Abstract
Several strains that grow on medium-chain-length alkanes and catalyze interesting hydroxylation and epoxidation reactions do not possess integral membrane nonheme iron alkane hydroxylases. Using PCR, we show that most of these strains possess enzymes related to CYP153A1 and CYP153A6, cytochrome P450 enzymes that were characterized as alkane hydroxylases. A vector for the polycistronic coexpression of individual CYP153 genes with a ferredoxin gene and a ferredoxin reductase gene was constructed. Seven of the 11 CYP153 genes tested allowed Pseudomonas putida GPo12 recombinants to grow well on alkanes, providing evidence that the newly cloned P450s are indeed alkane hydroxylases.
Many eubacteria are able to grow on linear alkanes by virtue of alkane hydroxylases (AHs) that activate alkanes to 1-alkanols. These are then further metabolized by alcohol and aldehyde dehydrogenases to fatty acids, which enter the central metabolism (34). AHs belong to several different oxygenase classes. Short-chain-length (C2 to C4)-alkane degraders possess enzymes related to the soluble and particulate methane monooxygenases (34), while integral membrane nonheme iron AHs related to AlkB of Pseudomonas putida GPo1 were found in medium-chain-length (MCL) (C5 to C11)- and especially in long-chain-length (LCL) (≥C12)-alkane-degrading Alpha-, Beta-, and Gammaproteobacteria and high-G+C gram-positive bacteria (34, 37).
Several strains in our collection of alkane degraders grow well on MCL alkanes but could not be shown to contain AlkB homologs that act on MCL alkanes (some of these strains do contain AlkB homologs that hydroxylate LCL alkanes); these strains include the following. (i) Gordonia sp. strain 7E1C (Rhodococcus rhodochrous NCIMB 12566) is of interest because it is able to oxidize substituted phenoxy propane to phenoxy propanoic acids when pregrown on n-alkanes (17). It is known to contain an alkane-inducible cytochrome P450 (2). (ii) For Rhodococcus erythropolis NRRL B-16531 and Q15, none of the integral membrane AHs cloned from these isolates could be shown to act on MCL alkanes (39), even though these and other R. erythropolis isolates grow well on such alkanes (38). (iii) Of several hexane-degrading strains isolated from a trickling-bed bioreactor (32, 39), only 2 of 15 strains tested contained AlkB homologs that oxidize MCL alkanes (J. B. van Beilen et al., unpublished data). This left 13 strains for which growth on hexane initially could not be attributed to known enzyme systems. One of these strains, Sphingomonas sp. strain HXN-200, contains a soluble alkane hydroxylase, which is proposed to be responsible for a range of useful hydroxylation and epoxidation reactions of cyclic compounds such as pyrrolidines, pyrrolidinones, azetidines, azetidinones, piperidines, and piperidinones (4-6, 24, 25). Mycobacterium sp. strain HXN-1500, a strain able to convert limonene to perillyl alcohol (35), was found to contain a soluble alkane hydroxylase that is closely related to the Acinetobacter sp. strain EB104 hexane hydroxylase (CYP153A1), the first member of a new cytochrome P450 family (26). Based on conserved sequence motifs in CYP153A1 and two CYP153 homologs (CYP153A2 and CYP153A4) in bacterial genome sequences, highly degenerate primers were designed to clone the Mycobacterium sp. strain HXN-1500 CYP153A6 gene and flanking regions. The CYP153A6 gene was subsequently functionally expressed in Pseudomonas putida GPo12 by using the broad-host-range vector pCom8, and the recombinant was used for the production of perillyl alcohol from limonene (35).
In this study, we used the same degenerate primers to carry out a search for CYP153 genes in alkane-degrading strains. It appeared that most of the hexane-degrading strains contained CYP153 sequences. Seven of the 11 full-length CYP153 genes tested could be functionally expressed in P. putida and allowed the host strain to grow on alkanes.
MATERIALS AND METHODS
Strains, growth media, and materials.
The strains and plasmids used or constructed for cloning and expression are listed in Table 1. Luria broth (LB) (30) and E2 medium (22), supplemented with carbon sources or antibiotics, were used throughout. All cultures were grown aerobically at 30°C or 37°C. Escherichia coli strains were transformed by electroporation (10). To select E. coli transformants, ampicillin was used at 100 μg/ml, tetracycline at 12.5 μg/ml, chloramphenicol at 25 μg/ml, and gentamicin at 10 μg/ml. Plasmids were transferred to P. putida GPo12(pGEc47ΔB) by triparental mating using E. coli CC118(pRK600) as a helper strain (9) and selection with 50 μg/ml of gentamicin. Bacterial strains were grown on alkanes as described previously (31, 32). P. putida GPo12(pGEc47ΔB) was tested for growth on 1-, 2-, and 3-octanol by placing a sterile Whatman paper filter with 50 μl of 1-, 2-, or 3-octanol in the lid of the petri dish.
TABLE 1.
Cloning strains, host strains, and plasmids used or constructed in this work
| Strain or plasmid | Relevant genotype or characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. coli JM101 | endA hsdR supR thi-1 Δ(lac-proAB) F′(traD36 proAB lacIq) lacZM15 | 40 |
| E. coli DH10B | Cloning strain | Gibco BRL |
| E. coli GEc137 | DH-1 thi fadR | 11 |
| P. putida GPo12 | GPo1 cured of the OCT plasmid | 31 |
| E. coli CC118(RK600) | Helper strain for triparental mating | 9 |
| Plasmids | ||
| pGEc47ΔB | pGEc47, deletion in alkB | 31 |
| pZErO2.1 | Cloning vector; Kmr | Invitrogen |
| pCR-TOPO2.1 | Cloning vector; Kmr, Apr | Invitrogen |
| pCom8 | Broad-host-range expression vector with PalkB; GmroriT alkS | 33 |
| pCom8-PFR1500 | pCom8 containing the HXN1500 CYP153A6, ferredoxin, and ferredoxin reductase | 35 |
| pCom8-PA7F200 | pCom8 containing the HXN200 CYP153A7 and ferredoxin | This study |
| pCom8-PA7F200R1500 | As pCom8-PA7F200, including the HXN1500 ferredoxin reductase | This study |
| pCom12-PA7F200R1500 | As pCom8-PA7F200R1500, with unique EcoRI and PacI cloning sites flanking the CYP153A7 gene | This study |
| pCom12-PxF200R1500 | As pCom8-PA7F200R1500, with the CYP153A1, -A5, -A7, -A8, -A11, -A12, -A13, -A14, -D2, or -D3 gene cloned between the EcoRI and PacI sites | This study |
PCR and gene cloning.
Chromosomal DNA was isolated from gram-positive and gram-negative strains as described by Desomer et al. (8). Restriction enzymes, T4 DNA ligase, and dideoxynucleotides were from Roche Molecular Biologics and were used as specified by the supplier. Oligonucleotides were synthesized by Microsynth, Switzerland. Taq DNA polymerase was obtained from Promega. PCRs were carried out using a Perkin-Elmer GeneAmp PCR system 9600. For PCR using the CYP153 primers (P450fw1 [GTSGGCGGCAACGACACSAC], based on the substrate-binding stretch GGNDTTRN in the I-helix, and P450rv3 [GCASCGGTGGATGCCGAAGCCRAA], based on the sequence ending with the heme-binding cysteine, HLSFGFGIHRC), the following program was used: 4 min at 95°C; 25 cycles of 45 s at 95°C, 1 min at 58°C, and 1 min at 72°C; 5 min at 72°C; and 4°C until further use. If multiple bands were present, PCR products were purified over a 1% agarose gel in Tris-borate-EDTA buffer, cut out from the gel, and isolated by electroelution (30). The PCR fragments were cloned in pCR-TOPO2.1 (Invitrogen). Plasmid DNA was isolated using the Roche Molecular Biologics High Pure plasmid isolation kit. Both strands of the inserts were sequenced on a Li-Cor 4000L sequencer using IRD800-labeled −40 forward (AGGGTTTTCCCAGTCACGACGTT) and −40 reverse (GAGCGGATAACAATTTCACACAGG) primers (MWG-Biotech) and the Amersham Thermosequenase cycle sequencing kit. 16S rRNA gene sequencing was done as described by Karlson et al. (20). The primers used for amplification were 16F27 and 16R1525, and the primers used for sequencing were 16F355, 16R519, and 16R1488 (15). Southern blotting and colony blotting were carried out as described before (30, 32).
To clone complete CYP153 genes and flanking DNA, the amplified CYP153 gene fragments were digoxigenin labeled and used as probes in Southern blotting to identify easy-to-clone restriction fragments. Chromosomal DNA fragments of appropriate size were isolated from preparative Tris-borate-EDTA agarose gels by electroelution, ligated in pZErO-2, and transformed into E. coli DH10B. Transformants containing CYP153 genes or gene fragments were identified by colony blotting. Both strands of the inserts were sequenced using clones made with the EZ::TN 〈 TET-1〉 kit (Epicenter) and primers described in the manual for the same kit.
Sequence analysis.
Nucleotide and amino acid sequences were analyzed and compared using LASERGENE Navigator from DNASTAR (Madison, Wisconsin). Nucleotide and amino acid sequences were compared with the sequence databases at NCBI by using BLAST (1). In all cases, except for CYP153A7, where an N-terminal sequence was available, the start codons of cloned genes and relevant genes in sequenced genomes were identified by sequence analysis. In almost all cases, the first potential ATG start codon of the open reading frame was preceded by a ribosomal binding sequence. If this was not the case, alternative start codons were identified. New P450 sequences were deposited in the cytochrome P450 database and named by David Nelson (http://drnelson.utmem.edu/CytochromeP450.html). Distance trees based on multiple sequence alignments were generated with ClustalX (16), using the neighbor-joining method with 1,000 bootstrap trials.
Expression of CYP153 genes in Pseudomonas.
The HXN200 CYP153A7 gene (ahpG1) and the downstream ferredoxin gene (ahpI1) were amplified with primers HXN200-P450Fdx-FW (GAATTCATTatgGAACATACAGGACAAA [EcoRI site 3 bases upstream of the ATG start codon]) and HXN200-P450Fdx-RV (GGATCCTTATCTGACTCTTGCGTCATC [BamHI site downstream of the ferredoxin gene]). The Mycobacterium sp. strain HXN1500 ferredoxin reductase gene (ahpH) was amplified with primers HXN1500-Red-FW2 (GCCAGATCTAaggaggGCTATatgATCCACACCGGCGTGACCGAA [BglII site and a new ribosome binding site upstream of the ATG start codon]) and 1500-FERred RV1 (AAGCTTTATCAGTGGGTTAGAG [HindIII site downstream of the stop codon]). (In all primer sequences, the introduced sites are underlined and other features are lowercase.) The PCR fragments were cloned in pCR-TOPO2.1 and sequenced to verify that no mutations had been introduced. The ahpG1-ahpI1 fragment was cloned between the EcoRI and BamHI sites of pCom8, resulting in construct pCom8-PA7F200. The EcoRI and BglII ahpG1-ahpI1 fragment and the BamHI and HindIII ahpH fragment were cloned together between the EcoRI and HindIII sites of pCom8, resulting in construct pCom8-PA7F200R1500.
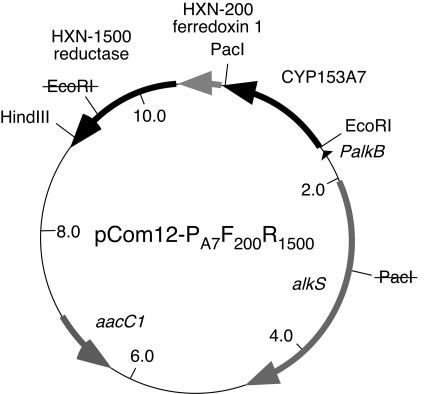
To facilitate cloning of further CYP153 genes in the same constructs, an EcoRI site in the HXN-1500 reductase gene and a PacI site in the alkS gene were removed by QuikChange mutagenesis (primers delEco [GGGTTTTCGACGCCATAAATTCGCGCGGCCGG] and delPac [CAGAGAAGTTAATAAAAGACCTATCTTCACAATCC]). Subsequently, a PacI site was introduced between the P450 and ferredoxin genes (newPac [CAACTAAATCCTTGTATCTTAATTAACCTTGTGCTTGCGCACTACGC]). The resulting vector, named pCom12-PA7F200R1500 (Fig. 1), allows the introduction of novel CYP153 genes as EcoRI-PacI or MfeI-PacI restriction fragments (MfeI has compatible sticky ends with EcoRI and was used in cases where the CYP153 gene contained an EcoRI site).
FIG. 1.
Map of pCom12-PA7F200R1500. The plasmid is based on pCom8, which contains the PalkB promoter, a gene encoding the positive regulator alkS of PalkB, a gentamicin resistance gene (aacC1), origins of replication for E. coli and Pseudomonas sp., and an origin of transfer to allow transfer by conjugation (33). In pCom12 vectors an EcoRI site is located between the ribosome binding site and the ATG start codon of the CYP153A7 gene (AATTggagaATTCATTatg, where the ribosome binding site and ATG are in lowercase and the EcoRI site is underlined). A PacI site is located between the CYP153A7 gene and the downstream ferredoxin gene. The lined-out PacI and EcoRI sites were removed by site-directed mutagenesis.
The four other HXN-200 CYP153 genes were amplified with primers that introduced an EcoRI site (CYP153A8 and CYP153D3) or MfeI site (CYP153D2 and CYP153A11) 3 bases upstream of the predicted translation start codon and a PacI site 2 to 20 bases downstream of the stop codon (for CYP153D2, forward primer GCAATTGCCCATGGCTACCGTCATCCG and reverse primer ATTAATTAAGGGCTCCTCATGCCCCTG; for CYP153A8, forward primer CGAAGAATTCGACATGGATACCGACATGG and reverse primer GCTTTTAATTAATCAGCCAATCAGCGGGCG; for CYP153D3, forward primer GGAATTCATCATGGCCAGCACCGCC and reverse primer CGTTAATTAACGTCTGGGCGGCAG; and for CYP153A11, forward primer GTCAATTGCATATGGCTACGCGATCGATGCAGTCCGGTCCGGATCGCGAAGAACCGG ACCGCCCTATTGCCG and reverse primer CTTAATTAAAAGATTGTGACCTTCCCTTGGTGC). The CYP153A11 forward primer also removed an internal MfeI site. The two Alcanivorax borkumensis AP1 CYP153 genes were amplified with primers AP1-FW (GCAATTGGACATGTCAACGAGTTCAAGTAC) and AP1-RV (CTTAATTAACCTTTCCGTTCTGGCGAGTC), the Mycobacterium marinum CYP153A14 gene was amplified with marFW (GGAATTCGACATGAGCAATATTCGCGAGGC) and marRV (CTTAATTAAGCACGGTCATGCGCCACC), and the Rhodopseudomonas palustris CYP153A5 gene was amplified with palFW (GGCAATTGACGATGCACGGCACCATCG) and palRV (TTTTAATTAAAGGTCATGAGACCCGGACC). All PCR fragments were first cloned in pCR-TOPO2.1, and sequenced. Subsequently, the EcoRI-PacI or MfeI-PacI inserts were cloned in pCom12, and the resulting plasmids were transferred to P. putida GPo12(pGEc47ΔB).
Nucleotide sequence accession numbers.
Accession numbers for CYP153 sequences are given in Table 2. The Gordonia sp. strain 7E1C 16S sequence received the accession number AJ784814.
TABLE 2.
Occurrence of CYP153 and AlkB-type alkane hydroxylases in alkane-degrading strainsa
| Organism | No. of CYP153s (evidenceb) | CYP153 gene accession no. | P450 namec | Flanking genesd | Functional expressione
|
AlkB homologs (evidencef) | Reference(s) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| C5 | C6 | C8 | C10 | |||||||
| Acinetobacter sp. strain EB104 | 1 (cloned) | AJ311718 | CYP153A1 | Fd, FdR | ++ | +++ | ++++ | +++ | 1 (cloned) | 26 |
| Alcanivorax borkumensis AP1 | 2 (cloned) | AJ844909AJ844908 | CYP153A13 | NDg | − | +/− | ++ | +++ | 2 (cloned) | 36 |
| CYP153A12 | ND | − | − | − | − | |||||
| Alcanivorax borkumensis SK2 | 2 (genome) | Unfinished genome | CYP153A13a | Fd, AlkJ, FdR | NTh | NT | NT | NT | 2 (genome) | 14 |
| CYP153A13b | Unrelated | NT | NT | NT | NT | |||||
| Bradyrhizobium japonicum USDA 110 | 3 (genome) | blr1853NP_768493 | CYP153A9 | Unrelated | NT | NT | NT | NT | 0 (genome) | 19 |
| blr7243NP_773883 | CYP153A4 | P450, unrelated | NT | NT | NT | NT | ||||
| blr7242NP_773882 | CYP153A3 | P450, unrelated | NT | NT | NT | NT | ||||
| Burkholderia fungorum LB400 | 1 (genome) | ZP_00028060 | CYP153A10 | Fd, FdR AlkJ | NT | NT | NT | NT | 1 (genome) | |
| Caulobacter crescentus CB15 | 1 (genome) | CC0063NP_418882 | CYP153A2 | Unrelated | NT | NT | NT | NT | 0 (genome) | 27 |
| Erwinia chrysanthemi 3937 | 1 (genome) | Unfinished genome | CYP153E1 | Unrelated | NT | NT | NT | NT | 0 (genome) | |
| Gordonia rubripertinctus 7E1C (NCIMB 12566) | 1 (PCR) | AJ833999 | Not named | ND | NT | NT | NT | NT | 0 (PCR negative) | 2, 21 |
| Hyphomonas neptunium ATCC 15444 | 1 (genome) | Unfinished genome | CYP153A15 | Unrelated | NT | NT | NT | NT | 0 (genome) | |
| Novosphingobium aromaticivorans Saro_178 | 2 (genome) | ZP_00096754 | CYP153D1 | Unrelated | NT | NT | NT | NT | 0 (genome) | |
| ZP_00094181 | CYP153C1 | Unrelated | NT | NT | NT | NT | ||||
| Mycobacterium marinum M | 1 (genome) | Unfinished genome | CYP153A14 | FdR, Fd, TnpA, IS | +++ | +++ | ++++ | ++++ | 2 (genome) | 29 |
| M. rhodesiae HXN-100 | 1 (PCR) | AJ833989 | Not named | ND | NT | NT | NT | NT | 0 (PCR negative) | 38; van Beilen, submitted for publication |
| M. septicum HXN-500 | 1 (PCR) | AJ833990 | Not named | ND | NT | NT | NT | NT | 0 (PCR negative) | 28; van Beilen, submitted |
| M. peregrinum HXN-600 | 1 (PCR) | AJ833991 | Not named | ND | NT | NT | NT | NT | 1 (PCR) | 28, 38 |
| M. abscessus HXN-1000 | 1 (PCR) | AJ833992 | Not named | ND | NT | NT | NT | NT | 1 (PCR) | 28, 38 |
| M. abscessus HXN-1200 | 1 (PCR) | AJ833993 | Not named | ND | NT | NT | NT | NT | 0 (PCR negative) | 28; van Beilen, submitted |
| Mycobacterium sp. strain HXN-1500 | 1 (PCR) | AJ783967 | CYP153A6 | Fd, FdR | ++ | +++ | ++++ | +++ | 0 (PCR negative) | 28, 35 |
| M. septicum HXN-1900 | 1 (PCR) | AJ833995 | Not named | ND | NT | NT | NT | NT | 0 (PCR negative) | 28; van Beilen, submitted |
| Mycobacterium paraffinicum ATCC 12670 | 1 (PCR) | AJ834000 | Not named | ND | NT | NT | NT | NT | NT | 7 |
| Oleomonas sagaranensis HXN-1400 | 1 (PCR) | AJ833994 | Not named | ND | NT | NT | NT | NT | 0 (PCR negative) | 28; van Beilen, submitted |
| R. erythropolis NRRL B-16531 | 2 (PCR) | AJ833997 | Not named | ND | ND | NT | NT | NT | 4 (cloned) | 17, 18, 39 |
| AJ833998 | Not named | ND | ND | NT | NT | NT | ||||
| R. erythropolis HXN-2000, 35-O, 42-O, 62-O, 23-D, and 50-V | 2 (PCR) | Identical to R. erythropolis NRRL B-16531 genes | Not named | ND | ND | NT | NT | NT | 3-5 (PCR) | 28, 38 |
| Rhodopseudomonas palustris CGA009 | 1 (genome) | ZP_00011192 | CYP153A5 | Unrelated | − | − | − | − | 0 (genome) | 23 |
| Sphingomonas sp. strain HXN-200 | 5 (cloned) | AJ850057 | CYP153A7 | Fd, AlkJ | + | ++ | +++ | ++ | 0 (PCR negative) | 25 |
| AJ850058 | CYP153A8 | Fd | − | − | − | − | ||||
| AJ850059 | CYP153A11 | Fd | ++ | ++ | ++ | ++ | ||||
| AJ850060 | CYP153D2 | Unrelated | − | +/− | ++ | +++ | ||||
| AJ850057 | CYP153D3 | Unrelated | − | − | − | − | ||||
Most strains shown here belong to an in-house collection of hydrocarbon-degrading bacteria described earlier (32, 35). The HXN strains are hexane-degrading bacteria isolated from a trickling-bed bioreactor that was operated for 3 years to study the removal of hexane from an airstream (28, 38).
Genome, the genome sequence contains one or more CYP153 sequences; PCR, the degenerate CYP153 or AlkB primers yielded one or more PCR fragments; cloned, the complete CYP153 genes were cloned and expressed in P. putida GPo12(pGEc47ΔB).
The CYP153 sequences as named by David Nelson.
Fd, ferredoxin; FdR, ferredoxin reductase; AlkJ, alcohol dehydrogenase; TnpA, transposase; IS, insertion sequence.
++++, rate of growth on E2 medium plates with pentane (C5), hexane (C6), octane (C8), or decane (C10) comparable to that of P. putida GPo1 grown with the same substrate; +++, ++, +, and +/−, lower growth rates.
See footnote b. PCR negative, PCR fragments were not obtained.
ND, not determined.
NT, not tested.
RESULTS
Amplification of CYP153 gene fragments and cloning of full-length CYP153 genes.
In our search for new alkane hydroxylases, we have identified several strains that grow well on MCL alkanes but do not contain AlkB homologs acting on MCL alkanes. PCR with degenerate CYP153 primers, developed to clone the Mycobacterium sp. strain HXN-1500 alkane hydroxylase (35), yielded PCR products of the expected length from most of these strains (designated “PCR” in Table 2). These include all mycobacteria, all R. erythropolis isolates, and several of the Proteobacteria. Only three target strains did not yield PCR fragments (van Beilen et al., unpublished data). Because the degenerate primers were biased for the amplification of high-G+C content CYP153 genes (initially only Mycobacterium sp. strain HXN-1500 was a target), unbiased and low-G+C primers were also designed and tested, but these did not yield additional PCR fragments. Sequencing showed that all PCR fragments encoded peptides with high sequence identity (>60%) to the corresponding regions of CYP153A1 and CYP153A6, indicating that internal segments of CYP153 genes have been amplified.
In the case of Sphingomonas sp. strain HXN-200, Southern blotting using a digoxigenin-labeled CYP153 PCR fragment amplified from HXN-200 chromosomal DNA gave multiple signals. Suitable restriction fragments were identified by Southern blotting and cloned. A 4.7-kb EcoRI fragment, a 4.4-kb BamHI fragment, and a 2.5-kb EcoRI fragment overlapped, resulting in a 7.2-kb sequence containing the CYP153A7 and CYP153D3 genes and a ferredoxin gene adjacent to the CYP153A7 gene. A 2.0-kb SacI fragment contained the CYP153A8 gene and a ferredoxin gene. The CYP153A11 gene and an adjacent ferredoxin gene were found on overlapping PstI 4.2-kb and SacI 3.2-kb fragments, and the CYP153D2 gene was found on overlapping PstI 4.4-kb and EcoRI 4.4-kb fragments. All sequences were submitted to GenBank and received the accession numbers shown in Table 2 and in Fig. S1 in the supplemental material. The N-terminal sequence of CYP153A7 corresponded to that of the N-benzylpyrrolidine-hydroxylating enzyme purified earlier from Sphingomonas sp. strain HXN-200 (3).
PCR using the degenerate CYP153 primers yielded one of two closely related DNA sequences (87% DNA sequence identity) with all R. erythropolis isolates. Subsequent Southern blotting using one of two sequences as a probe revealed that all seven tested R. erythropolis strains contain two closely related CYP153 genes (data not shown).
Alcanivorax borkumensis AP1 was included in this study because it was thought likely that it contains CYP153 sequences in addition to its two integral membrane AHs (36); a double alkane hydroxylase knockout mutant of the closely related isolate SK2 was still able to grow on alkanes (14), and the genome sequence of SK2 possesses two almost identical copies of a CYP153 gene (Alfred Pühler, personal communication). As A. borkumensis AP1 and A. borkumensis SK2 are closely related isolates (36), we used primers based on the sequence of the SK2 CYP153 genes to amplify the full-length AP1 CYP153 gene(s). Sequence comparisons of eight clones indicated that AP1 contains at least two CYP153 genes. The two cloned sequences (CYP153A12 and CYP153A13) had 94% DNA sequence identity to each other. The CYP153A13 gene sequence was practically identical to the two SK2 CYP153 gene sequences (99.6%). In total, we cloned 8 full-length CYP153 genes using PCR and Southern blotting, while an additional 11 full-length sequences were found in finished or unfinished microbial genome projects.
Sequence analysis of CYP153 genes and flanking DNA.
A distance tree was calculated based on the alignment of the CYP153 peptides encoded by the PCR fragments, the corresponding regions of CYP153 proteins encoded by bacterial genome sequences (designated “genome” in Table 2), and a number of more distantly related sequences such as P450cam (see Fig. S1 in the supplemental material). Most sequences belong to the CYP153A subfamily, while the remaining five CYP153 sequences belong to other subfamilies (C, D, and E). The most deeply branching sequence that belongs to the CYP153 family is that of the Novosphingobium aromaticivorans CYP153C1, which shows between 38 and 46% full-length amino acid sequence identity with other CYP153 sequences.
The flanking regions of five CYP153 genes encoded homologs of ferredoxin as well as ferredoxin reductase, the other two components of class I cytochrome P450 systems. In addition, ferredoxin genes were found downstream of three of the five HXN-200 CYP153 genes (Table 2).
The eight ferredoxins encoded near CYP153 genes all belonged to the [2Fe-2S] type, which is ubiquitous in plants, animals, and bacteria (12). A closely related example of these ferredoxins is putidaredoxin, which transfers electrons from putidaredoxin reductase to P450cam (CYP101) (13). The sequence identity among the CYP153-associated ferredoxins was typically higher than 40%, which was slightly higher than the sequence identities with putidaredoxin (>35%).
The five ferredoxin reductases were slightly more closely related to each other (42 to 59%) than to putidaredoxin reductase (41 to 51%) and were still quite closely related to rubredoxin reductase (34 to 37%), which acts on rubredoxin, a structurally unrelated electron transfer protein.
Functional expression of CYP153 genes.
Previously, we have obtained functional expression of all three components of the Mycobacterium sp. strain HXN-1500 CYP153A6 AH system in P. putida GPo12(pGEc47ΔB) and used the recombinant to produce perillyl alcohol in gram amounts (35). To prove that the newly cloned and identified CYP153 genes indeed encode AHs, we choose 11 genes for expression in the same host, taking the ability of recombinants to use alkanes as the sole C and energy source as evidence for functional expression and hydroxylation of alkanes by the expressed CYP153s.
The first CYP153 selected for heterologous expression was CYP153A7, the proposed N-benzylpyrrolidine-hydroxylating alkane hydroxylase isolated from Sphingomonas sp. strain HXN-200 (3). CYP153A7 was expressed using the broad-host-range expression vector pCom8 (33), which was used earlier to obtain functional heterologous expression of CYP153A6 (35) and a wide range of integral membrane AHs (31). Because we have not been able to identify and clone the HXN-200 ferredoxin reductase gene, we initially cloned the CYP153A7 gene (ahpG1) together with the downstream ferredoxin gene (ahpI1) in pCom8 (resulting in pCom8-PA7F200) and tested functional expression in the host strains E. coli GEc137(pGEc47ΔB) and P. putida GPo12(pGEc47ΔB). These hosts are recombinant strains containing all genes necessary for alkane utilization except a functional AH gene (31). As we obtained only weak growth on solid mineral salts medium with n-octane vapor as the C source, a plasmid that also included the Mycobacterium sp. strain HXN-1500 ferredoxin reductase gene (pCom8-PA7F200R1500) was constructed next. An E. coli recombinant containing this plasmid still showed very weak and difficult-to-reproduce growth on n-octane, but growth of the P. putida recombinant on n-octane was much stronger. In addition, after several rounds of reinoculation the growth rate increased significantly, eventually resulting in dense growth on minimal medium plates 24 h after inoculation. P. putida GPo12(pGEc47ΔB, pCom8-PA7F200R1500) grew in liquid mineral medium with n-octane as the sole C source with a doubling time of close to 3 hours. Passage over rich medium or storage of the recombinant strain did not result in loss of the phenotype, and resequencing revealed no changes in the insert of pCom8-PA7F200R1500.
To facilitate heterologous expression of other CYP153 genes, we constructed a derivative of pCom8-PA7F200R1500 that allowed easy insertion of P450 genes as EcoRI-PacI or MfeI-PacI fragments (pCom12-PA7F200R1500) (Fig. 1). We then cloned the Acinetobacter sp. strain EB014 CYP153A1; R. palustris CGA009 CYP153A5; Sphingomonas sp. strain HXN-200 CYP153A8, -A11, -D2, and -D3; A. borkumensis AP1 CYP153A12 and -A13; and M. marinum M CYP153A14 genes in pCom12 and transferred the resulting plasmids to P. putida GPo12(pGEc47ΔB). All recombinants were tested for growth on n-pentane, n-hexane, n-octane, and n-decane on solid medium. In addition to CYP153A6 and -A7, CYP153A1, -A11, -A13, -A14, and -D2 also allowed the respective recombinants to grow well on most of the alkanes (Table 2; Fig. 2). As was the case with the CYP153A6 and -A7 recombinant strains, some of the new recombinants initially did not grow well on alkanes, and several rounds of reinoculation were necessary to obtain fast growth on alkanes. CYP153A5, -A8, -A12, and -D3 did not allow the recombinants to grow on any of the alkanes tested. We determined CO difference spectra of cell extracts of these four recombinants but found no evidence for the presence of cytochrome P450 (data not shown). In all cases, plasmids were reisolated from the recombinants to verify their identity by restriction analysis and partial sequencing.
FIG. 2.
Growth on n-octane vapor, after 4 days, of P. putida GPo12(pGEc47ΔB, pCom12-PxF200R1500) recombinants expressing seven different CYP153 genes. (CYP153A1, -A6, -A7, -A11, -A13, -A14, or -D2). Recombinants containing CYP153A5, -A8, -A12, and -D3 did not grow on octane. In the absence of n-octane, none of the recombinants grew (not shown).
Culture supernatants of octane-grown P. putida recombinants containing CYP153A1, -A6, -A7, -A13, and -D2 were tested for the presence of metabolites. No products were found. As a first indication for terminal versus subterminal hydroxylation by the expressed enzymes, we tested the host for the ability to grow on 1-, 2-, and 3-octanol. Only 1-octanol supported dense growth on plates after 2 days. Plates incubated with 2- or 3-octanol remained empty after prolonged incubation. Moreover, P. putida (pGEc47ΔB, pCom8-PA6F200R1500) cultures grew well on n-octane in the presence of 0.1% 2-octanol, but the 2-octanol was not degraded.
DISCUSSION
In the course of our research on integral membrane nonheme iron AHs, we have identified several strains that grow well on MCL alkanes but do not contain integral membrane AHs that act on these alkanes (32, 39; van Beilen et al., unpublished data). Several of these strains are useful biocatalysts for hydroxylation and epoxidation reactions; others are (potential) pathogens or are useful in biodegradation. Using PCR with CYP153 primers, we now show that many of these strains (mycobacteria, rhodococci, and Proteobacteria) contain cytochrome P450 enzymes belonging to the CYP153 family. Expressed CYP153s enable P. putida recombinants to grow well on alkanes, which demonstrates that these CYP153s indeed act as alkane hydroxylases. As all original hosts are alkane degraders, the hydroxylation of simple alkanes and related compounds may well be the physiological function of CYP153 enzymes. It is interesting to note that, in contrast to the yeast microsomal cytochrome P450 AHs and the AlkB-type AHs, the CYP153s appear to be soluble enzymes, which facilitates their characterization (3, 35).
Two of the four CYP153 genes that did not enable the P. putida recombinants to grow on C5 to C10 n-alkanes (CYP153A8 and CYP153D3) were cloned from HXN-200, an organism containing at least five CYP153 sequences, suggesting (i) that these CYP153s act on substrates other than MCL alkanes (for example, long-chain-length alkanes, alkylbenzenes, or alicyclic compounds) or (ii) that the cloned CYP153 genes are pseudogenes (this is especially likely for CYP153A12 cloned from A. borkumensis AP1). More general explanations (also valid for CYP153A5) are that these CYP153s (i) require their cognate electron transfer proteins for functional expression (in the pCom12 expression vector, the HXN-200 ferredoxin I and the HXN-1500 ferredoxin reductase replace the physiological electron donors), (ii) do not fold properly or are not expressed in P. putida for other reasons, or (iii) hydroxylate alkanes at subterminal positions (see below). We are now testing these alternatives.
The presence of multiple AHs in one strain has previously been documented for the integral membrane AHs; a majority of strains actually contains more than one AH. In some cases, these AHs have different substrate ranges or different induction patterns (34). Most CYP153 genes were found in bacteria that do not possess AlkB-type enzymes acting on MCL alkanes. It is important to note that several of these bacteria do contain AlkB homologs acting on LCL alkanes; in these strains, the two different types of AHs hydroxylate different subsets of alkanes. We found one exception: A. borkumensis AP1 and SK2 possess AlkB and CYP153 enzymes that both act on MCL alkanes (36).
Because the P. putida host strain is able to grow only on 1-alkanols and not on 2- or 3-alkanols, 2-octanol is not degraded by P. putida GPo12(pGEc47ΔB), and subterminal oxidation products could not be detected in culture supernatants, we conclude that the expressed CYP153 enzymes hydroxylate mainly the terminal positions of alkanes.
In previous work, we have shown that integral membrane AHs related to the P. putida GPo1 AH are ubiquitous in alkane-degrading Alpha-, Beta-, and Gammaproteobacteria and the high-G+C gram-positive bacteria. Most of these AHs hydroxylate alkanes longer than C10. We now show that many alkane-degrading bacteria also contain cytochrome P450 AHs that hydroxylate C5 to C10 alkanes. These P450 enzymes appear to be responsible for several of the interesting hydroxylation and epoxidation reactions catalyzed by alkane-degrading bacteria, while in A. borkumensis, they are likely to contribute to oil degradation in polluted marine environments. Quite a few strains contain multiple CYP153 enzymes, and sometimes also integral membrane AHs, and are difficult to cultivate due to their hydrophobic nature and/or pathogenicity. Thus, with the construction of seven recombinant strains expressing various individual CYP153 enzymes, we have created a useful toolbox not only for biocatalysis but also for the detailed characterization of members of this new class of P450 enzymes.
Supplementary Material
Acknowledgments
This research was supported by the Swiss Priority Program in Biotechnology of the Swiss National Science Foundation.
We thank Alfred Pühler for access to the A. borkumensis SK2 genome sequence prior to publication, Lalita Ramakrishnan for a sample of M. marinum M chromosomal DNA, Carrie Harwood for sending us R. palustris CGA009, Otmar Asperger for Acinetobacter sp. strain EB104, and Karl-Heinrich Engesser for his kind gift of the 15 hexane-degrading HXN strains.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Cardini, G., and P. Jurtshuk. 1970. The enzymatic hydroxylation of n-octane by Corynebacterium sp. strain 7E1C. J. Biol. Chem. 245:2789-2796. [PubMed] [Google Scholar]
- 3.Chang, D. 2003. Regio- and stereoselective hydroxylations with Sphingomonas sp. HXN-200. Ph.D. thesis. Swiss Federal Institute of Technology, Zürich, Switzerland.
- 4.Chang, D., H.-J. Feiten, K.-H. Engesser, J. B. van Beilen, B. Witholt, and Z. Li. 2002. Practical syntheses of N-substituted 3-hydroxyazetidines and 4- hydroxypiperidines by hydroxylation with Sphingomonas sp. HXN-200. Org. Lett. 4:1859-1862. [DOI] [PubMed] [Google Scholar]
- 5.Chang, D., B. Witholt, and Z. Li. 2000. Preparation of (S)-N-substituted 4-hydroxy-pyrrolidine-2-ones by regio- and stereoselective hydroxylation with Sphingomonas sp. HXN-200. Org. Lett. 2:3949-3952. [DOI] [PubMed] [Google Scholar]
- 6.Chang, D. L., H. J. Feiten, B. Witholt, and Z. Li. 2002. Regio- and stereoselective hydroxylation of N-substituted piperidin-2-ones with Sphingomonas sp HXN-200. Tetrahedron Asymmetry 13:2141-2147. [Google Scholar]
- 7.Davis, J. B., H. H. Chase, and R. L. Raymond. 1956. Mycobacterium paraffinicum n. sp., a bacterium isolated from soil. Appl. Microbiol. 4:310-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desomer, J., M. Crespi, and M. Van Montagu. 1991. Illegitimate integration of non-replicative vectors in the genome of Rhodococcus fascians upon electrotransformation as an insertional mutagenesis system. Mol. Microbiol. 5:2115-2124. [DOI] [PubMed] [Google Scholar]
- 9.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eggink, G., R. G. Lageveen, B. Altenburg, and B. Witholt. 1987. Controlled and functional expression of Pseudomonas oleovorans alkane utilizing system in Pseudomonas putida and Escherichia coli. J. Biol. Chem. 262:17712-17718. [PubMed] [Google Scholar]
- 12.Grinberg, A. V., F. Hannemann, B. Schiffler, J. Muller, U. Heinemann, and R. Bernhardt. 2000. Adrenodoxin: structure, stability, and electron transfer properties. Proteins 40:590-612. [DOI] [PubMed] [Google Scholar]
- 13.Gunsalus, I. C., and G. C. Wagner. 1978. Bacterial P-450cam methylene monooxygenase components: cytochrome m, putidaredoxin, and putidaredoxin reductase. Methods Enzymol. 52:166-188. [DOI] [PubMed] [Google Scholar]
- 14.Hara, A., S.-H. Baik, K. Syutsubo, N. Misawa, T. H. M. Smits, J. B. van Beilen, and S. Harayama. 2004. Cloning and functional analysis of alkB genes in Alcanivorax borkumensis SK2. Environ. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
- 15.Hauben, L., L. Vauterin, J. Swings, and E. R. B. Moore. 1997. Comparison of 16S ribosomal DNA sequences of all Xanthomonas species. Int. J. Syst. Bacteriol. 47:328-335. [DOI] [PubMed] [Google Scholar]
- 16.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 17.Hou, C. T., M. A. Jackson, M. O. Bagby, and L. A. Becker. 1994. Microbial oxidation of cumene by octane-grown cells. Appl. Microbiol. Biotechnol. 41:178-182. [Google Scholar]
- 18.Iizuka, H., and K. Komagata. 1964. Microbiological studies on petroleum and natural gas. I. Determination of hydrocarbon-utilizing bacteria. J. Gen. Appl. Microbiol. 10:207-221. [Google Scholar]
- 19.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 20.Karlson, U., D. F. Dwyer, S. W. Hooper, E. R. B. Moore, K. N. Timmis, and L. D. Eltis. 1993. Two independently regulated cytochromes P-450 in a Rhodococcus rhodochrous strain that degrades 2-ethoxyphenol and 4- methoxybenzoate. J. Bacteriol. 175:1467-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kester, A. S., and J. W. Foster. 1963. Diterminal oxidation of long-chain alkanes by bacteria. J. Bacteriol. 85:859-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lageveen, R. G., G. W. Huisman, H. Preusting, P. E. F. Ketelaar, G. Eggink, and B. Witholt. 1988. Formation of polyester by Pseudomonas oleovorans: the effect of substrate on the formation and composition of poly-(R)-3-hydroxyalkanoates and poly-(R)-3-hydroxyalkenoates. Appl. Environ. Microbiol. 54:2924-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larimer, F. W., P. Chain, L. Hauser, J. Lamerdin, S. Malfatti, L. Do, M. L. Land, D. A. Pelletier, J. T. Beatty, A. S. Lang, F. R. Tabita, J. L. Gibson, T. E. Hanson, C. Bobst, J. Torres, C. Peres, F. H. Harrison, J. Gibson, and C. S. Harwood. 2004. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat. Biotechnol. 22:55-61. [DOI] [PubMed] [Google Scholar]
- 24.Li, Z., H.-J. Feiten, D. Chang, W. A. Duetz, J. B. van Beilen, and B. Witholt. 2001. Preparation of (R)- and (S)-N-protected-3-hydroxypyrrolidines by hydroxylation with Sphingomonas sp. HXN-200, a highly active, regio- and stereoselective, and easy to handle biocatalyst. J. Org. Chem. 66:8424-8430. [DOI] [PubMed] [Google Scholar]
- 25.Li, Z., H.-J. Feiten, J. B. van Beilen, W. Duetz, and B. Witholt. 1999. Preparation of optically active N-benzyl-3-hydroxypyrrolidine by enzymatic hydroxylation. Tetrahedron Asymmetry 10:1323-1333. [Google Scholar]
- 26.Maier, T., H.-H. Foerster, O. Asperger, and U. Hahn. 2001. Molecular characterization of the 56-kDa CYP153 from Acinetobacter sp. EB104. Biochem. Biophys. Res. Commun. 286:652-658. [DOI] [PubMed] [Google Scholar]
- 27.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. K. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plaggemeier, T. 2000. Elimination der schwer wasserlöslichen Modellabluftinhaltsstoffe n-Hexan und Toluol in Biorieselbettverfahren. Ph.D. thesis. Universität Stuttgart, Stuttgart, Germany.
- 29.Ramakrishnan, L., and S. Falkow. 1994. Mycobacterium marinum persists in cultured mammalian cells in a temperature-restricted fashion. Infect. Immun. 62:3222-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Smits, T. H. M., S. B. Balada, B. Witholt, and J. B. van Beilen. 2002. Functional analysis of alkane hydroxylases from gram-negative and gram-positive bacteria. J. Bacteriol. 184:1733-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smits, T. H. M., M. Röthlisberger, B. Witholt, and J. B. van Beilen. 1999. Molecular screening for alkane hydroxylase genes in Gram-negative and Gram-positive strains. Environ. Microbiol. 1:307-318. [DOI] [PubMed] [Google Scholar]
- 33.Smits, T. H. M., M. A. Seeger, B. Witholt, and J. B. van Beilen. 2001. New alkane-responsive expression vectors for E. coli and Pseudomonas. Plasmid 46:16-24. [DOI] [PubMed] [Google Scholar]
- 34.van Beilen, J. B., Z. Li, W. A. Duetz, T. H. M. Smits, and B. Witholt. 2003. Diversity of alkane hydroxylase systems in the environment. Oil Gas Sci. Technol. 58:427-440. [Google Scholar]
- 35.van Beilen, J. B., D. Lüscher, R. Holtacker, U. Bauer, B. Witholt, and W. A. Duetz. 2005. Biocatalytic production of perillyl alcohol from limonene using a novel Mycobacterium cytochrome P450 alkane hydroxylase expressed in P. putida. Appl. Environ. Microbiol. 71:1737-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Beilen, J. B., M. Marin, T. H. M. Smits, M. Röthlisberger, A. Franchini, B. Witholt, and F. Rojo. 2004. Characterization of two alkane hydroxylase genes from the marine hydrocarbonoclastic bacterium Alcanivorax borkumensis. Environ. Microbiol. 6:264-273. [DOI] [PubMed] [Google Scholar]
- 37.van Beilen, J. B., S. Panke, S. Lucchini, A. G. Franchini, M. Röthlisberger, and B. Witholt. 2001. Analysis of Pseudomonas putida alkane degradation gene clusters and flanking insertion sequences: evolution and regulation of the alk-genes. Microbiology 147:1621-1630. [DOI] [PubMed] [Google Scholar]
- 38.van Beilen, J. B., T. H. M. Smits, L. G. Whyte, S. Schorcht, M. Röthlisberger, T. Plaggemeier, K.-H. Engesser, and B. Witholt. 2002. Alkane hydroxylase homologues in Gram-positive strains. Environ. Microbiol. 4:676-682. [DOI] [PubMed] [Google Scholar]
- 39.Whyte, L. G., T. H. M. Smits, D. Labbé, B. Witholt, C. W. Greer, and J. B. van Beilen. 2002. Cloning and characterization of multiple alkane hydroxylase systems in Rhodococcus sp. strains Q15 and 16531. Appl. Environ. Microbiol. 68:5933-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.