Abstract
Real-time PCR (RT-PCR) technology was used for the specific detection and quantification of members of the family Geodermatophilaceae in stone samples. Differences in the nucleotide sequences of the 16S rRNA gene region were used to design a pair of family-specific primers that were used to detect and quantify by RT-PCR DNA from members of this family in stone samples from different geographical origins in Spain. These primers were applied later to identify by PCR-specific amplification new members of the family Geodermatophilaceae isolated from the same stone samples. The diversity and taxonomic position of the wild-type strains identified from ribosomal sequence analysis suggest the presence of a new lineage within the genus Blastococcus.
The microbial diversity in stones and monument surfaces has been extendedly studied according to different approaches (colony and cell morphology, physiology, protein patterns, and 16S rRNA gene sequences studies) and bacterial strains have been repeatedly described as isolated from different surfaces of stones and monuments (3, 25), karstic caves (5), mountain soils (12), and biodeteriorated wall paintings (6). These studies demonstrated the occurrence of different groups of heterotrophic bacteria, including many species of the order Actinomycetales. These actinomycetes demonstrated a great taxonomic diversity and, in spite of a predominance of isolates of the genus Streptomyces, members of the genera Nocardia, Rhodococcus, and Geodermatophilus have also been described (5).
The family Geodermatophilaceae (18) includes the three genera Geodermatophilus (10), Blastococcus (1), and Modestobacter (12). Some strains of the genus Geodermatophilus have been isolated from stone and monument surfaces in the Mediterranean basin, although many of them originate from extreme cryptolithic environments (3, 24). The presence and diversity of Geodermatophilaceae in the environment was previously described after a 16S rRNA gene restriction analysis confirmed the presence of three well-separated clusters of the genera Geodermatophilus, Blastococcus, and Modestobacter (24, 25).
In recent years, 16S rRNA gene PCR primers have been extensively used for the molecular identification of microorganisms at different taxonomic levels (11, 15, 19, 20) and more specifically of different families and genera of actinomycetes (13, 16, 17). Two probes were recently described for the specific detection of Geodermatophilaceae and Modestobacter in rock surfaces by using fluorescence in situ hybridization (22). Alternatively, the development of a real-time PCR (RT-PCR) with specific primers could provide a quantitative method to detect the presence of members of these taxa in any given environmental sample.
We have focused here on the development of family-specific primers to detect by RT-PCR the presence of members of the family Geodermatophilaceae in stones and associated saxicolous lichen samples. Two different geographical origins in Spain were sampled to evaluate the presence of this microbial group: the Central Mountain System in Madrid, with plutonic and metamorphic rocks covered with sedimentary rocks and the limestone outcrops from San Vicente beach in Mallorca island from Balearic islands. We discuss the occurrence of members of Geodermatophilaceae in both geological systems.
MATERIALS AND METHODS
Bacterial strains.
New isolates were isolated from stones from two Spanish mountain areas (Table 1). Isolates were grown at 28°C on YMG (0.4% glucose, 0.4% yeast extract, and 1.0% malt extract) agar or LM medium (10).
TABLE 1.
Geographical origin of stone samples used in this study, detection of Geodermatophilaceae using primers Geosp2 and Geosp1, and concentration of total DNA
| Sample | Geographical origin | Stone type | Total DNAb | RT-PCR amplificationa (Geosp2/Geosp1) |
|---|---|---|---|---|
| 1A | La Cabrera (Madrid) | Granite (stone) | 5.69 | 1 |
| 1B | La Cabrera (Madrid) | Granite (lichen) | 15.17 | 1 |
| 2A | La Cabrera (Madrid) | Granite (stone) | 29.58 | 1 |
| 2B | La Cabrera (Madrid) | Granite (lichen) | 59.28 | 1 |
| 3 | Cala S. Vicente (Mallorca) | Limestone | 9.40 | 0 |
| 4 | Cala S. Vicente (Mallorca) | Limestone | 69.99 | 1 |
| 5A | El Atazar (Madrid) | Slate (stone) | 41.99 | 1 |
| 5B | El Atazar (Madrid) | Slate (lichen) | 89.21 | 1 |
Amplification results: 1, positive amplification; 0, absence of amplification products.
Values are expressed as ng of DNA/mg of stone.
Isolation of stone-inhabiting bacteria.
Stone samples were taken from lichen colonized and lichen-free surfaces in each sampling site. At each site, two to five rock faces separated by 20 to 200 m and in different orientations were sampled. The stone surface was disinfected with 96% ethanol to reduce the influence of dust and airborne spores. Samples for isolation plates were prepared by chipping or scrapping with a sterile knife and chisel blade to remove small superficial fragments from 10- to 20-cm2 area from the outer layer of the field collected rock. Fragments were pulverized in a sterile mortar.
Genomic DNA extraction.
Total genomic DNAs from the type strains used in the present study were recovered and purified as previously described (8).
Stone DNA extraction.
Total DNA extraction from lichen-colonized and lichen-free stone surfaces was adapted from a previously described method (26). Stone particles were (250 mg) frozen in liquid nitrogen, ground, and suspended in 0.5 ml of 0.4% (wt/vol) skim milk solution (Difco). Stone particles were sedimented by centrifugation for 10 min at 17,500 × g. Supernatants (300 μl) were mixed with 200 μl of extraction buffer (0.66% sodium dodecyl sulfate, 0.31 M NaCl, 110 mM potassium acetate [pH 5.1]), vortexed, and added to 500 μl of water-saturated phenol. After extraction, supernatants (400 μl) were recovered by centrifugation (15 min, 17,500 × g), and total DNA was precipitated overnight at −20°C by addition of 1 ml of ethanol. After a 15-min centrifugation at 17,500 × g, DNA was washed with 75% cold ethanol and finally dissolved in 100 μl of sterile distilled water. DNA was quantified by spectrometry at 260 nm. The initial yields of total stone DNA is indicated in Table 1. The concentrations of total stone DNA from all of the samples were normalized to 1 ng/μ land serially diluted from 1 to 10−5 ng/μl.
Design of oligonucleotide primers.
Sequence comparison and analysis were carried out by using programs from the University of Wisconsin GCG package (version 7.2, 1994). GenBank 16S rRNA/DNA sequences were used to design the primers Geosp2 (5′-TCCAAGAAATTGGTGCTA-3′) and Geosp1 (5′-CAGTTGTKGCCCAGAGAC-3′; reverse primer) shown in Fig. 1. Alignments of the 16S region were performed by using the multiple alignment software CLUSTAL W (21) to determine the regions only conserved among Geodermatophilaceae species from which the family-specific primers were derived. The genus specificity of oligonucleotides was tested against all DNA sequences available in GenBank with the FASTA program. The melting temperature (Tm) was estimated by using the formulae of Thomas and Dancis and the Lathe formulae (20). Relative Tm values obtained using 0.3 M as a standard salt concentration helped to design pairs of primers with similar high melting temperatures. The presence of primer dimers was discarded by analysis of the melting curve in RT-PCR using iCycler IQ software (Bio-Rad Laboratories, Hercules, CA). The designed oligonucleotides were supplied by Eurogentec (Liege, Belgium).
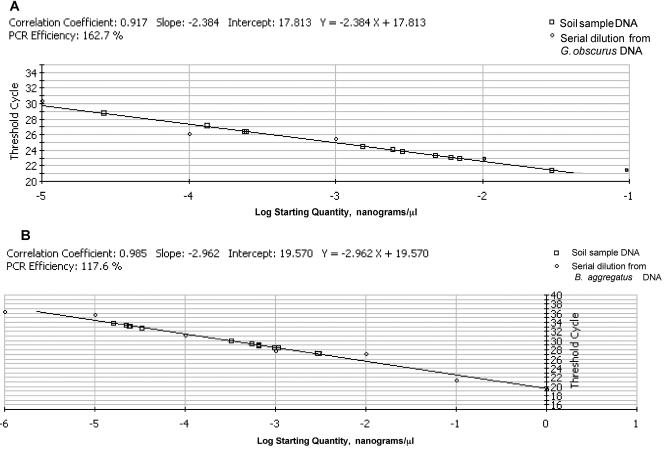
FIG. 1.
Standard curve for the log starting concentration of serial dilutions of DNA from Geodermatophilus obscurus DSM43160T (A) and Blastococcus aggregatus DSM4725T (B) versus the CT value.
RT-PCR amplification.
DNA preparations were used as a template DNA for the Taq polymerase. Reactions were performed in a final volume of 50 μl containing a 0.2 mM concentration of each of the four deoxynucleoside triphosphates (Roche), 0.1 μM concentrations of each primer, 10 μl of the extracted DNA (1/100 dilution) corresponding to 1 ng of total DNA, 0.5 U of Taq polymerase (Appligene) in its recommended reaction buffer, and 1 μl of a 1:10,000 dilution of SYBR Green Dye I. Controls without bacterial DNA were included for each PCR experiment. Amplifications were performed in a iCycler IQ detection system (Bio-Rad) according to the following profile: 40 cycles of 30 s at 93°C, 30 s at 49°C, and 2 min at 72°C, followed by 10 min at 72°C. Amplified products were confirmed by observation of a single melting peak and the presence of a unique band of the expected size in a 1.2% agarose gel stained with ethidium bromide.
Quantification of template DNA was performed by interpolation in a standard curve of threshold cycle (CT) values generated by amplification of known concentrations of genomic DNA of Geodermatophilus obscurus DSM43160T and Blastococcus aggregatus DSM4725T. Amplification efficiencies (E) were calculated by the formula E = 10(1/m) − 1, where m is the slope of the standard curve (7).
Calculation of 16S rRNA gene molecules.
To determine the number of molecules of 16S rRNA gene amplified by RT-PCR, we estimated the weight of the 587-bp Geodermatophilaceae-specific amplification fragment in 3.99 × 105 Da (587 bp × 680 Da/bp). To calculate the number of nanomoles per microgram, we used the following calculation: 103 ng/μg × [1 nmol/(3.99 × 105 ng)] = 2.5 × 10−3 nmol/μg. The number of molecules per microgram was determined as follows: [(6.022 × 1014 molecules)/nmol] × 2.5 × 10−3 nmol/μg = 1.505 × 1012 molecules/μg = 1.505 × 109 molecules/ng).
Cloning of PCR fragments from stone samples.
Amplification products with the primers Geosp2 and Geosp1 from stone samples and lichens were cloned by using the TOPO cloning Kit (Invitrogen Life Technologies).
DNA sequencing.
PCR primers 27f and 1525r (9) were used for the amplification of the 16S rRNA gene genes of the wild-type isolates. The 1,500 bp. PCR products were purified and used as a template in sequencing reactions with the primers 27f, 357f, 530f, 926f, 1392f, 1525r, 1110r, 685r, and 357r (9). Geodermatophilaceae amplification fragments from stone samples cloned into pCRTOPO vector were sequenced by using M13 forward and reverse primers (TOPO Cloning Kit; Invitrogen Life Technologies). Amplified and cloned DNA fragments were sequenced by using an ABI Prism Dye terminator cycle sequencing kit (Amersham Biosciences).
Sequence analysis.
Sequences were assembled by using the GCG Fragment Assembly System (Program Manual for the Wisconsin Package, version 8). Alignments of the 16S sequences were performed by using the multiple alignment program CLUSTAL W (22). The phylogenetic analysis was completed with 16S rRNA gene sequences of type species of actinomycetes available in GenBank. The data were resampled with 1,000 bootstrap replicates (4) by using the heuristic search option of PAUP (21). The percentage of bootstrap replicates that yielded each grouping was used as a measure of statistical confidence. A grouping found on 95% of the bootstrap replicates was considered statistically significant.
The GenBank accession numbers for the 16S rRNA gene sequences of Blastococcus sp. isolates CIBE-G1, CIBE-G2, CIBE-G3, CIBE-G4, CIBE-G5, CIBE-G6, CIBE-G7, CIBE-G8, CIBE-G9, CIBE-G10, CIBE-G11, CIBE-G12, CIBE-G13, and CIBE-G14 are AY903267 to AY903280, respectively. The accession numbers for the amplified partial 16S region from stone samples GEO1A-10, GEO1A-9, GEO1A-4, GEO1A-1, GEO1A-3, GEO1A-2, GEO1A-7, GEO1A-6, GEO1A-8, and GEO1A-5 are AY903281 to AY903290, respectively.
RESULTS AND DISCUSSION
PCR primers for the family Geodermatophilaceae.
The design of the two PCR specific primers for members of the family Geodermatophilaceae was based on the analysis of conserved sequences in the 16S rRNA gene in all members of the family. Primers Geosp2 (18 bp, forward) and Geosp1 (18 bp, reverse) hybridize, respectively, in positions 156 to 174 and positions 725 to 743 (2) of the 16S rRNA gene (see supplemental material). Sequence alignment shows a total identity with members of the genus Geodermatophilus. The species Modestobacter multiseptatus shows a single mismatch with one of the primers. Two Blastococcus species (Blastoccoccus aggregatus DSM4525T and Blastococcus saxobsidens BC444T, BC542, BC521, BC517, and BC512) show three mismatches with the sequence of the primer Geosp1 and one mismatch with the sequence of the primer Geosp2 (see supplemental material). This region is not so well conserved in other genera, where more than five mismatches can be observed. We have validated the primer pair Geosp2-Geosp1 with DNA extracted from three different subspecies of Geodermatophilus obscurus and from Blastococcus aggregatus DSM4525T obtaining in all cases the expected family-specific amplification fragment of 587 bp. No amplification was obtained when the primers were also tested with members ofthe families Pseudonocardiaceae, Micromonosporaceae, Actinosynnemataceae, Nocardiaceae, and Streptomycetaceae. Our data show that the primer pair Geosp2-Geosp1 allows the specific identification of all of the tested strains belonging to the family in spite of the small differences observed in their sequences.
Detection and quantification of Geodermatophilaceae DNA in stone surfaces by specific amplification.
The detection limits of the primers Geosp2 and Geosp1 was established in standard curves representing the threshold cycles (CT) versus the log of 10-fold serial dilutions (1 to 10−5 ng/μl) of DNA from Geodermatophilus obscurus DSM43160T and Blastococcus aggregatus DSM4725T (Fig. 1). A strong linear relationship between CT and the log of the starting copy number was demonstrated with both G. obscurus DSM43160T DNA and B. aggregatus DSM4725T DNA standard curves (correlation coefficients r2 ≥ 0.917 and r2 ≥ 0.985; amplification efficiencies [E] = 162.7% and [E] = 117.6%, respectively). The lowest estimated amount of DNA detected by PCR is in both cases in the range of 10−5 ng/μl. However, the CT values generated by amplification of the minimal level of DNA using both standard curves shows differences with lower values for G. obscurus DNA (CT = 30) than for B. aggregatus DNA (CT = 36) (Fig. 1).
The family-specific primers were applied to detect the presence of Geodermatophilaceae in DNA from stone samples obtained from two different geographic origins in Spain. Total DNAs were purified from the surface of five stone samples, including three lichen colonized and five lichen-free surfaces and diluted to a final concentration of 1 ng/μl. A specific amplification fragment was detected in all of the samples with the exception of the limestone sample three from Mallorca Island (Table 1). Serial dilutions of the template DNA samples (10−1 to 10−11 ng/μl) were tested in RT-PCR experiments to evaluate the concentration range of Geodermatophilaceae DNA in crude stone DNA extracts by extrapolation in both standard curves (Table 2). In all cases, the minimal concentration of total stone DNA required to obtain an amplification product was ca. 10−2 ng/μl, and no significant differences were observed between the stone DNAs and stone with lichen DNA samples (Table 2). The levels of Geodermatophilaceae DNA detected in total stone DNA samples as deduced from the quantitative amplification of serial dilutions show similar concentrations and range between 10−2 and 10−3 ng/μl.
TABLE 2.
RT-PCR quantification of Geodermatophilaceae DNA in serial dilutions of stone DNA samples using the primers Geosp2 and Geosp1a
| Sample | DNA quantification (ng/μl)
|
|||||
|---|---|---|---|---|---|---|
| Group A at dilution:
|
Group B at dilution:
|
|||||
| 10−1 | 10−2 | 10−3 | 10−1 | 10−2 | 10−3 | |
| 1A (stone) | 2.18 × 10−3 | 7.50 × 10−4 | NA | 3.22 × 10−3 | 3.43 × 10−4 | NA |
| 1B (lichen) | 9.70 × 10−3 | 9.52 × 10−5 | NA | 6.24 × 10−3 | 1.17 × 10−4 | NA |
| 2A (stone) | 1.66 × 10−2 | 8.40 × 10−4 | NA | 7.06 × 10−3 | 6.27 × 10−4 | NA |
| 2B (lichen) | 3.40 × 10−3 | 1.18 × 10−4 | NA | 3.25 × 10−3 | 8.42 × 10−5 | NA |
| 3 (limestone) | NA | NA | NA | NA | NA | NA |
| 4 (limestone) | 7.78 × 10−3 | 1.31 × 10−4 | NA | 3.74 × 10−3 | 4.55 × 10−4 | NA |
| 5A (stone) | 1.34 × 10−3 | NA | NA | 2.49 × 10−3 | NA | NA |
| 5B (lichen) | 1.46 × 10−3 | NA | NA | 1.17 × 10−3 | NA | NA |
Standard curves were established using serial dilutions of DNA from Geodermatophilus obscurus DSM43160T (group A) and DNA from Blastococcus aggregatus DSM4725T (group B). NA, no amplification.
The quantification data of Geodermatophilaceae DNA in stone DNA serial dilutions using G. obscurus and B. aggregatus DNA standard curves show results within the same concentration range. In both analysis, the minimal amount of Geodermatophilaceae DNA detected is ca. 10−4 ng/μl (Table 2). However, a higher concentration of Geodermatophilaceae DNA were observed in a 10−1 ng of DNA/μl dilution of stone sample 2A, where 1.66 × 10−2 and 7.06 × 10−3 ng/μl were estimated by using, respectively, the Geodermatophilus obscurus DSM43160T and Blastococcus aggregatus DSM4725T standard curves (Table 3).
TABLE 3.
Calculation of total 16S rRNA content and total cell counts per milligram of stone
| Sample | No. of 16S rRNA genes (molecules/mg of stone) | Total bacterial cells/mg of stone |
|---|---|---|
| 1A (stone) | 6.56 × 106 | 6.56 × 106-3.93 × 107 |
| 1B (lichen) | 2.92 × 107 | 2.92 × 107-1.75 × 108 |
| 2A (stone) | 5.01 × 107 | 5.01 × 107-3.06 × 108 |
| 2B (lichen) | 1.24 × 107 | 1.24 × 107-7.55 × 106 |
| 3 (limestone) | NAa | NA |
| 4 (limestone) | 2.34 × 107 | 2.34 × 107-1.40 × 108 |
| 5A (stone) | 4.04 × 106 | 4.04 × 106-2.42 × 107 |
| 5B (lichen) | 4.40 × 106 | 4.40 × 106-2.64 × 107 |
NA, no amplification.
When we compared the amplification data from crude DNA, the CT values generated by amplification of the minimal level of Geodermatophilaceae DNA using both standard curves show differences between both analysis, with lower values in G. obscurus curve (CT = 29 to 30) than in B. aggregatus curve (CT = 32 to 34). However, in spite of the lack of linearity observed in some DNA dilutions, in most of the cases DNA quantification data derived from the B. aggregatus DNA standard curve are smaller than those obtained from the same stone DNA dilutions with the G. obscurus standard curve. The lack of specificity using DNA from B. aggregatus can be due to the lack of total identity of the primer pair Geosp2-Geosp1 with the sequence of the 16S rRNA region of B. aggregatus (see supplemental material).
To evaluate the effect of nonspecific DNA on the detection level of our primers in amplification reactions with crude stone DNA preparations, we performed a competition experiment where Blastococcus aggregatus DSM4725T DNA was added to a DNA preparation of stone sample 3 where no amplification products were detected with primers Geosp1 and Geosp2. The presence of endogenous bacterial DNA in the stone sample was confirmed by RT-PCR experiments with the universal primers 1497f and 115r directed to conserved sequences at the end and beginning of the 16S and 23S rRNA genes, respectively (9). Serial dilutions of total stone DNA (10−1 ng/μl) containing B. aggregatus DNA (10−2 ng/μl) were tested in real-time PCR experiments to evaluate the concentration range of B. aggregatus DNA detected in these conditions. The minimal concentration of B. aggregatus DNA is 1.17 × 10−4 ng/μl, whereas this stone sample dilution contains 10−3 ng/μl B. aggregatus DNA showing the 10-fold reduction in the detection levels in crude stone samples (Table 4).
TABLE 4.
RT-PCR quantification of Geodermatophilaceae DNA in serial dilutions of stone DNA from sample 3A using the primer pairs 1497f-115r and Geosp2-Geosp1
| Sample | Primer pair | DNA concn (ng/μl) at dilution:
|
|||
|---|---|---|---|---|---|
| 10−1 | 10−2 | 10−3 | 10−4 | ||
| 3A | 1497f-115r | 5.67 × 10−1 | 4.57 × 10−2 | 7.98 × 10−3 | 5.17 × 10−4 |
| 3A | Geosp2-Geosp1 | NAa | NA | NA | NA |
| 3A + DNA | Geosp2-Geosp1 | 6.24 × 10−3 | 1.17 × 10−4 | NA | NA |
NA, no amplification.
16S rRNA content and total cell counts.
Our data show the relative low frequency of DNA sequences from members of Geodermatophilaceae in the stone samples studied, suggesting their minor presence in these environments. To evaluate the presence of Geodermatophilaceae cells in stones we have estimated that the number of 16S rRNA gene molecules in crude stone DNA preparations range from 106 to 107 molecules/ng of crude stone DNA (10−1 ng/μl) (Table 3). The highest 16S rRNA gene content was observed in stone sample 2A from La Cabrera, with 5.01 × 107 molecules/ng (Table 3).
The number of 16S rRNA genes per genome reported in Actinomycetales is still limited to a small number of genera, and it has been shown to vary from one to six copies as described in Streptomyces and Thermomonospora species (14, 27; rRNA Operon Copy Number Database [http://rrndb.cme.msu.edu]). Given the lack of information regarding the number of 16S rRNA genes per genome in Geodermatophilaceae, we have considered a range of one to six genes per genome to calculate the number of cells per milligram of stone (Table 3). Total cell counts in stone samples ranged from a high of 5.01 × 107 to 3.06 × 108 (sample 2A) to a low of 4.04 × 106 to 2.42 × 107 (sample 5B) cells/mg of stone using DNA from G. obscurus in the standard curve (dilution of 10−1 ng/μl) (Table 3).
These results confirm the ability of these pairs of primers to detect the presence of Geodermatophilaceae directly from crude stone DNA extracts prior to the microbial isolation, and this is the first study detecting and quantifying this taxon in the environment using quantitative PCR.
Cloning and sequence analysis of PCR products from stone DNA.
To confirm the specificity of the PCR primers, we cloned and sequenced the amplification fragments obtained by using the primers Geosp1 and Geosp2 from the stone sample 1A from La Cabrera Mountains. Ten transformants were randomly selected, and the sequences obtained from the 587-bp amplification fragments (positions 133 to 720 of 16S rRNA gene region) showed a high conservancy among the sequences, with nucleotide similarity ranging between 99.0 to 99.87%. FastA analysis of cloned sequences shows as closest match sequences of type species of the genera Geodermatophilus and Blastococcus (99% similarity), therefore validating the specificity of the primers for the detection of members of Geodermatophilaceae in stone samples.
Identification of wild-type isolates and evaluation of their diversity.
One of the purposes of the design of the primers was the rapid detection of members of the family among the strains that are isolated from the environment and that share morphological traits with the family Geodermatophilaceae. We have applied these pairs of primers to the rapid PCR identification of 14 wild-type strains isolated from stone sample 1A (La Cabrera Mountain, Madrid, Spain) that exhibited morphological characteristics of Geodermatophilaceae. All of the isolates were dark-brown pigmented and exhibited a sparse growth as irregularly shaped coccoid cell aggregates. The tentative taxonomic identification of the wild-type isolates was confirmed in all cases by real-time PCR with Geodermatophilaceae-specific primers (data not shown). To confirm the genus assignment of our isolates, we determined the almost complete nucleotide sequence of the 16S rRNA gene of the 14 strains. In all of the cases we found that the annealing sequence of each pair of primers was highly conserved among the strains, supporting the specificity of the primers. The alignment shows high homology with the sequence of the forward primer Geosp2 (one mismatch) and reverse primer Geosp1 (two mismatches) (see supplemental material). Among the strains assigned to the family Geodermatophilaceae the level of sequence similarity for the complete 16S rRNA gene sequence ranges from 98.9 to 99.86%, indicating the high relatedness of this group of isolates obtained from the same environment. For all of the isolates the closest sequence match was observed with species of the genus Blastococcus (sequence similarity levels 97.1 to 99.2%), and lower sequence homologies were observed with other members of the family, such as species of the genus Geodermatophilus (96.7 to 96.4%) and the genus Modestobacter (97.%).
The sequences from cloned amplification fragments of stone sample 1A DNA and the homologous region of the 16S rRNA gene of the 14 sequenced wild-type strains present a high sequence similarity that ranged between 98 and 96.5%. The high sequence conservation of the amplified region makes it impossible to distinguish at the genus level among members of the family, and therefore these results do not exclude the presence of members of other genera of the family in the stone sample.
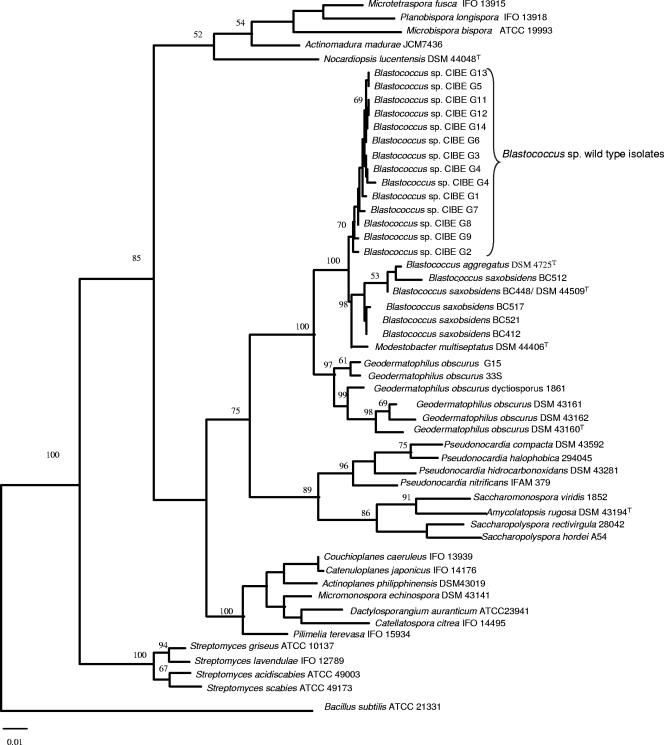
A phylogenetic tree based on complete 16S rRNA gene sequences of the wild-type isolates was built by using the maximum-parsimony method (Fig. 2) showing the inter- and intraspecific relationships of the wild-type strains to reference strains of the family Geodermatophilaceae, such as the genera Geodermatophilus, Blastococcus, and Modestobacter and other members of the order Actinomycetales. The topology of the tree confirms previous works where the monophyletic lineage of the family Geodermatophilaceae is supported by 100% of bootstrap value (24). All of the wild-type strains in our study cluster with 70% of bootstrap value and are closely related within the same group with species of the genus Blastococcus. This close relationship to members of this genus is highly supported by the bootstrapping values (100%). Nevertheless, the presence of this cluster clearly distinct from the cluster grouping the remaining species of Blastococcus highly supported by a 98% of bootstrap could suggest the existence of a new species within the genus Blastococcus that will have to be investigated further given the different morphological features exhibited in culture. Previous works on stones biodiversity have noted that, whereas strains of the genus Geodermatophilus are very rare, strains of Modestobacter are very common on stone surfaces from different climates and strains of the genus Blastococcus could be found on rock surfaces in the Mediterranean basin (25). Our data support the view that the interspecific diversity of the genus Blastococcus is higher than the number of species described at the moment, being the surface of rock samples and the microenvironment that is associated valuable sources for isolation of new members of the genus.
FIG. 2.
Phylogenetic tree showing the position of the wild-type strains of the family Geodermatophilaceae derived from 16S rRNA gene analysis by using the maximum-parsimony method in PAUP 4.0. The numbers above the branches indicate the bootstrap percentage from 1,000 replicates that was used as a measure of statistical confidence. GenBank accession numbers of sequences from all of the species included in the analysis are as follows: X92359 (Geodermatophilus obscurus G16), X92357 (Geodermatophilus obscurus DSM 43162), X92355 (Geodermatophilus obscurus DSM 43161), X92356 (Geodermatophilus obscurus DSM 43160), L46061 (Geodermatophilus obscurus dictyosporus 33S), L46020 (Geodermatophilus obscurus G15), Y18646 (Modestobaster multiseptatus DSM 44406), AJ315674 (Blastococcus saxobsidens BC412), AJ316573 (Blastococcus saxobsidens BC521), AJ316572 (Blastococcus saxobsidens BC517), AJ316571 (Blastococcus saxobsidens BC448/DSM 44509T), AJ316570 (Blastococcus saxobsidens BC512), X97888 (Nocardiopsis lucentesis U0297), D85497 (Streptoalloteichus hindustans IFO15115), X53191 (Kibdelosporangium aridum 2030), Z38017 (Saccharomonospora azurea K161), AF139830 (Saccharomonospora glauca KCTC 3673), Z38021 (Saccharomonospora viridis 1852), AF051342 (Amycolatopsis rugosa DSM 43194T), U9334 (Saccharopolyspora hirsuta ATCC 27865), X53197 (Saccharopolyspora hordei A54), AF061976 (Saccharopolyspora rectivirgula 28042), Z14111 (Pseudonocardia halophobica 294045), X55609 (Pseudonocardia nitrificans IFAM 379), AJ252826 (Pseudonocardia hidrocarboxidans DSM 43281), X76959 (Pseudonocardia compacta DSM 43592), X93187 (Actinoplanes philippinensis DSM 43019), D85479 (Couchioplanes caeruleus IFO 13939), D85476 (Catenuloplanes japonicus IFO 14176), X92597 (Micromonospora echinospora DSM 43141), U58528 (Dactylosporangium aurantiacum ATCC 23491), D85477 (Catellatospora citrea IFO 14495), D86946 (Pilimelia terevasa IFO 15934), X97888 (Nocardiopsis lucentesis DSM 44048T 14626), U48973 (Microtetraspora fusca IFO 13915), D85494 (Planobispora longispora IFO 13918), U83912 (Microbispora bispora ATCC 19993), U58257 (Actinomadura madurae JCM 7436), AB041132 (Streptomyces acidiscabies ATCC 49003), D63862 (Streptomyces scabies ATCC 49173), Y15501 (Streptomyces griseus ATCC 10137), D85116 (Streptomyces lavendulae IFO 12789), and AB018487 (Bacillus subtilis ATCC 21331).
In the present study we have presented the design and application of the new pair of family-specific primers as a simple method for the rapid identification of new members of the family Geodermatophilaceae using RT-PCR. Our results with reference strains, as well as the high degree of conservancy observed among the sequences of the wild-type isolates, validate the specificity of this new pair of primers. The genetic diversity of the wild-type strains obtained from stone samples supports the usefulness of these tools for the specific detection and tentative family assignment of new isolates of these taxa. Further studies with these primers could reveal the extreme value of these tools to determine the presence and distribution of new members of this family in alternative ecological niches not yet explored for this bacterial group.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ahrens, R., and G. Moll. 1970. Ein neues knospendes Bakterium aus der Ostsee. Arch. Microbiol. 70:243-265. [PubMed] [Google Scholar]
- 2.Brosius, J., T. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of an rRNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 3.Eppard, M., W. E. Krumbein, K. Koch, K. Rhiel, J. T. Staley, and E. Stackebrandt. 1996. Morphological, physiological, and molecular characterization of actinomycetes isolated from dry soil, stones, and monument surfaces. Arch. Microbiol. 166:12-22. [DOI] [PubMed] [Google Scholar]
- 4.Felsenstein, J. 1985. Confidence intervals on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 5.Groth, I., R. Vetterman, B. Schuetze, P. Schumann, and C. Sáiz-Jimenez. 1999. Actinomycetes in Karstic caves of northern Spain Altamira and Tito Bustillo. J. Microbiol. Methods 36:115-122. [DOI] [PubMed] [Google Scholar]
- 6.Gurtner, C., J. Heyrmann, G. Piñar, W. Lubitz, J. Swings, and S. Rölleke. 2000. Comparative analyses of the bacterial diversity on two different biodeteriorated wall paintings by DGGE and 16S rRNA gene sequence analysis. Int. Biodeterior. Biodegrad. 46:229-239. [Google Scholar]
- 7.Higuchi, C., R. Fockler, C. Dollinger, and R. Watson. 1993. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology 11:1026-1030. [DOI] [PubMed] [Google Scholar]
- 8.Innis, M. A., D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.). 1990. PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, Calif.
- 9.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 10.Luedemann, G. M. 1968. Geodermatophilus, a new genus of the Dermatophilaceae Actinomycetales. J. Bacteriol. 96:1848-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McVeigh, H. P., M. Divers, S. Warwick, J. Munro, and T. M. Embley. 1995. Exploration of Actinomycetes diversity using rRNA sequences, p. 253-260. In V. G. Debabov, Y. V. Dudnik, and V. N. Danilenko (ed.), Proceedings of the Ninth International Symposium on the Biology of the Actinomycetes. All-Russia Scientific Research Institute for Genetics and Selection of Industrial Microorganisms, Moscow, Russia.
- 12.Mevs, U., E. Stackebrandt, P. Schumann, C. A. Gallikowski, and P. Hirsch. 2000. Modestobacter mutiseptatus gen. nov., sp. nov., a budding actinomycete from soils of the Asgard Range Transatlantic Mountains. Int. J. Syst. Evol. Microbiol. 50:337-346. [DOI] [PubMed] [Google Scholar]
- 13.Morón, R., I. González, and O. Genilloud. 1999. New genus-specific primers for the PCR identification of members of the genera Pseudonocardia and Saccharopolyspora. Int. J. Syst. Bacteriol. 49:149-162. [DOI] [PubMed] [Google Scholar]
- 14.Omura, S., H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, M. Shinose, Y. Takahashi, H. Horikawa, H. Nazakawa, T. Osonoe, H. Kikuchi, T. Shiva, Y. Sakaki, and M. Hattori. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 98:12215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rheims, H., and E. Stackebrandt. 1999. Application of nested polymerase chain reaction for the detection of as yet uncultured organism of the class Actinobacteria in environmental samples. Environ. Microbiol. 1:137-143. [DOI] [PubMed] [Google Scholar]
- 16.Salazar, O., I. González, and O. Genilloud. 2002. New genus-specific primers for the PCR identification of new isolates of the genera Nocardiopsis and Saccharothrix. Int. J. Syst. Evol. Microbiol. 52:1411-1421. [DOI] [PubMed] [Google Scholar]
- 17.Salazar, O., R. Morón, and O. Genilloud. 2000. New genus-specific primers for the PCR identification of the genus Saccharomonospora and evaluation of the microbial diversity of wild-type isolates of Saccharomonospora detected from soil DNAs. Int. J. Syst. Evol. Microbiol. 50:2043-2055. [DOI] [PubMed] [Google Scholar]
- 18.Stackebrandt, E., F. A. Rainey, and N. L. Ward-Rainey. 1997. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Evol. Microbiol. 47:479-491. [Google Scholar]
- 19.Stackebrandt, E., D. Witt, C. Kemmerling, R. Kroppenstedt, and W. Liesack. 1991. Designation of streptomycete 16S and 23S rRNA-based target regions for oligonucleotide probes. Appl. Environ. Microbiol. 57:1468-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 21.Swofford, D. L. 1993. PAUP: phylogenetic analysis using parsimony, version 3.1.1. Laboratory of Molecular Systematics, Smithsonian Institute, Washington, D.C.
- 22.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urzì, C., V. La Cono, and E. Stackebrandt. 2004. Design of two oligonucleotide probes for the identification of Geodermatophilaceae strains using fluorescence in situ hybridization FISH. Environ. Microbiol. 6:678-685. [DOI] [PubMed] [Google Scholar]
- 24.Urzì, C., P. Salamone, P. Schumann, M. Rohde, and E. Stackebrandt. 2004. Blastococcus saxobsidens sp. nov., and emended description of the genus Bastococcus aggregatus Ahrens and Moll 1970. Int. J. Syst. Evol. Microbiol. 54:253-259. [DOI] [PubMed] [Google Scholar]
- 25.Urzì, C., L. Brusetti, P. Salamone, C. Sorlini, E. Stackebrandt, and C. Danffochio. 2001. Biodiversity of Geodermatophilaceae isolated from altered stones and monuments in the Mediterranean basin. Environ. Microbiol. 3:471-479. [DOI] [PubMed] [Google Scholar]
- 26.Volossiouk, T., E. J. Rob, and R. N. Nazar. 1995. Direct DNA extraction for PCR-mediated assays of soil organisms. Appl. Environ. Microbiol. 61:3972-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yap, W. H., and Z. Zhang. 1999. Distinct types of rRNA operons exist in genome of the actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon. J. Bacteriol. 181:5201-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.