed by an increase at 24 h. In both experiments, the majority of E. coli isolates were shed within 48 h but were still detected (using the cotton wool bud swab method) at day 14 and by direct contact at day 7. It was found that wiping the slugs directly onto the agar produced higher counts than the cotton wool bud swab method during the first 24 h (Fig. 2).
FIG. 2.
Carriage and transfer of nalidixic acid-resistant E. coli from the external surface of the slug via contact and the reduction over time determined by swabbing the surface with a cotton wool bud (a) and wiping the external surface of the slug across an agar plate (b).
E. coli concentration in slug feces after consumption of inoculated food.
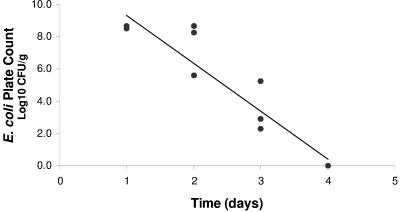
The highest concentrations of E. coli were found in feces during the first 2 days following ingestion of inoculated food (Fig. 3). The majority of E. coli isolates were excreted during the first 48 h and showed a significant linear log reduction (R2 = 0.8879, P < 0.000005) with a decimal reduction time of 0.34 days.
FIG. 3.
Reduction with time of E. coli numbers in slug feces after consumption of inoculated food (detection limit, 1 CFU/g).
Survival of E. coli in feces.
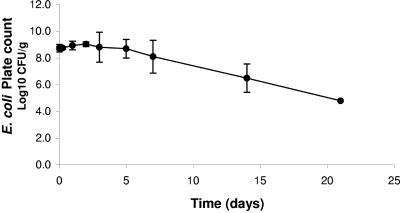
There was a small though insignificant increase in the E. coli concentration of inoculated slug feces (Fig. 4) during the first 2 days, from 8.7 to 9.0 log10 CFU g−1. This was followed by a highly significant (R2 = 0.999, P < 0.0005) log linear reduction from day 5, with a decimal reduction time of 4.15 days. Escherichia coli was still detected by direct plating at 21 days (the lack of results after day 21 was due to dehydration of the slug feces).
FIG. 4.
Mean E. coli concentration in inoculated slug feces and its reduction over time.
DISCUSSION
Fieldwork.
This study is the first to confirm (by field sampling, PCR, and VNTR analysis) that slugs can be carriers of the same pathogenic E. coli O157 found locally in ruminant feces. This provides evidence to support transfer of E. coli O157 from ruminant feces to slugs (pathways A and B) (Fig. 1). Previous studies with slugs have shown no relationship between E. coli carriage and potential local sources (10) or failed to detect pathogens (41). The prevalence of E. coli O157 in slugs in the field studied here appears low at 0.21%, but slug populations of 300 m −2 have been recorded in arable fields (54), which equates to 1.44 positive slugs m −2 using the prevalence value reported here. This prevalence may have been higher with increased rainfall during the testing period where E. coli O157 may have been distributed over a greater area, thus increasing the probability of slug contact. However, despite low rainfall (<1 mm per day), slugs tested here contained relatively high numbers of commensal E. coli isolates (approximately 1 log lower than the numbers recorded in sheep feces). The low prevalence of E. coli O157 in slugs is more likely due to low shedding rates of this pathogen in the ruminant reservoir where the concentrations in sheep fecal samples were ≤100 CFU g−1 (50). The prevalence of E. coli O157 in sheep here was estimated to be approximately 7% (50), which is similar to the 2003 Scottish Aberdeenshire value of 6.5% (33). In the present study, the pathogenic isolate of E. coli O157 found in the slugs was indistinguishable by VNTR from those carried by the sheep in the same field. Although the matching sheep isolate was recovered later than the isolate from the slug, it is likely that the strain was present in the sheep feces earlier but had not been sampled. Alternatively, it may have been sampled but the isolate not selected post-IMS for further analysis.
For contamination of vegetables by this vector to occur, the source of E. coli O157 (e.g., manure, feces, or runoff from pasture) and the vegetable site would need to be within the range of the slug. It has been reported previously (16) that this can be as much as 12 m per night for D. reticulatum. This distance may be extended in times of food deprivation (3) and during dispersal from areas of high population density (13). Laboratory experiments confirm that once a slug has become contaminated with E. coli via pathways A and B (contact and/or ingestion) (Fig. 1), it can persist for several days internally and externally, and therefore, there is sufficient time for an effective bacterial transfer (Fig. 1, pathways C and D).
Regardless of the sharp initial decline in E. coli numbers and high variation between individual slugs, the laboratory tests importantly demonstrate that external carriage of E. coli persists for relatively long time periods (Fig. 1, pathway A). Furthermore, E. coli can be readily transferred by contact (Fig. 1, pathway C). The results obtained at days 7 and 14 (Fig. 2) indicate sufficient time for pathogen transfer to a vegetable to take place.
The method of direct wiping of slugs to agar plates might be more realistic of how microorganisms transfer to vegetables. This method initially showed higher counts of E. coli than the swab method, probably because the swab failed to remove all bacteria present from the slug surface. Interestingly, there was a rise in counts at day 2 for both methods followed by a fall. It is unclear whether this is a real phenomenon, and further work would need to be done to verify this. The high variation between individuals may indicate a persistence of E. coli at different residual sites on the slug surface or simply slug-to-slug variation. Further work could be performed to determine the distribution of E. coli on the slug surface and to identify niches that E. coli preferentially colonizes.
The laboratory experiments demonstrate that slugs carry E. coli internally and excrete it in their feces following ingestion of contaminated food. This confirms pathways B and D (Fig. 1). It has also been reported that gastropods find mammalian feces an attractive food source (43), which together with the regular ingestion of potentially contaminated soil, demonstrates the potential of internal pathogen carriage. The majority of E. coli excretion by slugs occurred within the first 48 h, indicating direct passage through the intestines and agreeing with previous research (53). Escherichia coli excreted in the feces fell to below detection levels by day 4, and as slugs feed on average twice per day, there is sufficient time for them to travel to vegetables and contaminate the crop by excretion (Fig. 1, transfer pathway D). It has also been reported that other slug species (Arion ater) are stimulated to excrete while feeding (41), which would increase the likelihood of pathogen transfer to vegetables. The survival of E. coli in excreted slug feces was shown to persist for several days, where E. coli remained viable for >21 days in slug feces that was becoming dehydrated. A vegetable contaminated with slug feces, therefore, has the potential to harbor viable E. coli through harvest time and retail shelf life periods. The dehydration of feces during the later stages of the experiment also highlights the problem of applying laboratory data to the field.
The results of these experiments together with the field studies provide evidence that slugs have the potential to transfer E. coli O157 to vegetables via the pathways shown in Fig. 1. Care should be taken when considering the results of the laboratory studies, since commensal E. coli strains were used for ease of containment. It is likely, however, that toxigenic strains will have similar physiological characteristics. The likelihood of contaminated vegetables being ingested by the public will depend on the survival of E. coli on the crop. As it is advised that all fruit and vegetables be washed before consumption, research should investigate the removal of E. coli from the surface of vegetables at sites of both excretion and contact. Feces are more likely to be noticed by the consumer and the product discarded, whereas sites of contamination by contact would go unnoticed. Mucous deposited onto the vegetable by the slug may act as a beneficial substrate for bacterial growth and also afford protection against desiccation and sanitizing practices. Furthermore, it has been reported that E. coli will preferentially colonize cut edges on vegetables (40), e.g., in preparation of precut fruit and vegetables or at slug feeding sites. These sites will not only provide E. coli with some protection (from desiccation, washing, and sanitizing agents) but also offer entrance sites for internalization (42) and further reduce E. coli fatality by sanitary practices. The infective dose of E. coli O157 is thought to be low (9, 44); therefore, minimal survival in the vegetable has potential serious human health implications.
This study has found that slugs carry E. coli O157 in the farming environment, which is indistinguishable by VNTR from E. coli O157 in sheep feces in the same field. The research demonstrates that E. coli in D. reticulatum has a relatively long external and internal survival time and also shows the ability of E. coli to persist at length in excreted slug feces. The potential for vegetable contamination is dependent on a number of factors which include slug control, slug population dynamics, weather, harvest, and postharvest practices. This study provides evidence that slugs can act as vectors of E. coli O157 from an environmental source to fruit or vegetables.
Acknowledgments
The British Biotechnology Science Research Council is acknowledged for funding the Ph.D. studentship of E.L.S.
We thank Anne Margrete Urdahl (Norwegian School of Veterinary Science) for providing the E. coli O157 isolates from sheep and Ken Forbes and Lynn Thomson (Department of Medical Microbiology, Aberdeen University) for providing advice on PCR and VNTR techniques.
REFERENCES
- 1.Aarts, H. J. M., L. A. J. T. van Lith, W. F. Jacobs-Reitsma. 1995. Discrepancy between Penner serotyping and polymerase chain reaction fingerprinting of Campylobacter isolated from poultry and other animal sources. Lett. Appl. Microbiol. 20:371-374. [DOI] [PubMed] [Google Scholar]
- 2.Ackers, M. L., B. E. Mahon, E. Leahy, B. Goode, T. Damrow, P. S. Hayes, W. F. Bibb, D. H. Rice, T. J. Barret, L. Hutwagner, P. M. Griffin, and L. Slutsker. 1998. An outbreak of Escherichia coli O157:H7 infections associated with leaf lettuce consumption. J. Infect. Dis. 177:1588-1593. [DOI] [PubMed] [Google Scholar]
- 3.Airey, W. J. 1987. The influence of food deprivation on the activity of slugs. J. Molluscan Stud. 53:37-45. [Google Scholar]
- 4.Anonymous. 2001. Task Force on E. coli O157 (2001), final report. [Online.] Food Standards Agency (Scotland) and the Scottish Executive Health Department, Edinburgh, Scotland. http://www.food.gov.uk/multimedia/pdfs/ecolitaskfinreport.pdf.
- 5.Blackburn, C. D., and A. R. Davies. 1994. Development of antibiotic-resistant strains for the enumeration of Foodborne pathogenic bacteria in stored foods. Int. J. Food Microbiol. 24:125-136. [DOI] [PubMed] [Google Scholar]
- 6.Brackett, R. E. 1999. Incidence, contributing factors, and control of bacterial pathogens in produce. Postharvest Biol. Technol. 15:305-311. [Google Scholar]
- 7.Breuer, T., D. H. Benkel, R. L. Shapiro, W. M. Hall, M. M. Winnett, M. J. Linn, J. Neimann, T. J. Barrett, S. Dietrich, F. P. Downes, D. M. Toney, J. L. Pearson, H. Rolka, L. Slutsker, P. M. Griffin, and the Investigation Team. 2001. A multistate outbreak of Escherichia coli O157:H7 infections linked to alfalfa sprouts grown from contaminated seeds. Emerg. Infect. Dis. 7:977-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 1997. Outbreaks of Escherichia coli O157:H7 infection with eating alfalfa sprouts-Michigan and Virginia, June-July 1997. Morbid. Mortal. Wkly. Rep. 46:741-744. [PubMed] [Google Scholar]
- 9.Doyle, M. P., L. R. Beuchat, and T. J. Montville. 2001. Food microbiology: fundamentals and frontiers, 2nd ed. ASM Press, Washington, D.C.
- 10.Elliot, L. P. 1969. Certain bacteria, some of medical interest, associated with the slug Limax maximus. J. Invertebr. Pathol. 15:306-312. [DOI] [PubMed] [Google Scholar]
- 11.Fenlon, D. R., I. D. Ogden, A. Vinten, and I. Svoboda. 2000. The fate of Escherichia coli and E. coli O157 in cattle slurry after application to land. J. Appl. Microbiol. Symp. Suppl. 88:149S-156S. [DOI] [PubMed] [Google Scholar]
- 12.Graczyk, T. K., R. Knight, R. H. Gilman, and M. R. Cranfield. 2001. The role of non-biting flies in the epidemiology of human infectious disease. Microbes Infect. 3:231-235. [DOI] [PubMed] [Google Scholar]
- 13.Hamilton, P. A., and W. G. Wellington. 1981. The effects of food and sensitivity of the movement of Arion ater and Ariolimax columbianus (Pulmonata:Stylommatophora) between habitats. Res. Pop. Ecol. (Kyoto) 23:309-317. [Google Scholar]
- 14.Harein, P. K., E. De las Casas, B. S. Pomeroy, and M. D. York. 1970. Salmonella spp. and serotypes of Escherichia coli isolated from the lesser mealworm collected in poultry brooder houses. J. Econ. Entomol. 63:80-82. [DOI] [PubMed] [Google Scholar]
- 15.Hilborn, E. D., P. A. Mshar, T. R. Fiorentino, Z. F. Dembek, T. J. Barret, R. T. Howard, and M. L. Cartter. 2000. An outbreak of Escherichia coli O157:H7 infections and haemolytic syndrome associated with consumption of unpasteurized apple cider. Epidemiol. Infect. 124:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogan, J. M. 1985. The behaviour of the grey field slug (Deroceras reticulatum) with particular reference to control in winter wheat. Ph.D. thesis. Newcastle University, Newcastle upon-Tyne, United Kingdom.
- 17.Iwasa, M., S. Makino, H. Asakura, H. Kobori, and Y. Morimoto. 1999. Detection of Escherichia coli O157:H7 from Musca domestica (Diptera:Muscidae) at a cattle farm in Japan. J. Med. Entomol. 36:108-112. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs-Reitsma, W. F., A. W. van de Giessen, N. M. Bolder, and R. W. A. W. Mulder. 1995. Epidemiology of Campylobacter spp. at two Dutch broiler farms. Epidemiol. Infect. 114:413-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janisiewicz, W. J., W. S. Conway, M. W. Brown, G. M Sapers, P. Fratamico, and R. L. Buchanan. 1999. Fate of Escherichia coli O157:H7 on fresh-cut apple tissue and its potential for transmission by fruit flies. Appl. Environ. Microbiol. 65:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kudva, I. T., K. Blanch, and C. Hovde. 1998. Analysis of Escherichia coli O157:H7 survival in ovine or bovine manure and manure slurry. Appl. Environ. Microbiol. 64:3166-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kudva, I. T., P. G. Hatfield, and C. J. Hovde. 1996. Escherichia coli O157:H7 in microbial flora in sheep. J. Clin. Microbiol. 34:431-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau, M. M., and S. C. Ingham. 2001. Survival of faecal indicator bacteria in bovine manure incorporated into soil. Lett. Appl. Microbiol. 33:131-136. [DOI] [PubMed] [Google Scholar]
- 23.Lin, Z., H. Kurazono, S. Yamasaki, and Y. Takeda. 1993. Detection of various variant verotoxin genes in E. coli by PCR. Microbiol. Immunol. 37:543-548. [DOI] [PubMed] [Google Scholar]
- 24.Lindstedt, B. A., T. Vardund, and G. Kapperud. 2004. Multiple-locus variable-number tandem-repeats analysis of Escherichia coli O157 using PCR multiplexing and multi-colored capillary electrophoresis. J. Microbiol. Methods 58:213-222. [DOI] [PubMed] [Google Scholar]
- 25.Locking, M., L. Browning, A. Smith-Palmer, J. Cowden, and S. Brownlie. 2005. Gastro-intestinal and foodborne infections. HPS Wkly. Rep. 39: 2-3. [Online.] http://www.show.scot.nhs.uk/scieh/PDF/pdf2005/0501.pdf
- 26.MacFaddin, J. F. 1976. Biochemical tests for identification of medical bacteria, p. 99-108. Williams and Wilkins Company, Baltimore, Md.
- 27.McAllister, J. C., C. D. Steelman, and J. K. Skeeles. 1994. Reservoir competence of the lesser mealworm (Coleoptera: Tenebrionidae) for Salmonella typhimurium (Eubacteriales:Enterobacteriaceae). J. Med. Entomol. 31:369-372. [DOI] [PubMed] [Google Scholar]
- 28.Meng, J., and M. P. Doyle. 1998. Emerging and evolving microbial foodborne pathogens. Bull. Inst. Pasteur 96:151-164. [Google Scholar]
- 29.Michino, H., K. Araki, S. Minami, T. Nakayama, Y. Ejima, K. Hiroe, H. Tanaka, N. Fujita, S. Usami, M. Yonekawa, K. Sadamoto, S. Takaya, and N. Sakai. 1998. Recent outbreaks of infections caused by Escherichia coli O157:H7 in Japan, p. 73-81. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 30.Natvig, E. E., S. C. Ingham, B. H. Ingham, L. R. Cooperband, and T. R. Roper. 2002. Salmonella enterica serovar Typhimurium and Escherichia coli contamination of root and leaf vegetables grown in soils with incorporated bovine manure. Appl. Environ. Microbiol. 68:2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen, E. M., M. N. Skov, J. J. Madsen, J. Lodal, J. B. Jespersen, and D. L. Baggesen. 2004. Verocytotoxin-producing Escherichia coli in wild birds and rodents in close proximity to farms. Appl. Environ. Microbiol. 70:6944-6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogden, I. D., N. F. Hepburn, M. MacRae, N. J. C. Strachan, D. R. Fenlon, S. M. Rusbridge, and T. H. Pennington. 2002. Long term survival of Escherichia coli O157 on pasture following an outbreak associated with sheep at a scout camp. Lett. Appl. Microbiol. 34:100-104. [DOI] [PubMed] [Google Scholar]
- 33.Ogden, I. D., M. MacRae, and N. J. C. Strachan. 2005. Concentration and prevalence of Escherichia coli O157 in sheep faeces at pasture in Scotland. J. Appl. Microbiol. 98:646-651. [DOI] [PubMed] [Google Scholar]
- 34.Omisakin, F., M. MacRae, I. D. Ogden, and N. J. C. Strachan. 2003. Concentration and prevalence of Escherichia coli O157 in cattle faeces at slaughter. Appl. Environ. Microbiol. 69:2444-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Philips, C. A. 1999. The epidemiology, detection and control of Escherichia coli O157. J. Sci. Food Agric. 79:1367-1381. [Google Scholar]
- 36.Port, G., and A. Ester. 2002. Gastropods as pests in vegetable and ornamental crops in Western Europe, p. 337-351. In G. M. Barker (ed.), Molluscs as crop pests. CABI Publishing, Wallingford, United Kingdom.
- 37.Pritchard, G. C., S. Williamson, T. Carson, J. R. Bailey, L. Warner, G. Willshaw, and T. Cheasty. 2001. Wild rabbits—a novel vector for verocytotoxigenic Escherichia coli O157. Vet. Rec. 149:567. [PubMed] [Google Scholar]
- 38.Rahn, K., S. A. Renwick, R. P. Johnson, J. B. Wilson, R. C. Clarke, D. Alves, S. McEwen, H. Lior, and J. Spika. 1997. Persistence of Eschericha coli O157:H7 in dairy cattle and the dairy farm environment. Epidemiol. Infect. 119:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rangel, J. M., P. H. Sparling, C. Crowe, P. M. Griffin, and D. L. Swerdlow. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg. Infect. Dis. 11:603-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seo, K. H., and J. F. Frank. 1999. Attachment of Escherichia coli O157:H7 to lettuce leaf surface and viability in response to chlorine treatment as demonstrated by using confocal laser microscope. J. Food Prot. 62:3-9. [DOI] [PubMed] [Google Scholar]
- 41.Shrewsbury, J. F. D., and G. J. Barson. 1947. A contribution to the study of bacterial flora of Arion ater. Proc. Soc. Appl. Bacteriol. 2:70-76. [Google Scholar]
- 42.Solomon, E. B., Yaron, S., and K. R. Mathews. 2002. Transmission of Escherichia coli O157:H7 from contaminated manure and irrigation water to lettuce plant tissue and its subsequent internalization. Appl. Environ. Microbiol. 68:397-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Speiser, B. 2001. Food and feeding behaviour, p. 259-288. In G. M. Barker (ed.), The biology of terrestrial molluscs. CABI publishing, Oxon, United Kingdom.
- 44.Strachan, N. J. C., M. P. Doyle, F. Kasuga, O. Rotariu, and I. D. Ogden. 2005. Dose response modelling of Escherichia coli O157 incorporating data from foodborne and environmental outbreaks. Int. J. Food Microbiol. 103:35-47. [DOI] [PubMed] [Google Scholar]
- 45.Strachan, N. J. C., G. M. Dunn, and I. D. Ogden. 2003. E. coli O157: burger bug or environmental pathogen, p. 105. In Fifth International Symposium on Shiga Toxin (Verocytotoxin)-Producing Escherichia coli Infections. In Conference Ltd., Edinburgh, United Kingdom.
- 46.Strother, K. O., C. D. Steelman, and E. E. Gbur. 2005. Reservoir competence of lesser mealworm (Coleoptera:Tenebrionidae) for Campylobacter jejuni (Campylobacterales: Campylobacteraceae). J. Med. Entomol. 42:42-47. [DOI] [PubMed] [Google Scholar]
- 47.Tauxe, R. V. 1997. Emerging foodborne diseases: an evolving public health challenge. Emerg. Infect. Dis. 3:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thorpe, C. M. 2004. Shiga toxin producing Escherichia coli infection. Clin. Infect. Dis. 38:1298-1303. [DOI] [PubMed] [Google Scholar]
- 49.Trevena, W. B., G. A. Willshaw, T. Cheasty, C. Wray, and J. Gallacher. 1996. Verocytotoxin producing E. coli O157 infection associated with farms. Lancet 347:60-61. [DOI] [PubMed] [Google Scholar]
- 50.Urdahl, A. M., Strachan, N. J. C., Y. Wasteson, M. MacRae, and I. D. Ogden. 2005. A longitudinal study of E. coli O157 contamination of a watercourse by farm ruminants at pasture. In Proceedings of Society for Veterinary Epidemiology and Veterinary Medicine, Nairn, Scotland, United Kingdom, 30th March-1st April. [Online.] http://www.svepm.org.uk/posters/index.pfp?path=.%2F2005%2F.
- 51.Walker, A. J., D. M. Glen, and P. R. Shewry. 1999. Bacteria associated with the digestive system of the slug Deroceras reticulatum are not required for protein digestion. Soil Biol. Biochem. 31:1387-1394. [Google Scholar]
- 52.Wang, G., T. Zhao, and M. P. Doyle. 1996. Fate of enterohemorrhagic Escherichia coli O157:H7 in bovine feces. Appl. Environ. Microbiol. 62:2567-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watkins, B., and K. Simkiss. 1990. Interactions between soil bacteria and the molluscan alimentary tract. J. Molluscan Stud. 56:267-274. [Google Scholar]
- 54.Wilson, M. J., D. M. Glen, S. K. George, J. D. Pearce, and C. W. Wiltshire. 1994. Biological control of slugs in winter wheat using the rhabditid nematode Phasmarhabditis hermaphrodita. Ann. Appl. Biol. 125:377-390. [Google Scholar]
- 55.Yamada, G., S. Yonemoto, S. Matsumoto, N. Toda, and O. Ibusuki. 1960. Studies on slugs as vectors of pathogenic microbes. J. Osaka City Med. Cent. 2:707-717. [Google Scholar]