Abstract
Mutations in mexR yield a multidrug resistance phenotype in nalB mutants of Pseudomonas aeruginosa as a result of derepression of the mexAB-oprM multidrug efflux operon. MexR produced by several nalB strains carried single amino acid changes that compromised MexR stability or its ability to dimerize. Changes at residues L95 and R21, however, produced a stable MexR protein capable of dimerization and, thus, likely compromised DNA binding.
Bacterial multidrug efflux pumps play an important role in the antimicrobial resistance of gram-negative pathogens (15, 17), particularly Pseudomonas aeruginosa, where five multidrug efflux systems (MexAB-OprM, MexCD-OprJ, MexEF-OprN, MexXY-OprM, and MexJK-OprM) have been described to date (16, 18, 21). Of particular significance is the MexAB-OprM efflux system, which is expressed constitutively, thereby contributing to the well-known intrinsic resistance of this organism to multiple antimicrobials (11, 12, 14, 23), and is also hyperexpressed in nalB multidrug-resistant mutants as a result of mutations in the mexR gene (8, 19, 20, 24, 26). The mexR gene is transcribed divergently from the mexAB-oprM genes and encodes a repressor of the MarR family of regulators (19). MarR is responsible for regulating expression of the marRAB (mar) operon associated with a multiple antibiotic resistance (mar) phenotype in Escherichia coli (2). MexR has been shown to bind, possibly as a dimer, to two sites in the mexR-mexA intergenic region, overlapping promoters for mexR and mexAB-oprM (6), consistent with its observed repression of these promoters (19, 24). To elucidate the effect of mexR mutations on MexR repressor activity and thereby identify functionally important residues in the protein, the influence of mexR mutations in nalB strains on MexR production and dimerization was assessed.
Bacteria (Table 1) were cultivated at 37°C in Luria-Bertani (LB) broth (Difco) supplemented as needed with tetracycline (10 μg/ml for E. coli; 100 μg/ml for P. aeruginosa), ampicillin (50 μg/ml), chloramphenicol (30 μg/ml), and mercuric chloride (15 μg/ml). Mutated mexR genes were PCR amplified from several P. aeruginosa nalB strains using primers MEXRF1 (5′-GCGAGAATTCCGTTCGTTGCATAG-CGTTGTC-3′; Eco-RI site underlined) and MEXRB1 (5′-GCGAGAATTCCGAAGGCATTCG-CCAGTAAGC-3′; EcoRI site underlined). PCR mixtures (100 μl) contained 1 μg of chromosomal DNA (prepared as described elsewhere [4]), 2.5 U of Vent DNA polymerase (New England Biolabs, Mississauga, Ontario, Canada), a 0.2 mM concentration of each deoxyribonucleotide triphosphate, 2.5 mM MgSO4, a 0.3 μM concentration of each primer, 1× ThermoPol buffer (New England Biolabs), and 10% (vol/vol) dimethyl sulfoxide. Mixtures were heated for 2 min at 94°C followed by 30 cycles of 94°C for 30 s, 56°C for 1 min, and 72°C for 1 min, before finishing with 72°C for 10 min. PCR fragments were electrophoresed on and recovered from 1.2% (wt/vol) agarose gels using the Prep-a-Gene DNA purification kit (Bio-Rad Laboratories) as outlined by the manufacturer. Following digestion with EcoRI and cloning into pRK415 (Table 1), mexR sequences were validated by nucleotide sequencing using primer PMSLA25 (5′-GGATTCGTCTGTTGCAGG-3′). The cloned mexR genes were then mobilized into P. aeruginosa via triparental mating (25) with transconjugants selected on tetracycline (100 μg/ml) and imipenem (0.5 μg/ml; for counterselection). Wild-type and mutant mexR genes derived from nalB strains were also PCR amplified (as above) and cloned into the bacterial two-hybrid vectors pDP804 and pMS604 (Table 1). For cloning into pDP804, primers XhoILA7 (5′-GCAG-CTCGAGATGAACTACCCCGTGAATCC-3′; XhoI site underlined) and BglIILA11 (5′-GCGGAGATCTAATATCCTCAAGCGGTTGC-GCG-3′; BglII site underlined) were used, while for cloning into pMS604, primers BstEIILA8 (5′-GAGCGGTGACCATGAACTACCCC-GTGAATCC-3′; BstEII site underlined) and XhoILA12 (5′-GAGGCTCGAGAATATCCTCA-AGCGTTGCGCG-3′; XhoI site underlined) were used. Reaction mixtures were formulated and heated as above, with the exception that wild-type mexR was amplified from plasmid pRSP55 (20 ng). PCR products were purified as above, restricted with XhoI and BglII or BstEII and XhoI, and cloned into XhoI-BglII-digested pDP804 or BstEII-XhoI-digested pMS604, as appropriate. Following cloning, the mexR sequences were validated by nucleotide sequencing using primer PMSLA25. Plasmid DNAs were prepared using the Qiagen miniprep kit as recommended by the manufacturer.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsc | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | endA hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 deoR [φ80dlacΔ(lacZ)M15] | 4 |
| SU202 | lexA71::Tn5 sulA211 sulA::lacZ Δ(lacIPOZYA)169 F′ lacIqlacZΔM15::Tn9 | 5 |
| P. aeruginosa strains | ||
| K767 | PAO1 prototroph; MexRWTa | 14 |
| K1491 | K767 ΔmexR | 24 |
| OCR1 | K767 nalB; MexRR70W | 14 |
| PAO503 | met-9011; MexRWT | 9 |
| K1647 | PAO503 nalB [Pa10c]b; MexRA110T | B. Wretlind, Stockholm, Sweden |
| K1648 | PAO503 nalB [T3]; MexRG58E | 9 |
| K1649 | PAO503 nalB [T4]; MexRL95F | 9 |
| K1651 | PAO503 nalB [T6]; MexRT69I | 9 |
| K1652 | PAO503 nalB [T7]; MexRR21W | 9 |
| K1657 | PAO503 nalB [K3]; MexRR91H | 9 |
| K1658 | PAO503 nalB [K4]; MexRL13M | 9 |
| H103 | PAO1 prototroph; MexRWT | R. E. W. Hancock, Van- couver, Canada |
| K1462 | H103 nalB; MexRL57R | 24 |
| K1463 | H103 nalB; MexRT130P | 24 |
| Plasmids | ||
| pRK415 | Broad-host-range cloning vector; Tcr | 10 |
| pRSP55 | pRK415::mexR767 (wild-type MexR) | This study |
| pLK501 | pRK415::mexR1649 (L95F) | This study |
| pLK503 | pRK415::mexR1652 (R21W) | This study |
| pLK505 | pRK415::mexR1462 (L57R) | This study |
| pLK507 | pRK415::mexR1658 (L13M) | This study |
| pMS604 | LexA1-87WT-Fos zipper fusion; ori (ColE1) Tcr | 5 |
| pDP804 | LexA1-87408-Jun zipper fusion; ori (P15A) Apr | 5 |
| pRK2013 | Broad-host-range helper vector; Tra+ Kmr | 7 |
| pLK451 | pDP804::mexR767 (wild-type MexR) | This study |
| pLK452 | pMS604::mexR767 (wild-type MexR) | This study |
| pLK453 | pMS604::mexR1647 (A110T) | This study |
| pLK454 | pMS604::mexR1648 (G58E) | This study |
| pLK455 | pMS604::mexR1649 (L95F) | This study |
| pLK457 | pMS604::mexR1652 (R21W) | This study |
| pLK458 | pMS604::mexR1657 (R91H) | This study |
| pLK459 | pMS604::mexR1658 (L13M) | This study |
| pLK460 | pMS604::mexR1462 (L57R) | This study |
| pLK461 | pMS604::mexR1463 (T130P) | This study |
The MexR protein (wild-type or mutant) produced by each strain, including the nature of any mutation, is indicated.
Strain designation of Jalal et al. (9) is shown in square brackets.
The nalB strain from which the mexR gene was cloned is indicated by a subscript, and the mutant version of MexR that is produced is shown in parentheses.
Cell envelope proteins were prepared as described previously (22), except that 15 ml of cell culture was harvested in stationary phase. Soluble proteins were recovered by saving the supernatant following centrifugation of cell envelopes. Protocols for sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western immunoblotting have been described previously (22). For gels immunoblotted and developed with an anti-MexR antiserum, 2-mercaptoethanol (2.5% [vol/vol] final concentration) was included in the gel loading buffer (prepared as a 4× stock solution) and the gels were thicker (3 mm) than usual to permit more sample to be loaded. Antibody to MexR was prepared in New Zealand White rabbits (by L. Mutharia, University of Guelph, Guelph, Ontario, Canada) using purified histidine-tagged MexR (6) as antigen, and it was adsorbed against whole-cell lysates of P. aeruginosa K1491 prior to use. The protocol for antimicrobial susceptibility has been described elsewhere (22).
Two-hybrid systems are routinely used to assess protein-protein interactions, and a recently developed bacterial version of this system has proven useful in this regard (5). The system is based on LexA, a protein that binds only as a dimer to the sulA operator and in so doing downregulates transcription. Using vectors (pMS604 and pDP804) engineered to encode only the DNA-binding domain of LexA (hereafter called LexA′) and a reporter strain with a lacZ gene controlled by a sulA operator (E. coli SU202), one simply clones genes and sequences predicted to encode interacting proteins and domains as fusions to lexA′ and screens for repression of lacZ (i.e., reduction in β-galactosidase activity). Such repression requires dimerization of the LexA DNA-binding domain, which will occur only if the protein sequences fused to LexA′ are able to interact. Since the system was originally designed to assess interactions between heterologous proteins, the lexA′ sequence in one plasmid (pMS604) was altered such that its LexA′ bound to an altered operator sequence and the reporter strain carried a hybrid sulA operator, such that only LexA′ heterodimers would bind and repress lacZ. The two-hybrid vectors also contain sequences encoding Jun and Fos zipper motifs (known to interact) fused to lexA′, such that E. coli SU202 carrying these vectors demonstrates substantial repression of lacZ (Table 2). The unaltered vectors thus provide a positive control for the system, although the Jun and Fos zipper-encoding sequences will be disrupted upon cloning test sequences into the two-hybrid vectors (and, thus, lacZ repression will be dependent upon interaction of protein sequences encoded by the cloned DNA).
TABLE 2.
β-galactosidase activity of E. coli SU202 isolates expressing wild-type and mutant MexR proteins in the LexA-based two-hybrid systema
| Two-hybrid plasmid combination | β-galactosidase activityb (Miller units) | |
|---|---|---|
| pMS604 derivative | pDP804 derivative | |
| None | pDP804 | 1,171 ± 152 |
| pMS604 | None | 1,370 ± 99 |
| pMS604 | pDP804 | 96 ± 24 |
| pLK452 (mexRWT)c | pLK451 (mexR767; wild-type MexR) | 77 ± 12 |
| pLK453 (mexR1647; A110T) | pLK451 (mexR767; wild-type MexR) | 1,520 ± 197 |
| pLK454 (mexR1648; G58E) | pLK451 (mexR767; wild-type MexR) | 614 ± 82 |
| pLK455 (mexR1649; L95F) | pLK451 (mexR767; wild-type MexR) | 89 ± 19 |
| pLK457 (mexR1652; R21W) | pLK451 (mexR767; wild-type MexR) | 87 ± 21 |
| pLK458 (mexR1657; R91H) | pLK451 (mexR767; wild-type MexR) | 570 ± 25 |
| pLK459 (mexR1658; L13M) | pLK451 (mexR767; wild-type MexR) | 1,760 ± 212 |
| pLK460 (mexR1462; L57R) | pLK451 (mexR767; wild-type MexR) | 684 ± 53 |
| pLK461 (mexR1463; T130P) | pLK451 (mexR767; wild-type MexR) | 615 ± 32 |
| pLK455 (mexR1649; L95F) | pDP804 | 1,117 ± 65 |
| pLK457 (mexR1652; R21W) | pDP804 | 1,265 ± 96 |
E. coli SU202 harboring the indicated two-hybrid vectors was assayed for β-galactosidase activity as described in the text.
β-Galactosidase activities are the means of at least three independent determinations, with the standard deviations indicated.
The strain from which the mexR gene was cloned is identified by a subscript. Any changes in MexR encoded by these genes are also highlighted.
In utilizing this system to assess MexR dimerization, then, the mexR gene was cloned into pDP804 and pMS604, and E. coli SU202 isolates carrying these vectors was cultured overnight in antibiotic-supplemented LB broth and diluted 1:99 in fresh LB medium supplemented with appropriate antibiotics and isopropyl-β-d-thiogalactopyranoside (1 mM). Once cultures had reached an optical density at 600 nm of 0.4 to 0.8, β-galactosidase assays were performed as described by Miller (14a). As seen in Table 2, E. coli SU202 carrying one or the other of the two-hybrid vectors demonstrated substantial β-galactosidase activity, consistent with the absence of LexA′ dimerization. When wild-type mexR genes were cloned into both two-hybrid vectors, E. coli SU202 harboring the resultant plasmids (pLK451 and pLK452) showed a substantial decrease in β-galactosidase activity below that seen for the vectors-only positive control, indicating that LexA′ and, thus, MexR, was dimerizing. Dimerization has been reported for MarR, whose crystal structure is known (3), and the recent MexR crystal structure confirms the dimeric nature of MexR (13).
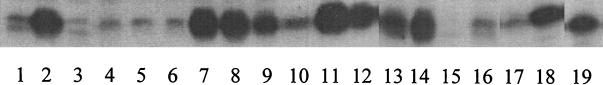
Mutations in mexR compromise MexR repressor activity, leading to MexAB-OprM hyperproduction in nalB mutants. Loss of activity might result, however, from protein instability, loss of dimerization, or defects in DNA binding. Initially, then, several nalB strains were screened for production of MexR. Intriguingly, although nalB strains show increased mexR gene expression owing to the characteristic mexR-negative autoregulation (19), many nalB strains produced little or no detectable MexR protein (Fig. 1). While this might be expected for mutants carrying frameshift mutations in mexR (see reference 24), those MexR proteins shown in Fig. 1 all carry single amino acid changes only. Some clustering of these mutations is evident (including residues 59 to 83), although the significance of this is not clear. Several nalB strains did, however, produce substantial levels of MexR, far above what was seen in wild-type strains (Fig. 1, lanes 1, 4, and 17) and comparable to levels seen in a nalC mutant (Fig. 1, lane 2) which hyperexpresses mexAB-oprM and mexR as a result of mutation(s) in an as-yet-unknown gene(s) (24). Thus, loss of repressor activity in these MexR proteins must be due to changes other than protein instability.
FIG. 1.
Expression of MexR in nalB mutants of P. aeruginosa. Soluble extracts of MexAB-OprM hyperexpressing multidrug-resistant strains of P. aeruginosa carrying mutations in mexR (and their parental strains) were electrophoresed on sodium dodecyl sulfate-polyacrylamide (15% [wt/vol]) gels and immunoblotted using antibodies to MexR. Lane 1, P. aeruginosa PAO1 strain K767 (MexRWT); lane 2, strain K1454 (nalC); lane 3, OCR1 (MexRR70W); lane 4, P. aeruginosa PAO503 (MexRWT); lane 5, K1655 (MexRR83H); lane 6, K1656 (MexRR83H); lane 7, K1657 (MexRR91H); lane 8, K1658 (MexRL13M); lane 9, K1647 (MexRA110T); lane 10, K1646 (MexRR59C); lane 11, K1648 (MexRG58E); lane 12, K1649 (MexRL95F); lane 13, K1651 (MexRT69I); lane 14, K1652 (MexRR21W); lane 15, K1653 (MexRL80P); lane 16, K1654 (MexRR70W); lane 17, P. aeruginosa PAO1 strain H103 (MexRWT); lane 18, K1462 (MexRL57R); lane 19, K1463 (MexRT130P). The impact of the nalB mutation on MexR in each strain is indicated in parentheses, with the exception of K1455, which is a nalC strain that hyperproduces MexAB-OprM as a result of an unknown mutation. The gel as presented is a composite assembled from lanes of the same gel.
To assess the possible impact of the mexR mutation in each of these mutants on protein dimerization, the corresponding genes were amplified by PCR and cloned into the two-hybrid vectors described above. As seen in Table 2, the bulk of the mutations yielding a stable MexR protein interfered with MexR-MexR interaction with, e.g., E. coli SU202 isolates expressing A110T (pLK453) or L13M (pLK459) MexR proteins demonstrating substantial levels of β-galactosidase activity (i.e., comparable to that seen for the single-vector negative controls). Changes at position 58 (G58E, pLK454), 91 (R91H, pLK458), 57 (L57R, pLK460), and 130 (T130P, pLK461) produced β-galactosidase activities that were half that of the negative controls though substantially above that seen for wild-type MexR (Table 2). Presumably, these MexR proteins interact only weakly or associate abnormally, such that LexA′ dimers are not positioned in such a way as to bind the sulA operator. Thus, while the aforementioned mutations compromise normal dimer activity by MexR, it is not at all clear that these changes identify residues directly involved in dimerization. The A110T change, for example, is far from sites of interaction between MexR monomers in the crystal structure (13), where any impact on dimerization or dimer structure would have to occur via conformational changes that interfere with dimerization (or formation of normal dimers).
Only the L95F (pLK455) and R21W (pLK457) changes had no apparent impact on MexR dimerization, with E. coli SU202 isolates expressing these MexR derivatives demonstrating β-galactosidase activities that were indistinguishable from that of the same strain expressing the wild-type proteins (Table 2). It is likely, therefore, that these mutations ultimately compromised DNA binding, though not necessarily because they define DNA-binding domain(s) of the protein. Indeed, while the L95F change occurs in a region of MexR implicated in DNA binding (13), residue 21 is far from the probable DNA-binding region of the protein. Still, it does occur in a region of MexR where the individual monomers interact with one another. The R21W change may, therefore, alter the positioning of the monomers with respect to one another, altering the proper spacing of the DNA-binding region of each monomer and compromising the ability of MexR dimer to bind to adjacent DNA-binding sites in the mexAB-oprM promoter (6).
Nonfunctional mutant MexR proteins that dimerize but are unable to bind DNA should be dominant over wild-type MexR and compromise, to some extent at least, wild-type MexR repressor activity when coexpressed in P. aeruginosa (as a result of forming heterodimers). Indeed, negative dominance of marR mutations was previously used to define residues implicated in the DNA binding of this repressor protein (1). Thus, the cloned mexR genes from nalB strains K1649 (pLK501; expresses MexRL95F) and K1652 (pLK503; expresses MexRR21W) were introduced into wild-type P. aeruginosa strain K767, and the impact on mexAB-oprM expression was assessed indirectly by measuring the impact on susceptibility to agents known to be substrates of this efflux system. Initial attempts at measuring the impact on expression directly, using Western immunoblotting with a MexB-specific antiserum, were unsuccessful, likely owing to a lack of sensitivity of the method. As seen in Table 3, P. aeruginosa K767 isolates carrying pLK501 or pLK503 showed increased resistance to representative antimicrobials relative to that of the vector control or strain K767 expressing wild-type MexR from a plasmid. In contrast, K767 carrying plasmid pLK505 (MexRL57R) or pLK507 (MexRL13M) did not show any change in drug susceptibility relative to the vector control. These data are consistent with the former but not the latter MexR derivatives negatively impacting the wild-type repressor activity of the chromosomally encoded MexR protein in strain K767, by the formation of heterodimers unable to bind to the mexAB-oprM promoter region. These data can be interpreted as suggesting a dimerization defect for the MexRL13M and MexRL57R derivatives. Had these variants retained dimerization ability but interfered with LexA′ binding to sulA (see above), thereby compromising LexA′-mediated repression of lacZ in the two-hybrid assay, one would have expected them to be also dominant over wild-type MexR and to enhance multidrug resistance. That they were not suggests that these changes compromised MexR dimerization. Recently, a plasmid-encoded MexRT69I variant (from nalB strain K1651) has been examined in the two-hybrid system and has also been shown to be competent for dimer formation (data not shown), although its dominance over wild-type MexR has yet to be assessed. Like residue L95, residue T69 occurs in a region of MexR implicated in DNA binding (D. Lim, personal communication).
TABLE 3.
Effect of cloned mexR mutant genes on the antibiotic susceptibility of wild-type P. aeruginosa isolatesa
| Plasmid | MexRb | MIC (μg/ml)c
|
||
|---|---|---|---|---|
| NOV | CAM | CAR | ||
| pRK415 | — | 256 | 16 | 64 |
| pRSP55 | Wild type | 128 | 8 | 32 |
| pLK501 | L95F | 512 | 64 | 256 |
| pLK503 | R21W | 512 | 64 | 256 |
| pLK505 | L57R | 256 | 32 | 64 |
| pLK507 | L13M | 128 | 16 | 64 |
The antibiotic susceptibilities of P. aeruginosa PAO1 strain K767 isolates carrying the indicated mexR plasmids were assessed as described in the text.
The alterations in the MexR proteins encoded by the various plasmids are highlighted; —, vector control (no plasmid-encoded MexR).
NOV, novobiocin; CAM, chloramphenicol; CAR, carbenicillin.
Acknowledgments
We thank M. Granger-Schnarr for providing the components of the bacterial two-hybrid system and B. Wretlind for providing nalB strains of P. aeruginosa.
This work was supported by an operating grant from the Canadian Cystic Fibrosis Foundation (CCFF). L.A. is supported by the Canadian Bacterial Diseases Network (one of the Networks of Centres of Excellence). R.S. is supported by a CCFF Postdoctoral Fellowship.
REFERENCES
- 1.Alekshun, M. N., Y. S. Kim, and S. B. Levy. 2000. Mutational analysis of MarR, the negative regulator of marRAB expression in Escherichia coli, suggests the presence of two regions required for DNA binding. Mol. Microbiol. 35:1394-1404. [DOI] [PubMed] [Google Scholar]
- 2.Alekshun, M. N., and S. B. Levy. 1999. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 7:410-413. [DOI] [PubMed] [Google Scholar]
- 3.Alekshun, M. N., S. B. Levy, T. R. Mealy, B. A. Seaton, and J. F. Head. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 8:710-714. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Short protocols in molecular biology, 2nd ed. John Wiley & Sons, Inc., New York, N.Y.
- 5.Dmitrova, M., G. Younes-Cauet, P. Oertel-Buchheit, D. Porte, M. Schnarr, and M. Granger-Schnarr. 1998. A new LexA-based genetic system for monitoring and analyzing protein heterodimerization in Escherichia coli. Mol. Gen. Genet. 257:205-212. [DOI] [PubMed] [Google Scholar]
- 6.Evans, K., L. Adewoye, and K. Poole. 2001. MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic region. J. Bacteriol. 183:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Figurski, D. H., and E. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jalal, S., and B. Wretlind. 1998. Mechanisms of quinolone resistance in clinical strains of Pseudomonas aeruginosa. Microbiol. Drug Resist. 4:257-261. [DOI] [PubMed] [Google Scholar]
- 9.Jalal, S., G. Wretlind, N. Gotoh, and B. Wretlind. 1999. Rapid identification of mutations in a multidrug efflux pump in Pseudomonas aeruginosa. APMIS 107:1109-1116. [DOI] [PubMed] [Google Scholar]
- 10.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 11.Köhler, T., M. Kok, M. Michea-Hamzehpour, P. Plesiat, N. Gotoh, T. Nishino, L. Kocjanici Curty, and J.-C. Pechere. 1996. Multidrug efflux in intrinsic resistance to trimethoprim and sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 40:2288-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, X.-Z., L. Zhang, R. Srikumar, and K. Poole. 1998. β-Lactamase inhibitors are substrates of the multidrug efflux pumps of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:399-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim, D., K. Poole, and N. Strynadka. Crystal structure of the MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J. Biol. Chem., in press. [DOI] [PubMed]
- 14.Masuda, N., and S. Ohya. 1992. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 36:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 15.Poole, K. 2000. Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob. Agents Chemother. 44:2233-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poole, K. 2001. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 3:255-264. [PubMed] [Google Scholar]
- 17.Poole, K. 2001. Multidrug resistance in gram-negative bacteria. Curr. Opin. Microbiol. 4:500-508. [DOI] [PubMed] [Google Scholar]
- 18.Poole, K., and R. Srikumar. 2001. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr. Top. Med. Chem. 1:59-71. [DOI] [PubMed] [Google Scholar]
- 19.Poole, K., K. Tetro, Q. Zhao, S. Neshat, D. Heinrichs, and N. Bianco. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito, K., H. Yoneyama, and T. Nakae. 1999. nalB-type mutations causing the overexpression of the MexAB-OprM efflux pump are located in the mexR gene of the Pseudomonas aeruginosa chromosome. FEMS Microbiol. Lett. 179:67-72. [DOI] [PubMed] [Google Scholar]
- 21.Schweizer, H. P. 2001. Triclosan: a widely used biocide and its link to antibiotics. FEMS Microbiol. Lett. 202:1-7. [DOI] [PubMed] [Google Scholar]
- 22.Srikumar, R., T. Kon, N. Gotoh, and K. Poole. 1998. Expression of Pseudomonas aeruginosa multidrug efflux pumps MexA-MexB-OprM and MexC-MexD-OprJ in a multidrug-sensitive Escherichia coli strain. Antimicrob. Agents Chemother. 42:65-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srikumar, R., X.-Z. Li, and K. Poole. 1997. Inner membrane efflux components are responsible for the β-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J. Bacteriol. 179:7875-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srikumar, R., C. J. Paul, and K. Poole. 2000. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 182:1410-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao, Q., X.-Z. Li, A. Mistry, R. Srikumar, L. Zhang, O. Lomovskaya, and K. Poole. 1998. Influence of the TonB energy-coupling protein on efflux-mediated multidrug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:2225-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziha-Zarifi, I., C. Llanes, T. Koehler, J.-C. Pechere, and P. Plesiat. 1999. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob. Agents Chemother. 43:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]