Abstract
The RuvABC proteins of Escherichia coli play an important role in the processing of Holliday junctions during homologous recombination and recombinational repair. Mutations in the ruv genes have a moderate effect on recombination and repair in wild-type strains but confer pronounced recombination deficiency and extreme sensitivity to DNA-damaging agents in a recBC sbcBC background. Genetic analysis presented in this work revealed that the ΔruvABC mutation causes an identical DNA repair defect in UV-irradiated recBC sbcBC, sbcBC, and sbcB strains, indicating that the sbcB mutation alone is responsible for the extreme UV sensitivity of recBC sbcBC ruv derivatives. In experiments with gamma irradiation and in conjugational crosses, however, sbcBC ΔruvABC and sbcB ΔruvABC mutants displayed higher recombination proficiency than the recBC sbcBC ΔruvABC strain. The frequency of conjugational recombination observed with the sbcB ΔruvABC strain was quite similar to that of the ΔruvABC single mutant, indicating that the sbcB mutation does not increase the requirement for RuvABC in a recombinational process starting from preexisting DNA ends. The differences between the results obtained in three experimental systems used suggest that in UV-irradiated cells, the RuvABC complex might act in an early stage of recombinational repair. The results of this work are discussed in the context of recent recombination models which propose the participation of RuvABC proteins in the processing of Holliday junctions made from stalled replication forks. We suggest that the mutant SbcB protein stabilizes these junctions and makes their processing highly dependent on RuvABC resolvase.
Homologous recombination is a fundamental cellular process that promotes genetic diversity and plays an important role in repairing various types of DNA damage. In addition, homologous recombination participates in DNA replication by promoting the restoration of collapsed replication forks (22, 26, 27, 35).
In wild-type Escherichia coli, most recombination events are initiated by the RecBCD enzyme (10, 30, 44). The RecBCD pathway is essential for repair of double-stranded DNA breaks; it also participates in recombination following conjugation, transduction, and transformation as well as in vegetative phage crosses. The RecBCD enzyme initiates recombination by unwinding and simultaneously degrading DNA from a double-stranded DNA end. Upon encountering a specific sequence named Chi, the 3′-5′ nuclease activity of the enzyme is attenuated, and a weaker nuclease activity of the opposite polarity, 5′-3′, is activated (1, 14). This nuclease modification allows the production of a long 3′-terminal single-stranded DNA (ssDNA) tail, which is used by RecA protein to invade a homologous duplex DNA molecule (2, 3, 13).
Strains lacking recBC function are recombination deficient and sensitive to UV light, ionizing radiation, and other DNA-damaging agents (9). The residual level of recombination in recBC null mutants can be restored to the wild-type level by extragenic suppressor mutations sbcB and sbcC(D). The sbcB mutations inactivate exonuclease I (ExoI), which digests ssDNA in the 3′-5′ direction (24). It is presumed that elimination of ExoI by an sbcB mutation might prolong the life of ssDNA molecules as potential substrates for recombination. The sbcC mutation is needed for full suppression of recBC and accumulates spontaneously during the growth of recBC sbcB cells (29).
The same effect was observed with mutations in another gene, sbcD, located immediately downstream of sbcC (17). It was recently shown that the SbcCD complex acts as a nuclease that cleaves hairpin structures formed at replication forks (12). In recBC sbcBC(D) mutants, recombination proceeds via the RecF pathway, which requires the products of several genes, including recQ, recJ, recF, recO, and recR (reviewed in references 11 and 30). The joint action of the RecQ helicase and the RecJ nuclease on a DNA duplex provides the recombinogenic 3′-ssDNA which is a substrate for the RecA protein. The RecF, RecO, and RecR proteins facilitate the loading of RecA onto the ssDNA.
Homologous pairing and strand exchange reactions catalyzed by RecA lead to the formation of a Holliday junction, a recombination intermediate that consists of two homologous DNA duplexes linked by a single-stranded crossover. During the final stage of recombination, Holliday junctions are processed into mature recombinant molecules. Three Ruv proteins involved in this process, RuvA, RuvB, and RuvC, have been characterized (49). RuvA specifically recognizes Holliday junctions and allows the RuvB helicase to bind. Together, RuvA and RuvB catalyze branch migration of recombination intermediates, leading to the extension of heteroduplex DNA (21, 45). Additional binding of the RuvC endonuclease allows the RuvABC complex to resolve Holliday junctions by nicking strands of like polarity (5, 20).
Cells carrying mutations in any of the ruv genes display moderate deficiencies in recombination and sensitivity to DNA-damaging agents (28, 43). This phenotype of ruv mutants is much more pronounced in a recBC sbcBC background, suggesting that the RuvABC complex is indispensable for recombination and repair in the RecF pathway (28). This difference in the requirement of two recombinational pathways for RuvABC is, however, poorly understood. The aim of this work was to examine the genetic basis for the extreme recombination deficiency of recBC sbcBC ruv mutants.
MATERIALS AND METHODS
Strains.
The E. coli strains used in this study are listed in Table 1. Most of them are derivatives of AB1157. New strains were constructed by P1 transduction, as described by Miller (36). To obtain sbcB15 sbcC201 strain LMM965, the parental strain JC7623 was first marked with recC266::Tn10 and then transduced to recBC+ with fuc3154::Tn10 kan. The Kmr Tcs transductants were checked for recBC+ phenotype by examining the efficiency of plating of the T42am phage (which is decreased about 1,000-fold in recBC+ cells in comparison to recB or recC mutants) (37). The same procedure was used with the rus-1 derivative of JC7623 to obtain sbcB15 sbcC201 rus-1 strain LMM967. To construct a strain that carries only sbcB15, the wild-type sbcC+ allele was cotransduced with phoR79::Tn10 into the sbcB15 sbcC201 strain LMM965. Strain LMM984 sbcB15 rus-1 was constructed in the same way, starting from the sbcB15 sbcC201 rus-1 strain LMM967. The sbcC+ phenotype of Tcr transductants was confirmed by the decreased efficiency of plating of λ phage carrying a 571-bp palindrome (17). λpal571 formed plaques on sbcC+ constructs LMM979 and LMM984 with about 300-fold-lower efficiency than on parental strains carrying mutations in sbcC.
TABLE 1.
E. coli strains
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| AB1157 | Wild type | 4 |
| LMM10 | ΔruvABC::cam | P1.JJC768 × AB1157 to Cmr UVs |
| JC5519 | recB21 recC22 | 4 |
| LMM20 | recB21 recC22 ΔruvABC::cam | P1.JJC768 × JC5519 to Cmr |
| JC7623 | recB21 recC22 sbcB15 sbcC201 | 4 |
| LMM864 | recB21 recC22 sbcB15 sbcC201 ΔruvABC::cam | P1.JJC768 × JC7623 to Cmr UVs |
| TNM734 | recB21 recC22 sbcB15 sbcC201 rus-1 | 33 |
| LMM867 | recB21 recC22 sbcB15 sbcC201 rus-1 ΔruvABC::cam | P1.JJC768 × TNM734 to Cmr |
| LMM963 | recB21 recC266::Tn10 sbcB15 sbcC201 | P1.N2103 × JC7623 to Tcr |
| LMM964 | recB21 recC266::Tn10 sbcB15 sbcC201 rus-1 | P1.N2103 × TNM734 to Tcr |
| LMM965 | fucP3154::Tn10 kan recB+ recC+ sbcB15 sbcC201 | P1.CAG12115 × LMM963 to Kmr Tcs T42r |
| LMM966 | fucP3154::Tn10 kan recB+recC+sbcB15 sbcC201 ΔruvABC::cam | P1.JJC768 × LMM965 to Cmr UVs |
| LMM967 | fucP3154::Tn10 kan recB+recC+sbcB15 sbcC201 rus-1 | P1.CAG12115 × LMM964 to Kmr Tcs T42r |
| LMM969 | fucP3154::Tn10 kan recB+recC+sbcB15 sbcC201 rus-1 ΔruvABC::cam | P1.JJC768 × LMM967 to Cmr |
| LMM979 | fucP3154::Tn10 kan recB+recC+sbcB15 sbcC+phoR79::Tn10 | P1.K797 × LMM965 to Tcr λpalr |
| LMM983 | fucP3154::Tn10 kan recB+recC+sbcB15 sbcC+phoR79::Tn10 ΔruvABC::cam | P1.JJC768 × LMM979 to Cmr UVs |
| LMM984 | fucP3154::Tn10 kan recB+recC+sbcB15 sbcC+phoR79::Tn10 rus-1 | P1.K797 × LMM967 to Tcr λpalr |
| LMM986 | fucP3154::Tn10 kan recB+recC+sbcB15 sbcC+phoR79::Tn10 rus-1 ΔruvABC::cam | P1.JJC768 × LMM984 to Cmr |
| N2364 | sbcC201 | 29 |
| LMM971 | sbcC201 ΔruvABC::cam | P1.JJC768 × N2364 to Cmr UVs |
| LMM995 | recB21 recC22 sbcB15 sbcC201 zef-3129::Tn10 | P1.CAG12099 × JC7623 to Tcr |
| LMM997 | ΔxonA300::cat | P1.STL2694 × AB1157 to Cmr |
| LMM1001 | sbcB15 zef-3129::Tn10 | P1.LMM995 × LMM997 to Tcr Cms |
| LMM1005 | sbcB15 zef-3129::Tn10 ΔruvABC::cam | P1.JJC768 × LMM1001 to Cmr UVs |
| LMM1015 | ruvB71::kan | P1.N4680 × AB1157 to Kmr UVs |
| LMM1017 | ΔxonA300::cat ruvB71::kan | P1.N4680 × LMM997 to Kmr UVs |
| LMM1018 | sbcB15 zef-3129::Tn10 ruvB71::kan | P1.N4680 × LMM1001 to Kmr UVs |
| LMM1032 | recJ2052::Tn10 kan | P1.STL113 × AB1157 to Kmr |
| LMM1033 | recB21 recC22 sbcB15 sbcC201 recJ2052::Tn10 kan | P1.STL113 × JC7623 to Kmr UVs |
| LMM1034 | sbcB15 zef-3129::Tn10 recJ2052::Tn10 kan | P1.STL113 × LMM1001 to Kmr |
| LMM1035 | sbcB15 zef-3129::Tn10 recJ2052::Tn10 kan ΔruvABC::cam | P1.STL113 × LMM1005 to Kmr |
| JJC768 | ΔruvABC::cam | B. Michel |
| N2103 | recC266::Tn10 | R. G. Lloyd |
| CAG12115 | fucP3154::Tn10 kan | CGSC 7428a |
| K797 | phoR79::Tn10 | CGSC 6456a |
| STL2694 | ΔxonA300::cat | 46 |
| CAG12099 | zef-3129::Tn10 | CGSC 7397a |
| N4680 | ruvB71::kan | R. G. Lloyd |
| STL113 | recJ2052::Tn10 kan | 46 |
| Hfr3000 | Hayes PO1 proAB+ | 4 |
Strain supplied from the E. coli Genetic Stock Center by M. Berlyn.
Media, growth conditions, and irradiation.
Bacteria were grown in Luria-Bertani (LB) medium and on LB plates (36). When required, LB plates were supplemented with the appropriate antibiotics: tetracycline, 10 μg/ml; kanamycin, 25 μg/ml; and chloramphenicol, 15 μg/ml.
In all experiments, bacteria were grown from a single colony in LB medium at 37°C until they reached an optical density at 600 nm of 0.2. For UV experiments, serial dilutions of bacterial cultures were plated on LB plates and irradiated with a dose of UV (254-nm) light of 10 J/m2. The dose rate was 0.25 J/m2/s. In experiments with gamma irradiation, bacteria were first exposed to a dose of 200 Gy and then plated for colonies. γ-Irradiation was carried out at 0°C from a 60Co source at a dose rate of 12.5 Gy/s. Colonies of survivors were scored after 24 to 48 h of incubation at 37°C.
Conjugational crosses.
Hfr crosses were performed as described by Miller (36). Inheritance of the chromosomal Pro+ marker was assayed. Donor (Hfr3000) and recipient strains were grown to an optical density at 600 nm of 0.2 at 37°C before being mixed at a ratio of 1:10. Matings were allowed for 30 min. proAB+ recombinants were selected on M9 plates (36) supplemented with glucose (0.4%), vitamin B1 (1 μg/ml), and all required amino acids (100 μg/ml) except proline. Streptomycin (100 μg/ml) was also added to the plates to counterselect donors.
RESULTS AND DISCUSSION
recBC sbcBC mutants of E. coli display recombinational repair proficiency similar to that of rec+ sbc+ cells (18) (compare strains JC7623 and AB1157 in Tables 2 and 3), suggesting that the two recombinational pathways (RecBCD and RecF) are equally efficient in repairing DNA strand breaks. Current models of homologous recombination and recombinational repair imply that the final stage of this process (i.e., the resolution of Holliday intermediates by the RuvABC complex) is common to both pathways (23, 26). However, it was shown that mutations in the ruv genes more strongly affect recombination and repair in recBC sbcBC mutants than in wild-type cells (28) (Tables 2 and 3).
TABLE 2.
Survival of different ruv derivatives after UV irradiationa
| Strain | Relevant genotype | Avg survival ratio ± SD at 10 J/m2 |
|---|---|---|
| AB1157 | Wild type | 0.89 ± 0.04 |
| LMM10 | ΔruvABC | 0.051 ± 0.002 |
| JC7623 | recBC sbcBC | 0.73 ± 0.02 |
| LMM864 | recBC sbcBC ΔruvABC | 0.0004 ± 0.00003 |
| JC5519 | recBC | 0.046 ± 0.012 |
| LMM20 | recBC ΔruvABC | 0.035 ± 0.004 |
| LMM965 | sbcBC | 0.69 ± 0.04 |
| LMM966 | sbcBC ΔruvABC | 0.0003 ± 0.0001 |
| LMM979 | sbcB | 0.84 ± 0.05 |
| LMM983 | sbcB ΔruvABC | 0.00039 ± 0.00008 |
| N2364 | sbcC | 0.83 ± 0.05 |
| LMM971 | sbcC ΔruvABC | 0.023 ± 0.003 |
| LMM984 | sbcB rus-1 | 0.89 ± 0.06 |
| LMM986 | sbcB rus-1 ΔruvABC | 0.68 ± 0.06 |
| LMM997 | ΔxonA | 0.90 ± 0.04 |
| LMM1015 | ruvB | 0.045 ± 0.004 |
| LMM1017 | ΔxonA ruvB | 0.041 ± 0.005 |
| LMM1018 | sbcB ruvB | 0.00062 ± 0.00011 |
| LMM1032 | recJ | 0.54 ± 0.09 |
| LMM1033 | recBC sbcBC recJ | 0.00005 ± 0.00002 |
| LMM1034 | sbcB recJ | 0.54 ± 0.07 |
| LMM1035 | sbcB recJ ΔruvABC | 0.00028 ± 0.00009 |
In all strains listed, the designation sbcB corresponds to the sbcB15 mutation. Values are averages ± standard deviations of results from three independent experiments. Ratios of the number of colonies on irradiated plates to that on nonirradiated plates are given.
TABLE 3.
Survival of different ΔruvABC derivatives after γ-irradiationa
| Strain | Relevant genotype | Avg survival ratio ± SD at 200 Gy |
|---|---|---|
| AB1157 | Wild type | 0.64 ± 0.07 |
| LMM10 | ΔruvABC | 0.067 ± 0.020 |
| JC5519 | recBC | 0.0007 ± 0.0001 |
| LMM20 | recBC ΔruvABC | 0.0004 ± 0.00005 |
| JC7623 | recBC sbcBC | 0.70 ± 0.18 |
| LMM864 | recBC sbcBC ΔruvABC | 0.00002 ± 0.00001 |
| LMM965 | sbcBC | 0.48 ± 0.12 |
| LMM966 | sbcBC ΔruvABC | 0.0003 ± 0.0001 |
| LMM979 | sbcB | 0.51 ± 0.05 |
| LMM983 | sbcB ΔruvABC | 0.0011 ± 0.0006 |
| N2364 | sbcC | 0.48 ± 0.02 |
| LMM971 | sbcC ΔruvABC | 0.033 ± 0.006 |
| TNM734 | recBC sbcBC rus-1 | 0.60 ± 0.05 |
| LMM867 | recBC sbcBC rus-1 ΔruvABC | 0.37 ± 0.02 |
| LMM967 | sbcBC rus-1 | 0.44 ± 0.05 |
| LMM969 | sbcBC rus-1 ΔruvABC | 0.37 ± 0.02 |
| LMM984 | sbcB rus-1 | 0.73 ± 0.09 |
| LMM986 | sbcB rus-1 ΔruvABC | 0.49 ± 0.09 |
In all strains listed, the designation sbcB corresponds to the sbcB15 mutation. Values are averages ± standard deviations of results from at least three independent experiments. Ratios of the number of colonies formed by irradiated cells to that formed by nonirradiated cells are given.
Since recBC sbcBC mutations inactivate three recombination-associated enzymes (RecBCD, ExoI, and SbcCD), we considered the possibility that some of these mutations could be responsible for the extreme repair deficiency that is typical of recBC sbcBC ruv strains. In order to test this possibility, we constructed a series of strains in which the ruvABC deletion was combined with particular mutations from the recBC sbcBC strain JC7623 (Table 1). The recombination proficiency of these strains was tested by measuring their survival after UV irradiation. The result obtained with a recBC ΔruvABC mutant was almost identical to those observed with recBC and ΔruvABC single mutants; UV survival of all three mutants was decreased about 20-fold in comparison to the wild-type strain (Table 2). This result shows that the combination of these mutations is not responsible for the extreme UV sensitivity of recBC sbcBC ΔruvABC cells. In contrast, the sbcBC ΔruvABC mutant LMM966 was 2,300-fold and 3,000-fold more sensitive to UV than its parental sbcBC strain LMM965 and wild-type strain AB1157, respectively. This high UV sensitivity was almost identical to that of the recBC sbcBC ΔruvABC strain LMM864.
In further analysis, we wanted to examine the contribution of particular sbc mutations to the UV sensitivity of the sbcBC ΔruvABC strain. We found that the sbcB ΔruvABC mutant LMM983 was as UV sensitive as sbcBC ΔruvABC and recBC sbcBC ΔruvABC strains (Table 2). On the other hand, the sbcC ΔruvABC strain LMM971 displayed moderate UV sensitivity similar to that of the ΔruvABC single mutant. Therefore, we concluded that the sbcB15 mutation alone is responsible for the extreme repair deficiency observed with ruv derivatives of the recBC sbcBC strain JC7623. In other words, in the presence of the sbcB15 mutation, recombinational repair of UV-induced DNA lesions relies on the RuvABC complex much more than in an sbcB+ background.
The RuvABC complex has been shown to promote two reactions in vitro, a Holliday junction branch migration reaction catalyzed by RuvAB, and a junction resolution reaction catalyzed by RuvC (49). To determine which of the two activities of RuvABC is critical for the repair proficiency of an sbcB mutant, we studied the effect of a rus-1 mutation on the UV survival of the sbcB ΔruvABC strain. The rus-1 mutation was previously shown to induce synthesis of the RusA protein, which can substitute for RuvABC in the process of Holliday junction resolution but does not catalyze branch migration (8, 32, 42). Our results showed that both sbcB rus-1 strain LMM984 and sbcB rus-1 ΔruvABC strain LMM986 were as UV resistant as the wild-type strain (Table 2). We therefore inferred that Holliday junction resolution is indispensable for successful recombinational repair in the sbcB mutant of E. coli.
The results described above suggest that interfering with ExoI function makes DNA repair more dependent on the RuvABC complex. The sbcB15 mutation is known to abolish the activity of ExoI but does not prevent synthesis of the mutant protein (38). Although it is generally assumed that the effect of the sbcB15 mutation on DNA recombination is due to inactivation of ExoI, some results suggest that SbcB15 protein additionally modulates the recombinational process by binding to 3′ DNA ends (6, 39). This binding could protect 3′ ends from the action of other nucleases that have affinity for the same DNA substrate.
To get further insight into the functional relationship between ExoI and RuvABC in UV repair, we tested the effect of the ΔxonA300 mutation on DNA repair in a strain deficient for Ruv activity. The ΔxonA300 mutation eliminates the whole sbcB coding sequence, leading to complete absence of ExoI protein (39). As shown in Table 2, the ΔxonA ruvB mutant LMM1017 was indistinguishable from the xonA+ ruvB strain LMM1015; both strains displayed moderate UV sensitivity typical of single ruv mutants. However, when ΔxonA was replaced with the sbcB15 mutation from JC7623 (see strains LMM1001 and LMM1018 in Table 1), the presence of the ruvB mutation dramatically increased UV sensitivity (Table 2). These results indicate that elimination of ExoI activity by itself is not responsible for the extreme DNA repair defect observed with sbcB15 ruv mutants. It is possible that the presence of the mutant SbcB protein modifies recombinational repair in such a way that it requires Holliday junction resolution as an obligatory step.
The pronounced recombinational deficiency of recBC sbcBC ruv mutants in comparison to rec+ sbc+ ruv strains suggests that an unknown intrinsic property of the RecF pathway makes recombination more dependent on Ruv functions. However, our results with sbcBC ruv and sbcB ruv strains indicate that a similar dependence on Ruv exists even in the presence of functional RecBCD enzyme. One possible explanation for these results is that the sbcB mutation redirects recombination from the RecBCD to the RecF pathway, thereby making the latter pathway dominant. To test this possibility, we compared the effect of a recJ mutation on UV survival in wild-type, recBC sbcBC, sbcB, and sbcB ΔruvABC backgrounds.
The RecJ protein is known to be required for the efficient initiation of recombination in the recBC sbcBC background (i.e., on the RecF pathway) (11). Consistent with the previous results of other authors (31), a recJ mutation caused strongly pronounced UV sensitivity in a recBC sbcBC strain while having only a slight effect in the wild-type background (compare strains LMM1032 and LMM1033 in Table 2). The repair proficiency of the sbcB recJ mutant was the same as that of the recJ single mutant and about 104-fold higher than that observed with the recBC sbcBC recJ mutant. These results indicate that in recBC+ sbcB15 strains, the RecBCD pathway remains fully active in recombinational repair. The sbcB recJ ΔruvABC strain LMM1035 displayed extreme UV sensitivity quite similar to that of the recBC sbcBC ΔruvABC, sbcBC ΔruvABC, and sbcB ΔruvABC mutants (Table 2). A similar set of results was obtained with strains containing a recF mutation instead of recJ (data not shown). It seems, therefore, that the ruv-associated UV repair defect observed in recBC sbcBC, sbcBC, and sbcB backgrounds depends directly on the sbcB mutation, regardless of the recombinational pathway that is active in the cell.
It was recently shown that the RuvABC complex has a dual repair function: in addition to its well-known role in the postsynaptic stage of recombinational repair, it also participates in the removal of stalled replication forks. It was proposed that replication forks arrested at different obstacles in DNA (e.g., DNA-bound proteins and DNA secondary structures) could be reversed and transformed into Holliday structures and then cleaved by the action of the RuvABC proteins (41). The double-stranded DNA ends thus created could be used by RecBCD enzyme to initiate homologous recombination, leading to replication fork restoration. A similar scenario was shown to occur in UV-irradiated cells, in which replication forks are frequently arrested at unrepaired pyrimidine dimers (34). Moreover, it was proposed that the majority of double-strand breaks (DSBs) in UV-irradiated cells arise from RuvABC action on reversed replication forks.
In light of these results, the strong requirement for RuvABC resolvase observed in UV-irradiated sbcB15 mutants could reflect the need for Holliday junction resolution in either the presynaptic or postsynaptic stage of recombinational repair. To discriminate between these two possibilities, we first examined the survival of different ΔruvABC and sbcB15 derivatives after exposure to gamma irradiation. It is known that γ-irradiation produces various types of DNA damage, including single- and double-strand DNA breaks, oxidized purine and pyrimidine bases, abasic sites, and DNA-protein crosslinks (reviewed in references 47 and 48). Among the lesions listed, DSBs are considered the primary cause of cellular lethality after ionizing radiation (16, 19). DSBs can arise directly from the radiation energy deposited in the DNA molecule or indirectly following attack on the DNA of free radicals produced from water. In addition, a significant number of DSBs appear during attempted excision repair at sites of clustered DNA lesions (7). Also, they can result from DNA polymerase running into unrepaired single-strand breaks (25). In all cases mentioned, DSBs are subject to recombinational repair, during which the RuvABC complex is expected to act only at the postsynaptic stage.
The results of experiments with γ-irradiation are presented in Table 3. The dose of 200 Gy moderately reduced the survival of ΔruvABC strain LMM10 while having a much stronger effect on the recBC mutant JC5519. The difference in survival between the two strains was almost 100-fold. These results show that in the wild-type background, the RecBCD enzyme has a more important role in γ-irradiation repair than the RuvABC complex. The recBC ΔruvABC double mutant LMM20 was as sensitive as the recBC single mutant. The lack of additivity of the recBC and ΔruvABC mutations indicates that the RuvABC proteins act in the RecBCD pathway during repair of γ-irradiation-induced DSBs. Also, these results show that in wild-type cells, the majority of DSB repair after ionizing radiation does not depend on RuvABC.
As expected, the ΔruvABC mutation had a much more profound effect in the recBC sbcBC than in the wild-type background; the recBC sbcBC ΔruvABC mutant was about 3,000-fold more sensitive to γ-irradiation than its rec+ sbc+ ΔruvABC counterpart (Table 3). Also, the sbcBC ΔruvABC and sbcB ΔruvABC mutants showed significantly higher γ-irradiation sensitivity (220-fold and 60-fold, respectively) than the single ΔruvABC mutant. The pronounced repair defect of the three sbcB ruv derivatives was rectified by the rus-1 mutation. Taken together, these results indicate that the sbcB15 mutation generally increases the requirement for RuvABC resolvase in γ-irradiation repair. However, the 15-fold and 55-fold-higher γ-irradiation resistance of the sbcBC ΔruvABC and sbcB ΔruvABC strains, respectively, in comparison to the recBC sbcBC ΔruvABC mutant indicates that the RecBCD function and, to a lesser extent, also the SbcC function in the former strains decrease the necessity for RuvABC in γ-irradiation repair. This finding suggests that, even in the presence of the sbcB mutation, the postsynaptic stage of DSB repair is (at least partly) independent of RuvABC proteins.
To further study the role of the RuvABC complex in the postsynaptic stage of recombination in the sbcB15 background, we investigated conjugational recombination in different ΔruvABC and/or sbcB15 derivatives. During conjugational crosses, a single strand of Hfr DNA is transferred to the F− recipient, where it provides a template for DNA synthesis (15). When mating terminates, the transferred DNA is released as a linear fragment that is subjected to recombinational exchanges with the circular recipient chromosome. The vast majority of recombinants in such crosses arise from recombinational events initiated at the ends of donor DNA by the RecBCD enzyme (10, 44). The RuvABC complex acts late in this process by cleaving the Holliday junctions formed after RecA-mediated DNA strand exchange.
As shown in Table 4, the ΔruvABC mutation mildly decreased the frequency of conjugational recombination. In contrast, recombination was drastically reduced in the recBC recipient. Introducing the ΔruvABC mutation into the recBC background had no additional effect on recombination frequency, indicating that the RecBCD and RuvABC proteins act in the same recombinational pathway. The recombination frequency of the recBC sbcBC ΔruvABC mutant was approximately 10-fold lower than that of the single ΔruvABC mutant, quite a modest difference in comparison with those obtained with the same strains after UV and γ-irradiation (compare results in Tables 2, 3, and 4).
TABLE 4.
Conjugational recombination with different ΔruvABC recipient strains
| Recipient strain | Relevant genotypea | Relative viabilityb | Avg relative yield of recombinantsc ± SD |
|---|---|---|---|
| AB1157 | Wild type | 1 | 1 |
| LMM10 | ΔruvABC | 0.62 ± 0.02 | 0.3 ± 0.12 |
| JC5519 | recBC | 0.29 ± 0.03 | 0.0032 ± 0.0013 |
| LMM20 | recBC ΔruvABC | 0.25 ± 0.04 | 0.0026 ± 0.0007 |
| JC7623 | recBC sbcBC | 0.59 ± 0.07 | 1 |
| LMM864 | recBC sbcBC ΔruvABC | 0.14 ± 0.01 | 0.022 ± 0.007 |
| LMM965 | sbcBC | 0.72 ± 0.08 | 1 |
| LMM966 | sbcBC ΔruvABC | 0.21 ± 0.03 | 0.063 ± 0.004 |
| LMM979 | sbcB | 0.92 ± 0.09 | 0.77 ± 0.18 |
| LMM983 | sbcB ΔruvABC | 0.25 ± 0.03 | 0.13 ± 0.02 |
| N2364 | sbcC | 1 | 1 |
| LMM971 | sbcC ΔruvABC | 0.68 ± 0.11 | 0.24 ± 0.05 |
In all strains listed, the designation sbcB corresponds to the sbcB15 mutation.
Viability is given relative to the number of CFU per milliliter in the cultures of the control recipient strain AB1157, which averaged 5.7 × 107.
Yields of recombinants are relative to the control strain AB1157 and have been corrected for any deficiency in the viability of the recipient cells. The average yield for the control strain AB1157 was 6.4 × 105 per ml of the mating mixture. Values are averages ± standard deviations of results from three independent experiments.
Also in contrast to the radiation experiments, the recombination proficiency of the recBC sbcBC ΔruvABC mutant was almost 10-fold higher than that of the recBC or recBC ΔruvABC strain. These differences between conjugational experiments on the one hand and radiation experiments on the other most probably reflect the differences between the DNA substrates present in the experimental systems used. As in the γ-irradiation experiment, the sbcBC ΔruvABC and sbcB ΔruvABC mutants displayed recombination frequencies somewhat higher than that of the recBC sbcBC ΔruvABC mutant. Furthermore, the sbcB ΔruvABC cells had slightly lower recombination than the ΔruvABC single mutant. Taking into account all results described, it appears that the sbcB15 mutation alone does not significantly change the role of the RuvABC complex in conjugational crosses and, by inference, does not affect its role in the postsynaptic stage of recombination in general. However, we cannot exclude the possibility that the sbcB15 mutation to a certain extent alters the postsynaptic stage of recombination in a recBC sbcC background.
To summarize, our results obtained in γ-irradiation and conjugational experiments suggest that the postsynaptic stage of recombination in the sbcB15 single mutant is largely independent of RuvABC proteins. Therefore, the extreme DNA repair defect that was observed with the sbcB15 ΔruvABC mutant after UV irradiation must be attributed to a block in an earlier phase of recombination. These results are in accord with the model which proposes that in UV-irradiated cells, the RuvABC complex acts on Holliday junctions made from reversed replication forks. Perhaps the SbcB15 protein alters the processing of such Holliday junctions and makes their removal more dependent on the action of the RuvABC resolvase. The same model could explain the increased requirement for RuvABC in γ-irradiated sbcB15 cells.
Although γ-radiation is often used as a test system to study recombinational repair of DSBs, the repair of other γ-irradiation-induced lesions could also require the action of recombination proteins. It is possible that some types of base damage that appear after ionizing radiation have the same effect on DNA replication as that proposed for UV-induced pyrimidine dimers, the reversal of replication forks. If so, in γ-irradiated E. coli cells, the RuvABC complex would be expected to act both during the repair of DSBs and on removal of regressed replication forks. This could explain our finding that the recBC sbcBC ΔruvABC mutant is more sensitive to γ-irradiation than the recBC ΔruvABC mutant (Table 3); in addition to a block in DSB repair, the first strain could also suffer from defective processing of regressed replication forks.
Concluding remarks.
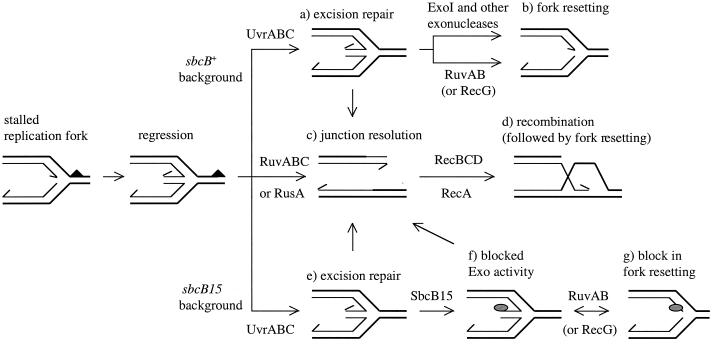
The recent model of recombinational repair in UV-irradiated cells (34) proposes several pathways for the processing of Holliday junctions formed by replication fork regression. In addition to resolution by the RuvABC complex (Fig. 1c), a Holliday junction could be transformed back to a replication fork by simple forward branch migration (catalyzed by RuvAB or RecG helicase) or by exonucleolytic degradation of the double-stranded DNA arm extruded upon replication fork regression (Fig. 1b). Previous studies by McGlynn and Lloyd (34) suggest that the RecBCD enzyme has no access to the double-stranded DNA tail made by fork reversal. Thus, exonucleolytic degradation of this tail must rely on other nucleases. However, RecBCD was proposed to have a key role in DNA repair following RuvABC-mediated fork cleavage (34) (Fig. 1c and d).
FIG. 1.
Effect of the SbcB15 protein on the repair of replication forks stalled at pyrimidine dimers. The upper part of the figure represents molecular events occurring in UV-irradiated wild-type (sbcB+) cells. The lower part pictures reactions occurring in the sbcB15 background. Reactions shown in the middle part of the figure are common to both backgrounds. The solid triangle and the shaded oval represent a pyrimidine dimer and the SbcB15 protein, respectively. The directions of DNA synthesis within replication forks are indicated by arrows. Details of the model are discussed in the text.
Our results suggest that in the presence of the sbcB15 mutation, RuvABC-mediated cleavage of Holliday junctions is the principal pathway for the repair of blocked replication forks. The simplest explanation for this finding would be that inactivation of sbcB function prevents exonucleolytic cleavage of the duplex DNA end formed upon replication fork reversal. However, the absence of any additional effect of the ΔxonA mutation in the ruv background suggests that ExoI does not play a crucial role in replication fork resetting. It is more likely that the sbcB15 mutation modulates the repair of blocked replication forks in an indirect way, possibly through the binding of the mutant SbcB15 protein to the duplex DNA end, which could prevent the action of other nucleases (Fig. 1f).
Such an explanation implies that the extruded duplex arm of a reversed replication fork must expose a single-stranded 3′ end as the substrate for the SbcB protein. The free 3′ end could arise from DNA unwinding catalyzed by RecQ or some other recombination-associated helicase. The binding of SbcB15 to the 3′ end of a duplex arm might affect not only the exonucleolytic processing of the regressed fork, but perhaps also the restoration of the replication fork by forward branch migration (Fig. 1g). In the latter case, SbcB15 may prevent completion of forward branch migration and/or interfere with the restart of DNA replication from the restored fork-like structure. In such a situation, the RuvABC-mediated cleavage of the regressed fork followed by recombinational repair would be the only way to circumvent the block in DNA synthesis and to obtain viable chromosomes.
The cleavage of a regressed replication fork produces a DNA duplex which can be used by either the RecBCD or the RecF pathway to initiate recombinational repair and to restore a replication fork (6). The relatively high UV resistance obtained with sbcB15 recJ (Table 2) and sbcB15 recF (not shown) mutants suggests that the SbcB15 protein does not interfere with RecBCD-dependent recombination. This implies that SbcB15 does not prevent the binding of RecBCD to duplex DNA. Perhaps RecBCD is capable of removing SbcB15 from the 3′ end either immediately upon binding to DNA or later, during its temporary exonucleolytic activity on the 3′ strand.
As already mentioned, upon encountering a Chi sequence, RecBCD enzyme changes its mode of action from a powerful DNA degradase to a recombinase that produces a recombinogenic 3′ ssDNA tail. At the same time, RecBCD loads the RecA protein onto the exposed ssDNA, which enables the subsequent synaptic stage of recombination (2, 3). It was also shown that the RecBCD-mediated loading of the RecA protein protects the 3′ ssDNA tail from digestion by ExoI (2). Therefore, it is very likely that the activity of the RecBCD enzyme prevents repeated binding of the SbcB15 protein in a similar way once it has been removed from the 3′ DNA end. In such conditions, the reactions initiated by RecBCD can proceed normally to form a D-loop that serves as a substrate for PriA-mediated replication restart (40) (Fig. 1c and d).
Our results obtained in conjugal crosses indicate that the postsynaptic stage of recombination is more dependent on the RuvABC proteins in the recBC sbcBC mutant than in the wild-type cells. The higher necessity for RuvABC in the recBC sbcBC background can be attributed not solely to sbcB15 but rather to a joint effect of all the mutations present. We speculate that in the absence of a functional RecBCD enzyme, the mutant SbcB15 protein binds to a recombinogenic 3′ DNA end, thereby making the final stage of recombination more dependent on the RuvABC resolvase. Such binding could occur if we assume that RecFOR-catalyzed loading of the RecA protein is not as efficient as that catalyzed by RecBCD. Perhaps SbcB15 bound to a 3′ DNA end interferes with DNA synthesis within the D-loop, which makes the recombination intermediate unstable and calls for Holliday junction resolution. However, the present results do not offer clear evidence for this assumption, and further work will be needed to address this issue.
Acknowledgments
We thank Mary Berlyn, Robert G. Lloyd, Bénédicte Michel, and Susan T. Lovett for bacterial strains and Mary Sopta for correction of the English text. We are also grateful to our colleagues Erika Salaj-Šmic and Ivana Ivančić-Baće for helpful discussions.
This work was supported by the Croatian Ministry of Science and Technology (grants 00981002 and 098426).
REFERENCES
- 1.Anderson, D. G., and S. C. Kowalczykowski. 1997. The recombination hot spot χ is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev. 11:571-581. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, D. G., and S. C. Kowalczykowski. 1997. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a chi-regulated manner. Cell 90:77-86. [DOI] [PubMed] [Google Scholar]
- 3.Arnold, D. A., and S. C. Kowalczykowski. 2000. Facilitated loading of RecA protein is essential to recombination by RecBCD enzyme. J. Biol. Chem. 275:12261-12265. [DOI] [PubMed] [Google Scholar]
- 4.Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 5.Bennett, R. J., H. J. Dunderdale, and S. C. West. 1993. Resolution of Holliday junctions by RuvC resolvase: cleavage specificity and DNA distortion. Cell 74:1021-1031. [DOI] [PubMed] [Google Scholar]
- 6.Bidnenko, V., M. Seigneur, M. Penel-Colin, M.-F. Bouton, S. D. Ehrlich, and B. Michel. 1999. sbcB sbcC null mutations allow RecF-mediated repair of arrested replication forks in rep recBC mutants. Mol. Microbiol. 33:846-857. [DOI] [PubMed] [Google Scholar]
- 7.Blaisdell, J. O., and S. S. Wallace. 2001. Abortive base-excision repair of radiation-induced clustered DNA lesions in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:7426-7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, S. N., S. D. Vincent, and R. G. Lloyd. 1998. Recognition and manipulation of branched DNA by the RusA Holliday junction resolvase of Escherichia coli. Nucleic Acids Res. 26:1560-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, A. J. 1973. Recombination deficient mutants of E. coli and other bacteria. Annu. Rev. Genet. 7:67-86. [DOI] [PubMed] [Google Scholar]
- 10.Clark, A. J. 1991. rec genes and homologous recombination proteins in Escherichia coli. Biochimie 73:523-532. [DOI] [PubMed] [Google Scholar]
- 11.Clark, A. J., and S. J. Sandler. 1994. Homologous genetic recombination: the pieces begin to fall into place. Crit. Rev. Microbiol. 20:125-142. [DOI] [PubMed] [Google Scholar]
- 12.Connelly, J. C., L. A. Kirkham, and D. R. F. Leach. 1998. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosome (SMC) family protein that cleaves hairpin DNA. Proc. Natl. Acad. Sci. USA 95:7969-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon, D. A., and S. C. Kowalczykowski. 1991. Homologous pairing in vitro stimulated by the recombination hotspot, Chi. Cell 66:361-371. [DOI] [PubMed] [Google Scholar]
- 14.Dixon, D. A., and S. C. Kowalczykowski. 1993. The recombination hotspot, Chi, is a regulatory sequence that acts by attenuating the nuclease activity of the Escherichia coli RecBCD enzyme. Cell 73:87-96. [DOI] [PubMed] [Google Scholar]
- 15.Firth, N., K. Ippen-Ihler, and R. A. Skurray. 1996. Structure and function of the F factor and mechanism of conjugation, p. 2377-2401. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 16.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, D.C.
- 17.Gibson, F. P., D. R. F. Leach, and R. G. Lloyd. 1992. Identification of sbcD mutations as cosuppressors of recBC that allow propagation of DNA palindromes in Escherichia coli K-12. J. Bacteriol. 174:1222-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horii, Z.-I., and A. J. Clark. 1973. Genetic analysis of the RecF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J. Mol. Biol. 80:327-344. [DOI] [PubMed] [Google Scholar]
- 19.Iliakis, G. 1991. The role of DNA double strand breaks in ionizing radiation-induced killing of eukaryotic cells. Bioessays 13:641-648. [DOI] [PubMed] [Google Scholar]
- 20.Iwasaki, H., M. Takahagi, T. Shiba, A. Nakata, and H. Shinagawa. 1991. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 10:4381-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwasaki, H., M. Takahagi, T. Shiba, and H. Shinagawa. 1992. Escherichia coli RuvA and RuvB proteins specifically interact with Holliday junctions and promote branch migration. Genes Dev. 6:2214-2220. [DOI] [PubMed] [Google Scholar]
- 22.Kogoma, T. 1997. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61:212-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kowalczykowski, S. C., D. A. Dixon, A. K. Eggleston, S. D. Laurer, and W. M. Rehrauer. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58:401-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kushner, S. R., H. Nagaishi, A. Templin, and A. J. Clark. 1971. Genetic recombination in Escherichia coli: the role of exonuclease I. Proc. Natl. Acad. Sci. USA 68:824-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuzminov, A. 1995. Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 16:373-384. [DOI] [PubMed] [Google Scholar]
- 26.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuzminov, A. 2001. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. USA 98:8461-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd, R. G., F. E. Benson, and C. E. Shurvinton. 1984. Effect of ruv mutations on recombination and DNA repair in Escherichia coli K12. Mol. Gen. Genet. 194:303-309. [DOI] [PubMed] [Google Scholar]
- 29.Lloyd, R. G., and C. Buckman. 1985. Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J. Bacteriol. 164:836-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lloyd, R. G., and K. B. Low. 1996. Homologous recombination, p. 2236-2255. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 31.Lovett, S. T., and A. J. Clark. 1984. Genetic analysis of the recJ gene of Escherichia coli K-12. J. Bacteriol. 157:190-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahdi, A. A., G. J. Sharples, T. N. Mandal, and R. G. Lloyd. 1996. Holliday junction resolvases encoded by homologous rusA genes in Escherichia coli K-12 and phage 82. J. Mol. Biol. 257:561-573. [DOI] [PubMed] [Google Scholar]
- 33.Mandal, T. N., A. A. Mahdi, G. J. Sharples, and R. G. Lloyd. 1993. Resolution of Holliday intermediates in recombination and DNA repair: indirect suppression of ruvA, ruvB, and ruvC mutations. J. Bacteriol. 175:4325-4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGlynn, P., and R. G. Lloyd. 2000. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell 101:35-45. [DOI] [PubMed] [Google Scholar]
- 35.Michel, B. 2000. Replication fork arrest and DNA recombination. Trends Biochem. Sci. 25:173-178. [DOI] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Oliver, B. D., and E. B. Goldberg. 1977. Protection of parental T4 DNA from a restriction exonuclease by the product of gene 2. J. Mol. Biol. 116:877-881. [DOI] [PubMed] [Google Scholar]
- 38.Phillips, G. J., D. C. Prasher, and S. R. Kushner. 1988. Physical and biochemical characterization of cloned sbcB and xonA mutations from Escherichia coli K-12. J. Bacteriol. 170:2089-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Razavy, H., S. K. Szigety, and S. M. Rosenberg. 1996. Evidence for both 3′ and 5′ single-strand DNA ends in intermediates in Chi-stimulated recombination in vivo. Genetics 142:333-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandler, S. J., and K. J. Marians. 2000. Role of PriA in replication fork reactivation in Escherichia coli. J. Bacteriol. 182:9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seigneur, M., V. Bidnenko, S. D. Ehrlich, and B. Michel. 1998. RuvAB acts at arrested replication forks. Cell 95:419-430. [DOI] [PubMed] [Google Scholar]
- 42.Sharples, G. J., S. N. Chan, A. A. Mahdi, M. C. Whitby, and R. G. Lloyd. 1994. Processing of intermediates in recombination and DNA repair: identification of a new endonuclease that specifically cleaves Holliday junctions. EMBO J. 13:6133-6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shurvinton, C. E., and R. G. Lloyd. 1982. Damage to DNA induces expression of the ruv gene of Escherichia coli. Mol. Gen. Genet. 185:352-355. [DOI] [PubMed] [Google Scholar]
- 44.Smith, G. R. 1991. Conjugal recombination in E. coli: myths and mechanisms. Cell 64:19-27. [DOI] [PubMed] [Google Scholar]
- 45.Tsaneva, I. R., B. Müller, and S. C. West. 1992. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell 69:1171-1180. [DOI] [PubMed] [Google Scholar]
- 46.Viswanathan, M., and S. T. Lovett. 1998. Single-strand DNA-specific exonucleases in Escherichia coli: roles in repair and mutation avoidance. Genetics 149:7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wallace, S. S. 1998. Enzymatic processing of radiation-induced free radical damage in DNA. Radiat. Res. 150:S60-S79. [PubMed] [Google Scholar]
- 48.Ward, J. F. 1988. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog. Nucleic Acid Res. Mol. Biol. 35:95-125. [DOI] [PubMed] [Google Scholar]
- 49.West, S. C. 1997. Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet. 31:213-244. [DOI] [PubMed] [Google Scholar]