Abstract
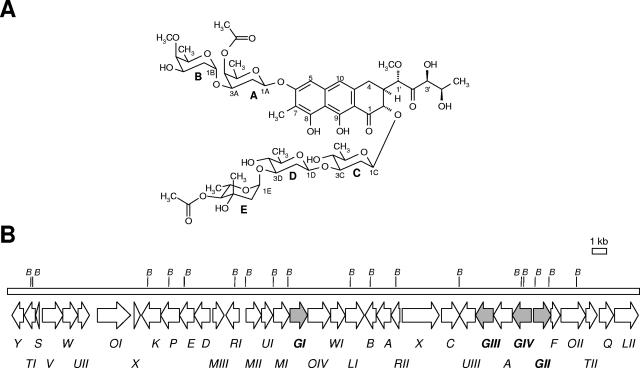
Chromomycin A3 is an antitumor drug produced by Streptomyces griseus subsp. griseus. It consists of a tricyclic aglycone with two aliphatic side chains and two O-glycosidically linked saccharide chains, a disaccharide of 4-O-acetyl-d-oliose (sugar A) and 4-O-methyl-d-oliose (sugar B), and a trisaccharide of d-olivose (sugar C), d-olivose (sugar D), and 4-O-acetyl-l-chromose B (sugar E). The chromomycin gene cluster contains four glycosyltransferase genes (cmmGI, cmmGII, cmmGIII, and cmmGIV), which were independently inactivated through gene replacement, generating mutants C60GI, C10GII, C10GIII, and C10GIV. Mutants C10GIV and C10GIII produced the known compounds premithramycinone and premithramycin A1, respectively, indicating the involvement of CmmGIV and CmmGIII in the sequential transfer of sugars C and D and possibly also of sugar E of the trisaccharide chain, to the 12a position of the tetracyclic intermediate premithramycinone. Mutant C10GII produced two new tetracyclic compounds lacking the disaccharide chain at the 8 position, named prechromomycin A3 and prechromomycin A2. All three compounds accumulated by mutant C60GI were tricyclic and lacked sugar B of the disaccharide chain, and they were named prechromomycin A4, 4A-O-deacetyl-3A-O-acetyl-prechromomycin A4, and 3A-O-acetyl-prechromomycin A4. CmmGII and CmmGI are therefore responsible for the formation of the disaccharide chain by incorporating, in a sequential manner, two d-oliosyl residues to the 8 position of the biosynthetic intermediate prechromomycin A3. A biosynthetic pathway is proposed for the glycosylation events in chromomycin A3 biosynthesis.
Chromomycin A3 (Fig. 1A) is an antitumor drug produced by Streptomyces griseus and other streptomycete species. It belongs to the class of antitumor compounds of the aureolic acid family (22), which inhibit growth and multiplication of several tumor cell lines. The antitumor properties are ascribed to their inhibitory effects on replication and transcription processes during macromolecular biosynthesis by interacting, in the presence of Mg2+, with GC-rich nucleotide sequences located in the minor groove of DNA. In this respect, they have been shown to prevent resistance to other antitumor agents by a number of mechanisms, including downregulation of proteins, such as MDR1 (16, 28). Chromomycin and the closely related compound mithramycin were also found to stimulate K562 cell erythroid differentiation (3), to be potent inhibitors of neuronal apoptosis (7), and to have antiviral activity against human immunodeficiency virus type 1 (2).
FIG. 1.
(A) Chemical structure of chromomycin A3. Sugars: A, 4-O-acetyl-d-oliose; B, 4-O-methyl-d-oliose; C and D, d-olivose; E, 4-O-acetyl-l-chromose B. (B) Genetic organization of the chromomycin biosynthesis gene cluster. Glycosyltransferase genes are in gray. B, BamHI sites.
Structurally, chromomycin A3 is related to mithramycin. Both consist of a tricyclic chromophore (aglycone) with two aliphatic side chains attached at C-3 and C-7. However, they differ in glycosylation pattern. While chromomycin A3 contains a trisaccharide of d-olivose (sugar C), d-olivose (sugar D), and 4-O-acetyl-l-chromose B (sugar E) and a disaccharide of 4-O-acetyl-d-oliose (sugar A) and 4-O-methyl-d-oliose (sugar B), mithramycin contains a trisaccharide of d-olivose, d-oliose, and d-mycarose and a disaccharide of d-olivose, attached at positions 2 and 6 of the aglycone, respectively.
In chromomycin, the carbohydrate moieties and the acetyl groups which decorate these sugars are major structural contributors to the biological activity. Thus, it has been shown that the acetyl groups in sugars A and E of chromomycin contribute distinctively in the DNA complex formation by providing additional H bond acceptor groups that interact with the 2-amino groups of G-bases, thus adding more specificity to DNA binding (6). Also, it has been recently demonstrated that elimination of acetyl groups leads to the generation of a chromomycin derivative with a significant decreased antitumor activity (14).
We have recently reported the cloning and characterization of the chromomycin A3 biosynthetic gene cluster (15). Because mithramycin and chromomycin share the same aglycone but differ in four of the five sugars attached to the aglycone, we were interested in characterizing the chromomycin glycosyltransferase genes. These genes together with those of the mithramycin cluster may serve as genetic tools for further studies on the generation of novel aureolic acid group derivatives with potential antitumor activity. Here we report the identification and functional characterization of the glycosyltransferase genes from this cluster (cmmGI, cmmGII, cmmGIII, and cmmGIV). We also show that inactivation of these glycosyltransferase genes leads to the accumulation of a variety of compounds, five of which are new chromomycin derivatives with antitumor activity.
MATERIALS AND METHODS
Microorganisms, culture conditions, and plasmids.
Streptomyces griseus subsp. griseus ATCC 13273, a chromomycin A3 producer, was used as donor of chromosomal DNA. For sporulation on solid medium, it was grown at 30°C on plates containing A medium (8). For growth in liquid medium, the organism was grown either on TSB medium (Trypticase soy broth; Oxoid) or in R5A medium (8). Escherichia coli ET12567(pUB307) (10) was used as a donor for intergeneric conjugation. When plasmid-containing clones were grown, the medium was supplemented with the appropriate antibiotics: 5 or 25 μg/ml thiostrepton for liquid or solid cultures, respectively; 100 μg/ml ampicillin; 25 μg/ml apramycin, 20 μg/ml tobramycin; 25 μg/ml kanamycin; 25 μg/ml chloramphenicol. pUC18, pHZ1358 (10), pUK21 (30), pEM4 (19), and pIJ2925 (10), were used for subcloning. pEM4T was constructed by subcloning a blunt-ended XbaI-HindIII fragment containing the oriT from pHZ1358 into the HindIII site made blunt of pEM4.
DNA manipulation and analysis.
Plasmid DNA preparations, restriction endonuclease digestions, alkaline phosphatase treatments, DNA ligations, Southern hybridization, and other DNA manipulations were performed according to standard techniques for Escherichia coli (24) and Streptomyces (10). Intergeneric conjugation from E. coli ET12567(pUB307) to S. griseus subsp. griseus was performed as described previously (10). Computer-aided database searching and sequence analysis were carried out using the University of Wisconsin Genetics Computer Group program package and the BLAST program (1).
Generation of mutants.
(i) cmmGI.
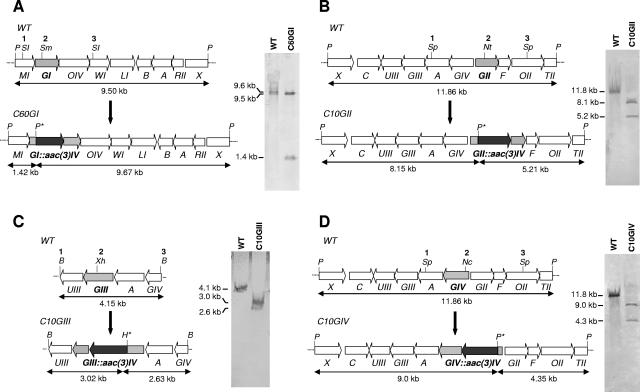
A 3.5-kb SacI DNA fragment (sites 1 and 3 in Fig. 2A) containing cmmGI, cmmOIV, the 3′ end of cmmMI and the 5′end of cmmWI was cloned into the same site of pUK21, previously digested with StuI and SmaI and religated to eliminate these two restriction sites. Then, a 1.5-kb BamHI (blunt ended)-SmaI fragment containing an apramycin resistance (Amr) cassette, the aac(3)-IV gene was inserted into a unique SmaI site (site 2 in Fig. 2A) located within the cmmGI gene and in the same direction of transcription of cmmGI to avoid polar mutations in downstream genes. The insert was rescued by digestion with SpeI (using these sites from the polylinker of pUK21) and subcloned into the XbaI site of pIJ2925. Finally, the whole insert was rescued as a BglII fragment (using these sites from the polylinker of pIJ2925) and subcloned into the BamHI site of pHZ1358, generating the final construct pC60GI.
FIG. 2.
Insertional inactivation of glycosyltransferase genes. Organization of chromosomal regions and Southern analysis of mutants C60GI (A), C10GII (B), C10GIII (C), and C10GIV (D). B, BamHI; H, HindIII; Nc, NcoI; Nt, NotI; P, PstI; SI, SacI; Sm, SmaI; Sp, SphI; Xh, XhoI; aac3(IV), apramycin resistance gene; WT, wild type. Restriction sites indicated with an asterisk are located in the aac(3)-IV gene and were used to verify the gene replacement events in the respective mutants.
(ii) cmmGII.
A 5-kb SphI fragment (sites 1 and 3 in Fig. 2B), containing cmmGIV, cmmGII, cmmF, and the 5′ ends of cmmA and cmmOII, was subcloned into the same site of pUC18, generating pUF1. Then, a 1.5-kb BamHI-HindIII fragment (both sites blunt ended) containing the aac(3)-IV gene was inserted into a unique NotI site (made blunt) (site 2 in Fig. 2B) located within the cmmGII gene and in the same direction of transcription of cmmGII. The insert was rescued as an XbaI-HindIII fragment (using these sites from the polylinker of pUC18) and subcloned into the same sites of pUK21. Finally, the whole insert was rescued as an XbaI fragment (using these sites from the polylinker of pUK21) and subcloned into the same site of pHZ1358, generating the final construct pC10GII.
(iii) cmmGIII.
A 4.2-kb BamHI fragment (sites 1 and 3 in Fig. 2C), containing cmmUIII, cmmGIII, cmmA and the 3′ end of cmmGIV, was subcloned into the same site of pUC18. Then, a 1.5-kb BamHI (blunt ended)-SmaI fragment containing the aac(3)-IV gene was inserted into a unique blunt-ended XhoI site (site 2 in Fig. 2C), located within the cmmGIII gene and in the same direction of transcription of cmmGIII. Finally, the whole insert was rescued as a BamHI fragment and subcloned into the same site of pHZ1358, generating the final construct pC10GIII.
(iv) cmmGIV.
To inactivate this gene, plasmid pUF1 (see above) was used. First, a 1.5-kb BamHI-HindIII fragment (both sites blunt ended) containing the aac(3)-IV gene was inserted into the unique NcoI site blunt ended (site 2 in Fig. 2D), located within the cmmGIV gene and in the same direction of transcription of cmmGIV, generating pUGIVA. Then, the whole insert in pUGIVA was rescued as a SpeI fragment (using these from the polylinker of pUK21) and subcloned into the XbaI site of pHZ1358, generating the final construct pC10GIV.
pC60GI, pC10GII, pC10GIII, and pC10GIV were independently introduced by intergeneric conjugation into S. griseus. Apramycin-resistant, thiostrepton-sensitive transconjugants were selected from each conjugation experiment for further characterization.
PCR amplification and gene expression.
Genes cmmGI, cmmGII, cmmGIII, and cmmGIV were amplified using the oligoprimers listed in Table 1. PCR conditions were as follows: 100 ng of template DNA was mixed with 30 pmols of each primer and 2 units of Pfx DNA Polymerase (New England Biolabs) in a total reaction volume of 50 μl containing 2 mM of each deoxynucleoside triphosphate, 10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris-HCl (pH 8.8), 2 mM MgSO4, and 0.1% Triton X-100. The polymerization reaction was performed in a thermocycler (MinyCycler; MJ Research) under the following conditions: initial denaturation of 2 min at 94°C; 30 cycles of 15 s at 94°C, 30 s at 55°C, and 1 min at 68°C; after the 30 cycles, an extra extension step of 5 min at 68°C was added.
TABLE 1.
Oligoprimers used for PCR amplification of cmmGI, cmmGII, cmmGIII, and cmmGIV
| Gene | Primer | Nucleotide sequence (5′-3′)a |
|---|---|---|
| cmmGI | Forward | GGACTAGTCCCACGGAGTTTCCTATG |
| Reverse | GCTCTAGAGTTAACTCAGTCTCCG GTGTCCG | |
| cmmGII | Forward | GGACTAGTGCTAGCGGAGCCCTCG CCTGTCATG |
| Reverse | GCTCTAGACTAACCGGCGCATCGCCG | |
| cmmGIII | Forward | GGACTAGTGTTAACGTGGAGAGGT GAGTGGCATG |
| Reverse | GCTCTAGACCTAGGTCAGGGCCCG GTCCTGGC | |
| cmmGIV | Forward | GGACTAGTCCTAGGCTCTGGGTGC CACTGAAAG |
| Reverse | GCTCTAGAGCTAGCTCATCCTGCC TCTTTCGTC |
Sequences for restriction sites are underlined. SpeI, 5′-ACTAGT-3′; XbaI, 5′-TCTAGA-3′; HpaI, 5′-GTTAAC-3′; NheI, 5′-GCTAGC-3′; AvrII, 5′-CCTAGG-3′.
After PCR amplification, all amplicons were independently subcloned as XbaI-SpeI fragments into the XbaI site of pEM4. Then, in the case of cmmGI and cmmGIII, the fragments were rescued as BamHI fragments and subcloned into the same site of pEM4T, generating the final constructs pEM4T-GI and pEM4T-GIII, respectively. For cmmGII and cmmGIV, the fragments were rescued as PstI-EcoRI fragments, subcloned into the same sites of pUK21, and then rescued as Ecl136II-EcoRV fragments to subclone into the blunt-ended EcoRI site of pEM4T, generating the final constructs pEM4T-GII and pEM4T-GIV, respectively. In all final constructs, the glycosyltransferase genes were under the control of the ermE*p.
HPLC analysis and purification of the new compounds.
High-performance liquid chromatography (HPLC) analysis of chromomycin-related compounds was performed as previously described (11). Strains were grown on R5A solid medium for 5 days at 28°C. In the case of mutants C10GII, C10GIII, and C10GIV, the R5A medium was supplemented with 2 μg/ml chromomycin A3, final concentration. For purification of compounds produced by mutants C60GI and C10GII, 130 plates and 100 plates were used, respectively. Agar cultures were removed from the plates and extracted with ethyl acetate as previously described (15). The new compounds were purified by chromatography in a μBondapak C18 radial compression cartridge (PrepPak cartridge, 25 by 100 mm; Waters). Extracts from both strains were first chromatographed with a mobile phase composed of acetonitrile and 0.1% trifluoroacetic acid (TFA) in water (60:40), at 10 ml/min. Compound PC-A3 produced by strain C10GII was repurified by chromatography with methanol and 0.1% TFA in water (77:33). Compounds PC-A4, PC-A4B, and PC-A4C produced by strain C10GI were rechromatographed with methanol and 0.1% TFA in water (70:30). After every step of purification, the compounds were desalted and concentrated by solid-phase extraction (15). The purified compounds were finally redissolved in a small volume of tert-butanol and lyophilized.
Physicochemical analysis.
Initial characterization of the new compounds was performed by HPLC/mass spectrometry (MS) using a Water Alliance 2695 HPLC system, equipped with a Waters 2996 photodiode array detector and a Micromass ZQ2000 mass spectrometer, controlled by MassLynx 4.0 software. An atmospheric pressure chemical ionization probe was used for ionization with the following parameters: solvent A, 0.1% formic acid in H2O; solvent B, acetonitrile; flow rate, 0.5 ml/min. From 0 to 10 min, a mixture of 75% solvent A and 25% solvent B was shifted to 100% solvent B with a linear gradient. From 10 to 12 min, 100% solvent B was used. From 12 to 14 min, 100% solvent B was shifted to 75% solvent A and 25% solvent B with a linear gradient. From 14 to 15 min, a mixture of 75% solvent A and 25% solvent B was used. A Waters Symmetry C18 column was used, 4.6 by 50 mm, with a particle size of 5 μm. The column temperature was 23°C, and the detection wavelength was 452 nm.
High-resolution mass spectra (fast atom bombardment [FAB] or electrospray ionization mode) were acquired with a Finnigan MAT LCQ mass spectrometer. Spectra were recorded on a Varian CARY50 spectrophotometer, and the infrared (IR) spectra were obtained from pure samples pressed in KBr disks using a Bio-Rad FTS3000MX FT IR. All nuclear magnetic resonance (NMR) data were recorded on a Varian Inova 400 instrument at a B0 of 9.4 T, in CDCl3 and acetone-d6 (see Tables SA1 to SA3 in the supplemental material). Chemical shifts (see data below) are reported in ppm relative to internal tetramethylsilane. All NMR assignments were based on chemical shift rules and on comparison with data for chromomycin A3 and premithramycin A3. Assignments were further confirmed and refined by two-dimensional NMR, such as H,H-COSY, HSQC, and HMBC, wherever needed.
Antibacterial and antitumor activities.
Antibacterial activity was tested against Micrococcus luteus as described previously (31). The antitumor activity of the compounds was tested against a variety of tumor cell lines. Quantitative measurement of cell growth and viability was carried out by using the colorimetric sulforhodamine B assay (27).
RESULTS
Identification of the chromomycin glycosyltransferase genes.
Chromomycin A3 contains five deoxysugars attached to the aglycone, forming a trisaccharide and a disaccharide chain. Analysis of the chromomycin gene cluster of S. griseus subsp. griseus revealed the existence of only four genes, named cmmGI, cmmGII, cmmGIII, and cmmGIV (Fig. 1B), which code for proteins which show high similarities to glycosyltransferases. Three of these genes are located at the right of the cluster and are divergently transcribed. cmmGIII and cmmGIV are transcribed in the same direction, while cmmGII, located immediately upstream of cmmGIV, is transcribed in the opposite direction. The fourth glycosyltransferase gene, cmmGI, is located in the middle of the cluster, about 11 kb apart from the other three glycosyltransferase genes. All chromomycin glycosyltransferases contain the conserved regions COG1819 (glycosyl transferases, related to UDP-glucorunosyltransferase), pfam06722 (conserved region of unknown function within bacterial glycosyltransferases) (amino acids 174 to 270 in CmmGI, 172 to 264 in CmmGII, 186 to 281 in CmmGIII, 191 to 287 in CmmGIV), and pfam03033 (glycosyltransferase family 28 N-terminal domain, which includes the acceptor binding site) (amino acids 3 to 112 in CmmGI and CmmGII, 3 to 148 in CmmGIII, and 3 to 153 in CmmGIV); CmmGI also contains the conserved domain pfam04101 (glycosyltransferase family 28 C-terminal domains which include the nucleotide binding site, amino acids 215 to 367).
Generation and characterization of S. griseus mutants with inactivated glycosyltransferase genes.
To assign each glycosyltransferase to a specific glycosylation step, all glycosyltransferase genes were independently inactivated. This was carried out by replacing the wild-type copy of the gene in the S. griseus chromosome by an in vitro-mutated one. To do that, several constructs in the unstable plasmid pHZ1358 were generated. In these constructs, each glycosyltransferase gene was interrupted by an apramycin resistance cassette, which was inserted in the direction of transcription of the targeted gene, at unique restriction sites within the corresponding coding regions (Fig. 2). In this way, plasmids pC60GI, pC10GII, pC10GII, and pC10GIV were constructed for the inactivation of cmmGI, cmmGII, cmmGIII, and cmmGIV, respectively. These constructs were introduced into S. griseus by intergeneric conjugation from E. coli, and transconjugants were selected for resistance to apramycin and then further tested for their susceptibility to thiostrepton as a consequence of a double crossover. One mutant was selected for each mutated gene, which were named C60GI (mutant in cmmGI), C10GII (mutant in cmmGII), C10GIII (mutant in cmmGIII), and C10GIV (mutant in cmmGIV). Southern hybridization analyses were carried out to verify the occurrence of the double recombination events (Fig. 2). These Southern analyses confirmed that, in all mutants, the replacement events had occurred. In trans complementation of each mutant was done to confirm that no other gene except the targeted gene was affected. This was carried out by introducing, in each mutant, a plasmid containing the wild-type copy of the appropriate glycosyltransferase gene under the control of the erythromycin resistance gene promoter. Recovery of chromomycin A3 production at levels similar to those of the wild-type strain was achieved for each mutant (data not shown).
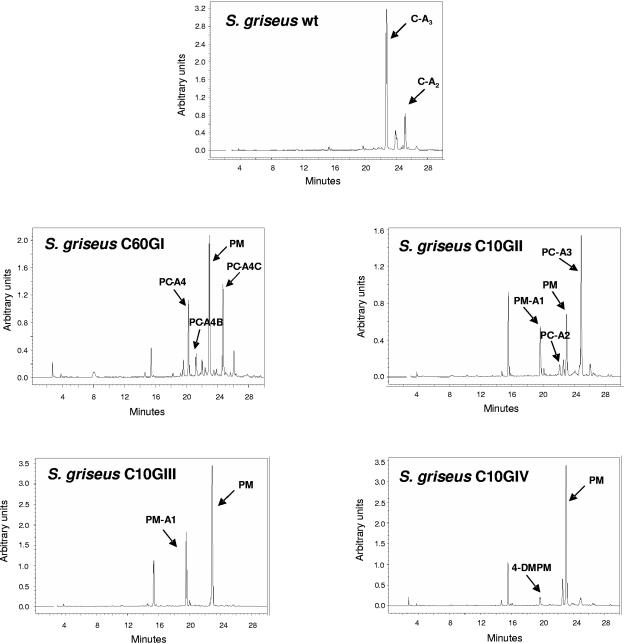
The different mutants were further characterized for production of chromomycin (or derivatives) by HPLC analysis. No chromomycin A3 was detected in extracts of the different mutants, indicating the involvement of the glycosyltransferase genes in chromomycin biosynthesis. Surprisingly, in three of four mutants (C10GII, C10GIII, and C10GIV), no peaks were detected whose absorption spectra suggested a structural relationship to chromomycins. It has been described for the tylosin biosynthetic pathway in Streptomyces fradiae that glycosylated macrolides control tylactone biosynthesis and that no tylosin biosynthetic intermediates were detected when glycosylation steps were affected. However, the production of tylosin intermediates was restored after the addition of glycosylated tylosin precursors (5). Based on this finding, we tested the possibility that the addition of small amounts of chromomycin A3 to cultures of the different mutants could induce biosynthesis of chromomycin derivatives. Using such culture conditions, we were able to detect peaks with absorption spectra corresponding to chromomycin derivatives. This indicates that, as it happens in the tylosin pathway (5), the biosynthesis of chromomycin is also stimulated by glycosylated biosynthesis intermediates.
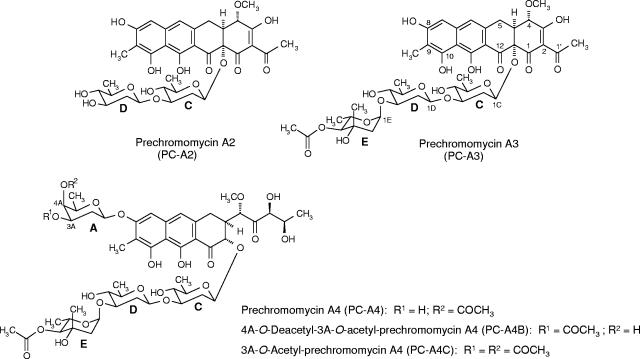
Analysis by HPLC of cultures of mutant C60GI (Fig. 3) showed four major peaks. Determination of the absorption spectra of the different peaks indicated that one of them would correspond to a tetracyclic intermediate, since it showed a UV absorption spectrum characteristic of tetracyclic premithramycinone type compounds, which are known as intermediates of the biosynthesis of mithramycin, another aureolic acid compound. This particular peak showed the same retention time and mass (m/z ion 415) as premithramycinone. Premithramycinone (PM) is a tetracyclic compound with no sugar attached (23). The other three compounds showed HPLC mobilities different from chromomycin A3 but the same UV absorption spectra, suggesting that they were all chromomycins with the typical tricyclic chromophore characteristic of aureolic acids. Structure elucidation of these compounds, later named prechromomycin A4 (PC-A4), 4A-O-deacetyl-3A-O-acetyl-prechromomycin A4 (PC-A4B), and 3A-O-acetyl-prechromomycin A4 (PC-A4C) (see below), showed that all of them lacked the 4-O-methyl-d-oliose residue (sugar B) of the disaccharide chain (Fig. 4). PC-A4B also differs from chromomycin A3 in the position of the acetyl residue at sugar A, being in this case attached at C-3 instead of at C-4. The third compound, PC-A4C (Fig. 4), possesses two acetyl groups at sugar A: one at its normal 4A position and the other at the 3A position. The absence of sugar B in all of these compounds indicates that the cmmGI product is a glycosyltransferase involved in the transfer of sugar B during chromomycin A3 biosynthesis.
FIG. 3.
HPLC analysis of cultures of wild-type (wt) S. griseus subsp. griseus and glycosyltransferase gene mutants. C-A2, chromomycin A2; C-A3, chromomycin A3; 4-DMPM, 4-demethyl-premithramycinone; PC-A2, prechromomycin A2; PC-A3, prechromomycin A3; PC-A4, prechromomycin A4; PC-A4B, 4A-O-deacetyl-3A-O-acetyl-prechromomycin A4; PC-A4C, 3A-O-acetyl-prechromomycin A4; PM, premithramycinone; PM-A1, premithramycin A1.
FIG. 4.
Chemical structures of the novel compounds accumulated by mutants C60GI and C10GII.
When the cmmGII gene was mutated (mutant C10GII), three major peaks and several minor peaks were detected by HPLC (Fig. 3). One of the major peaks corresponded to PM. A second major peak showed the same retention time and mass (545 g/mol) as a pure sample of premithramycin A1 (PM-A1) (8), a compound with the tetracyclic premithramycinone aglycone and a d-olivosyl residue attached at the 12a position but lacking all other sugar residues. The major compound produced by this mutant turned out to be a new compound and was named prechromomycin A3 (PC-A3) (Fig. 4). Structure elucidation of this compound showed that it corresponds to a tetracyclic compound lacking the disaccharide chain normally found at the 8 position but containing the complete chromomycin trisaccharide chain linked at the 12a position. Noticeably, the 4E-OH group of sugar E is acetylated, although the acetylation is normally carried out as the last biosynthetic step in chromomycin biosynthesis, i.e., after the completion of the glycosylation and methylation steps. Also the structure of one of the minor peaks could be determined as prechromomycin A2 (PC-A2), also a new compound. This contains the same 9-C-methylated aglycone as PC-A3 and a disaccharide chain linked to the 12a position, consisting of two d-olivoses. The fact that the complete trisaccharide chain is present in the major metabolite prechromomycin A3 allows the conclusion that CmmGII cannot be involved in the trisaccharide chain formation but must contribute to the formation of the disaccharide chain. Since the role of CmmGI in the disaccharide formation (transfer of sugar B) was deduced from the above-described experiment, CmmGII has to be responsible for the transfer of sugar A.
The two other glycosyltransferase genes (cmmGIII and cmmGIV) were also found to be involved in the biosynthesis of chromomycin A3. HPLC analysis of cultures of mutant C10GIII revealed two major (known) compounds (Fig. 3): premithramycin A1 (PM-A1) and PM. Consequently, CmmGIII is a glycosyltransferase responsible for the transfer of the second d-olivosyl residue (sugar D) of the trisaccharide chain, since premithramycin A1 only has the first deoxysugar of the trisaccharide chain attached. HPLC analysis of cultures of mutant C10GIV (Fig. 3) showed two peaks: the major peak corresponded to PM and the very minor peak had the same retention time and mass (401 g/mol) as the previously identified 4-demethyl-premithramycinone (22). This is also a tetracyclic compound with no sugar attached. This allows us to conclude that CmmGIV is the glycosyltransferase responsible for the attachment of the first sugar (sugar C) of the trisaccharide chain to premithramycinone.
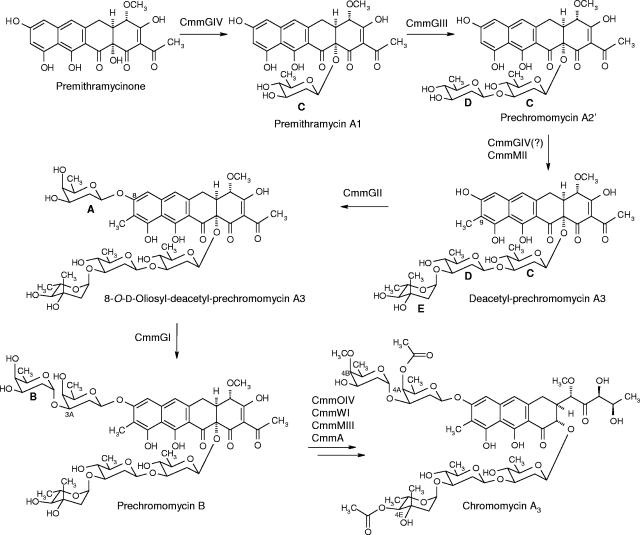
From all these experiments, it can be confirmed that the cmmGI, cmmGII, cmmGIII, and cmmGIV genes code for glycosyltransferases that are involved in chromomycin biosynthesis. CmmGIV and CmmGIII are involved in the formation of the trisaccharide chain, through the sequential transfer of the first and second d-olivosyl residues (sugars C and D, respectively) to the 12a and 3C positions, respectively, of the tetracyclic aglycone premithramycinone and the monosaccharidal premithramycin A1, respectively, while CmmGII and CmmGI are responsible for the formation of the disaccharide chain by incorporating, in a sequential manner, two d-oliosyl residues to the 8 and 3A positions, respectively, of trisaccharidal (deacteyl-prechromomycin A3) and tetrasaccharidal (dideacetyl-prechromomycin A4) biosynthetic intermediates of the chromomycin pathway, respectively (Fig. 5).
FIG. 5.
Glycosylation steps during the biosynthesis of chromomycin A3. The structures of premithramycinone and premithramycin A1 have been previously described (8, 23).
Isolation and structure elucidation of new compounds.
The three new compounds produced by mutant C60GI, as well as the two new compounds produced by C10GII, were purified by preparative HPLC from cultures grown on R5A solid medium. In the case of C10GII, the medium was supplemented with 2 μg/ml chromomycin A3 to stimulate metabolite production (see above). The yields for the different compounds were as follows (in mg/liter): 16.6 for PC-A4, 2.73 for PC-A4B, 7.5 for PC-A4C, 17 for PC-A3, and 6.2 for PC-A2.
The structures of the new tricyclic prechromomycins (PC-A4, PC-A4B, and PC-A4C) were identified by comparing their NMR data to those of chromomycin A3, and those of the tetracyclic prechromomycins were identified by comparison with the known tetracyclic intermediates of the mithramycin biosynthetic pathway (particularly premithramycinone and premithramycins A1, A2, and A3).
PC-A2.
The NMR data (see Tables SA1 and SA2 in the supplemental material) reveal an additional d-olivose moiety (see the coupling constants in the 1H NMR spectrum in Table SA2 in the supplemental material) compared to premithramycin A1 (8) and an additional C-methyl group attached to the 9 position of the aglycone (δ 2.11). The downfield-shifted 3C signal in the 13C NMR spectrum (δ 81.6 versus δ 71.2 in PM-A1) indicates that this second olivose is attached at the 3C position. Comparing the NMR data of PC-A2 with its diastereoisomer premithramycin A2 (PM-A2) (8) reveals that the only significant difference was found in the H,H-coupling pattern of the second sugar (particularly 3D-H, 4D-H, and 5D-H) due to the inverted stereochemistry at 4D position. The rest of the NMR data as well as the UV and IR data are virtually identical to those of PM-A2. The high-resolution FAB MS of PC-A2 confirms its molecular formula of C35H40O15.
PC-A3.
The NMR data for PC-A3 (see Tables SA1 and SA2 in the supplemental material) reveal a strong similarity to premithramycin A3 (both contain the same aglycone and a trisaccharide chain linked at the 12a position from which the first sugar moiety, a d-olivose, sugar C, is identical in both compounds). Only the data for the last two sugars of the trisaccharide chain (sugars D and E) differ. Comparison with the NMR data of chromomycin A3 reveals that PC-A3 contains the same trisaccharide chain as chromomycin A3. High-resolution FAB MS confirms the molecular formula of C44H54O19.
PC-A4, PC-A4B, PC-A4C.
The NMR data (see Table SA3 in the supplemental material) were compared with those for chromomycin A3, revealing that, in all three compounds, the sugar B signals were missing. The 4A-H (δ 4.98) signal in the 1H NMR spectrum of PC-A4 is upfield shifted, but not its 3A-H (δ 3.38) signal, like in chromomycin A3, due to the acetyl group linked to 4A-OH. In contrast, the 3A-H (δ 4.93) signal, but not the 4A-H (δ 3.88) signal, is upfield shifted in PC-A4B, and both 3A-H (δ 4.99) and 4A-H (δ 4.84) appear upfield shifted in PC-A4C.
The mass spectra of PC-A4 and PC-A4B show identical molecular ions, and that of PC-A4C is increased by 42 units due to the additional acetyl group. The mass spectra are consistent with the molecular formulae of C50H70O23 (for both PC-A4 and PC-A4B) and C52H72O24 (for PC-A4C).
Biological activity.
The biological activity of three of the new compounds (PC-A3, PC-A4, and PC-A4C) was tested and compared to the activity shown by the original compound chromomycin A3. Antibiotic activity tests against Micrococcus luteus showed that the new compounds possess a very low antibiotic activity (MIC, 40 μg/ml; MIC for chromomycin A3, 2.5 μg/ml). The compounds were also tested for antitumor activity against a panel of human tumor cell lines (Table 2). Compilation of the average 50% growth inhibition values showed that all three compounds were active, although the activity levels were lower than those of chromomycin A3. It is noticeable that PC-A3 retains antitumor activity in spite of being a tetracyclic compound and lacking the disaccharide chain. This was quite unexpected, since in the case of mithramycin, tetracyclic precursors did not show antitumor activity (18) (see also Discussion below).
TABLE 2.
Antitumor activity tests of chromomycin derivatives generated in this work in comparison with chromomycin A3
| Tumor type | Cell line | GI50a (μM) of:
|
|||
|---|---|---|---|---|---|
| Chromomycin A3 | Prechromomycin A4 | Prechromomycin A4C | Prechromomycin A3 | ||
| Prostate | DU-145 | 0.002 | 0.203 | 0.025 | 0.209 |
| LN-caP | 0.00025 | 0.011 | 0.0023 | 0.120 | |
| Ovarian | IGROV | 0.002 | 0.245 | 0.107 | 0.425 |
| Breast | SK-BR-3 | 0.0039 | 0.435 | 0.230 | 0.495 |
| Melanoma | SK-MEL-28 | 0.0019 | 0.239 | 0.075 | 0.238 |
| NSCL | A549 | 0.014 | 0.267 | 0.216 | 0.274 |
| Pancreas | PANC-1 | 0.002 | 0.261 | 0.109 | 0.224 |
| Colon | HT29 | 0.0047 | 0.540 | 0.226 | 0.339 |
| LOVO | 0.0035 | 0.194 | 0.096 | 0.201 | |
| Cervix | HELA | 0.0017 | 0.222 | 0.098 | 0.218 |
GI50, 50% growth inhibition.
DISCUSSION
Many natural products are glycosylated compounds in which the aglycone is decorated with saccharide moieties of variable chain lengths. Often these saccharide moieties participate in the molecular recognition of the cellular target, and therefore, these sugars are important for the biological activity of the compounds. Chromomycin A3 contains five deoxysugars, as a disaccharide and a trisaccharide chain, attached to the aglycone. The chromomycin gene cluster has been previously identified and characterized (15), and it has been shown to contain four putative glycosyltransferase genes. To confirm the role of each glycosyltransferase gene in the biosynthesis of chromomycin A3, they were independently inactivated by gene replacement, and most of the compounds produced by each mutant were isolated and characterized. This allowed us to assign each glycosyltransferase gene (and its product) to a specific glycosylation step and to propose the putative substrate on which the different glycosylation events take place (Fig. 5). Glycosyltransferases CmmGIV and CmmGIII are involved in trisaccharide formation, being responsible for the sequential transfer of sugars C and D, respectively, to the C-12a position of the tetracyclic polyketide-derived intermediate premithramycinone. Formation of the disaccharide chain is carried out by CmmGII and CmmGI: sugar A is transferred by CmmGII, most probably to deacetyl-prechromomycin A3, and sugar B is transferred by CmmGI, most likely to the product resulting from the previous reaction, 8-O-d-oliosyl-deacetyl-prechromomycin A3. In conclusion, CmmGI and CmmGII play roles in chromomycin biosynthesis similar to those of MtmGI and MtmGII for the closely related mithramycin biosynthesis. In both cases, these glycosyltransferases are involved in the formation of the disaccharide chain attached at the 6 position of the final molecules, and most recently, a tetracyclic intermediate with an sugar pattern analogous to that of PC-A4 (containing sugars C, D, E, and A) was identified for mithramycin biosynthesis (17). However, this was not oxidatively cleaved to a tricyclic compound by oxygenase MtmOIV. The fact that tricyclic compounds lacking the fifth deoxysugar are produced by mutant C60GI suggests that oxygenase CmmOIV, which most probably is involved in the fourth ring scission, has a greater degree of substrate flexibility than MtmOIV, its counterpart of the mithramycin pathway, since CmmOIV can apparently also recognize incompletely glycosylated biosynthetic intermediates as substrates.
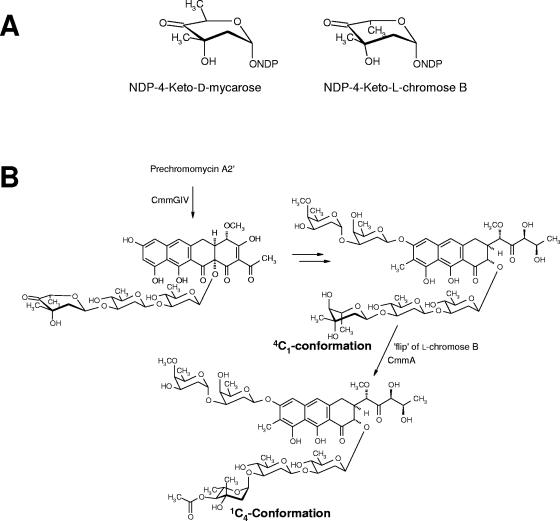
Although chromomycin A3 contains five deoxysugars, there are only four glycosyltransferase genes in the cluster. Transfer of the third sugar in the trisaccharide (l-chromose B residue, sugar E) could be carried out either by a not yet identified glycosyltransferase located outside of the cluster or, alternatively, by one of the four glycosyltransferases characterized in this work. However, CmmGI and CmmGII are excluded for this role, since mutations of the respective genes do not affect the incorporation of l-chromose B, leaving either CmmGIII or CmmGIV as candidates. Some other antibiotic gene clusters also have fewer glycosyltransferase genes than expected, for example, the mithramycin (4, 8), landomycin (29), and aclacinomycin (12) gene clusters. In the latter case, it was shown that the transfer of the second and the third deoxysugar of the trisaccharide chain was carried out by the same glycosyltransferase (12). Like chromomycin, mithramycin possesses a trisaccharide in the 2 position; however, this consists of different sugars, namely a d-olivose (sugar C), a d-oliose (sugar D), and a d-mycarose (sugar E). Evidence was found that sugar E of the mithramycin trisaccharide chain is transferred by the same glycosyltransferase that is responsible for the attachment of the first sugar, namely MtmGIV, and that both sugars are transferred as 4-ketosugars (20). Such a scenario could also be envisioned for chromomycin biosynthesis, since the proposed sugar donor substrates, NDP-4-keto-d-mycarose and NDP-4-keto-l-chromose B, respectively, only differ with respect to the configuration of C-5, when shown in a certain half-chair conformation (Fig. 6A). Thus, it is well possible for the chromomycin biosynthesis that l-chromose B is transferred by CmmGIV, which is closely related to MtmGIV. Alternatively, it is also possible that l-chromose B is transferred as a fully made sugar by CmmGIV (or CmmGIII).
FIG. 6.
(A) Chemical structures of NDP-4-ketosugars. (B) Proposed events for the incorporation of l-chromose B during the biosynthesis of chromomycin A3.
The last steps in chromomycin A3 biosynthesis involve an O-methylation and two O-acetylations of three of the sugar moieties (14). Both acetylation events are catalyzed by CmmA, an acetyltransferase that acts as the last enzyme of the chromomycin pathway incorporating acetyl residues first to the 4-hydroxygroup of sugar E and then to the 4-OH of sugar A (14). These decoration reactions convert a compound with relatively low biological activity into a very potent one. The sugars of chromomycin, particularly sugar E in its 1C4 conformation is essential for the DNA-binding properties of chromomycin A3 (6, 13, 14, 15, 25, 26). In context with the above discussed glycosylation events, it is possible that sugar E is initially attached as ketosugar, then reduced to an l-sugar in an unusual 4C1 conformation, and eventually flipped into the final 1C4 conformation that is also further stabilized by the bulky 4E-acetyl residue (Fig. 6B). All compounds produced by mutants C60GI and C10GII that contain the third sugar of the trisaccharide chain are acetylated in the 4E position, and all compounds produced by C60GI are also acetylated in the 3A and/or 4A position. The 4E-acetylation in PC-A3 (with a tetracyclic aglycone) is most likely the reason why this compound shows biological activity in contrast to the tetracyclic intermediates of the mithramycin pathway, where even the fully glycosylated premithramycin B (the last intermediate of the mithramycin pathway) shows no biological activity and no DNA interaction whatsoever (18, 21). PC-A4B has one of its acetyl groups located at 3A instead of at the normal 4A position. This is possibly a result from a chemical rearrangement, since O-acetyl substituents in sugars have the tendency to migrate, especially between two neighboring OH groups of the same sugar moiety (reference 9 and references therein). Once migrated from the 4A to the 3A position, acetyltransferase CmmA could reacetylate the now free 4A-OH group, resulting in PC-A4C, in which acetyl groups were found in both the 3A and 4A positions. However, the CmmA acetyltransferase also shows a certain degree of substrate flexibility, given the fact that it can utilize compounds with either a tricyclic or a tetracyclic aglycone and that it also can acetylate biosynthetic intermediates with incomplete sugar patterns. Thus, alternatively to the chemical rearrangement mentioned above, it also could be argued that CmmA is able to acetylate two different positions in sugar A.
We have previously shown that inactivation of selected genes of the chromomycin biosynthesis pathway leads to the generation of new compounds with antitumor activity (14, 15). With this work, we have shown that inactivation of two of the four glycosyltransferase genes (cmmGI and cmmGII) leads to the generation of four new chromomycin derivatives possessing antitumor activity. Interestingly, the most active compound among the three tested was PC-A4C, which contains two O-acetyl residues in sugar A. This suggests that the presence of an extra acetyl group in chromomycin derivatives increases the antitumor activity. It has been reported that acetyl groups add more specificity to the DNA binding by providing an additional H bond acceptor interacting with the 2-amino groups of G bases (6, 13, 25, 26). In this context, we have previously shown that the presence of both acetyl groups in sugars A and E of chromomycin is absolutely required for a high biological activity (14). The generation of the new chromomycins described here opens up the possibility to further examine structure-activity relationships of chromomycins regarding glycosylation and sugar acetylation patterns. Also, the characterization of the chromomycin glycosyltransferases as well as the construct of mutants in these glycosyltransferase genes will be of great interest for future attempts to generate chromomycin/mithramycin derivatives via genetic engineering combining chromomycin/mithramycin glycosyltransferase genes.
Supplementary Material
Acknowledgments
This work was supported by grants of the Spanish Ministry of Education and Science to C.M. (BMC2002-03599), the U.S. National Institutes of Health (CA 91901) to J.R., and the Plan Regional de Investigación del Principado de Asturias to J.A.S. (GE-MED01-05). N.M. was the recipient of a predoctoral fellowship of the Plan Regional de Investigación del Principado de Asturias.
We thank Pharmamar S.A. for helping us with the antitumor assays. We also acknowledge the University of Kentucky's core NMR facility and thank J. Goodman from the University of Kentucky mass spectrometry center for the high-resolution mass spectra.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bianchi, N., C. Rutigliano, M. Passadore, M. Tomassetti, L. Pippo, C. Mischiati, G. Feriotto, and R. Gambari. 1997. Targeting of the HIV-1 long terminal repeat with chromomycin potentiates the inhibitory effects of a triplex-forming oligonucleotide on Sp1-DNA interactions and in vitro transcription. Biochem. J. 326:919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianchi, N., F. Osti, C. Rutigliano, F. G. Corradini, E. Borsetti, M. Tomassetti,C. Mischiati, G. Feriotto, and R. Gambari. 1999. The DNA-binding drugs mithramycin and chromomycin are powerful inducers of erythroid differentiation of human K562 cells. Br. J. Haematol. 104:258-265. [DOI] [PubMed] [Google Scholar]
- 4.Blanco, G., E. Fernández, M. J. Fernández, A. F. Braña, U. Weiβbach, E. Künzel, J. Rohr, C. Méndez, and J. A. Salas. 2000. Characterization of two glycosyltransferases involved in early glycosylation steps during biosynthesis of the antitumor polyketide mithramycin by Streptomyces argillaceus. Mol. Gen. Genet. 262:991-1000. [DOI] [PubMed] [Google Scholar]
- 5.Butler, A. R., S. A. Flint, and E. Cundliffe. 2001. Feedback control of polyketide metabolism during tylosin production. Microbiology 147:795-801. [DOI] [PubMed] [Google Scholar]
- 6.Chakrabarti, S., D. Bhattacharyya, and D. Dasgupta. 2001. Structural basis of DNA recognition by anticancer antibiotics, chromomycin A(3), and mithramycin: roles of minor groove width and ligand flexibility. Biopolymers 56:85-95. [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee, S., K. Zaman, H. Ryu, A. Conforto, and R. R. Ratan. 2001. Sequence-selective DNA binding drugs mithramycin A and chromomycin A3 are potent inhibitors of neuronal apoptosis induced by oxidative stress and DNA damage in cortical neurons. Ann. Neurol. 49:345-354. [PubMed] [Google Scholar]
- 8.Fernández, E., U. Weissbach, C. S. Reillo, A. F. Braña, C. Méndez, J. Rohr, and J. A. Salas. 1998. Identification of two genes from Streptomyces argillaceus encoding glycosyltransferases involved in transfer of a disaccharide during biosynthesis of the antitumor drug mithramycin. J. Bacteriol. 180:4929-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kabel, M. A., P. de Waard, H. A. Schols, and A. G. J. Voragen. 2003. Location of O-acetyl substituents in xylo-oligosaccharides obtained from hydrothermally treated Eucalyptus wood. Carbohydr. Res. 338:69-77. [DOI] [PubMed] [Google Scholar]
- 10.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 11.Lozano, M. J., L. L. Remsing, L. M. Quirós, A. F. Braña, E. Fernández, C. Sánchez, C. Méndez, J. Rohr, and J. A. Salas. 2000. Characterization of two polyketide methyltransferases involved in the biosynthesis of the antitumor drug mithramycin by Streptomyces argillaceus. J. Biol. Chem. 275:3065-3074. [DOI] [PubMed] [Google Scholar]
- 12.Lu, W., C. Leimkuhler, M. Oberthür, D. Kahne, and C. T. Walsh. 2004. AknK is an L-2-deoxyfucosyltransferase in the biosynthesis of the anthracycline aclacinomycin A. Biochemistry 43:4548-4558. [DOI] [PubMed] [Google Scholar]
- 13.Majee, S., R. Sen, S. Guha, D. Bhattacharyya, and D. Dasgupta. 1997. Differential interactions of the Mg2+ complexes of chromomycin A3 and mithramycin with poly(dG-dC) x poly(dC-dG) and poly(dG) x poly(dC). Biochemistry 36:2291-2299. [DOI] [PubMed] [Google Scholar]
- 14.Menéndez, N., M. Nur-e-Alam, A. F. Braña, J. Rohr, J. A. Salas, and C. Méndez. 2004. Tailoring modification of deoxysugars during biosynthesis of the antitumor drug chromomycin A3 by Streptomyces griseus subsp. griseus. Mol. Microbiol. 53:903-915. [DOI] [PubMed] [Google Scholar]
- 15.Menéndez, N., M. Nur-e-Alam, A. F. Braña, J. Rohr, J. A. Salas, and C. Méndez. 2004. Biosynthesis of the antitumor chromomycin A3 in Streptomyces griseus: analysis of the gene cluster and rational design of novel chromomycin analogues. Chem. Biol. 11:21-32. [DOI] [PubMed] [Google Scholar]
- 16.Mir, M. A., S. Majee, S. Das, and D. Dasgupta. 2003. Association of chromatin with anticancer antibiotics, mithramycin and chromomycin A3. Bioorg. Med. Chem. 11:2791-2801. [DOI] [PubMed] [Google Scholar]
- 17.Nur-e-Alam, M., C. Méndez, J. A. Salas, and J. Rohr. 2005. Elucidation of the glycosylation sequence of mithramycin biosynthesis: isolation of 3A-deolivosylpremithramycin B and its conversion to premithramycin B by glycosyltransferase MtmGII. Chembiochem 6:632-636. [DOI] [PubMed] [Google Scholar]
- 18.Prado, L., E. Fernandez, U. Weissbach, G. Blanco, L. M. Quiros, A. F. Braña, C. Mendez, J. Rohr, and J. A. Salas. 1999. Oxidative cleavage of premithramycin B is one of the last steps in the biosynthesis of the antitumor drug mithramycin. Chem. Biol. 6:19-30. [DOI] [PubMed] [Google Scholar]
- 19.Quirós, L. M., I. Aguirrezabalaga, C. Olano, C. Méndez, and J. A. Salas. 1998. Two glycosyltransferases and a glycosidase are involved in oleandomycin modification during its biosynthesis by Streptomyces antibioticus. Mol. Microbiol. 28:1177-1186. [DOI] [PubMed] [Google Scholar]
- 20.Remsing, L. L., J. Garcia-Bernardo, A. M. Gonzalez, E. Künzel, U. Rix, A. F. Braña, D. W. Bearden, C. Méndez, J. A. Salas, and J. Rohr. 2002. Ketopremithramycins and ketomithramycins, four new aureolic acid-type compounds obtained upon inactivation of two genes involved in the biosynthesis of the deoxysugar moieties of the antitumor drug mithramycin by Streptomyces argillaceus, reveal novel insights into post-PKS tailoring steps of the mithramycin biosynthetic pathway. J. Am. Chem. Soc. 124:1606-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Remsing, L. L., H. R. Bahadori, G. M. Carbone, E. M. McGuffie, C. V. Catapano, and J. Rohr. 2003. Inhibition of c-src transcription by mithramycin: structure-activity relationships of biosynthetically-produced mithramycin analogues using the c-src promoter region as target. Biochemistry 42:8313-8324. [DOI] [PubMed] [Google Scholar]
- 22.Rohr, J., C. Méndez, and J. A. Salas. 1999. The biosynthesis of aureolic acid group antibiotics. Bioorg. Chem. 27:41-54. [Google Scholar]
- 23.Rohr, J., U. Weissbach, C. Beninga, E. Künzel, K. Siems, K. U. Bindseil, F. Lombó, L. Prado, A. F. Braña, C. Méndez, and J. A. Salas. 1998. The structure of premithramycinone and demethylpremithramycinone, plausible early intermediates of the aureolic acid group antibiotic mithramycin. Chem. Commun. 1998:437-438. [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Silva, D. J., and D. E. Kahne. 1993. Studies of the 2:1 chromomycin A3-Mg2+ complex in methanol: role of the carbohydrates in complex formation. J. Am. Chem. Soc. 115:7962-7970. [Google Scholar]
- 26.Silva, D. J., R. Goodnow Jr., and D. Kahne. 1993. The sugars in chromomycin A3 stabilize the Mg+2 dimer complex. Biochemistry 32:463-471. [DOI] [PubMed] [Google Scholar]
- 27.Skehan, P., R. Storeng, D. Scudiero, A. Monks, J. McMahon, D. Vistica, J. T. Warren, H. Bokesch, S. Kenney, and M. R. Boyd. 1990. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 82:1107-1112. [DOI] [PubMed] [Google Scholar]
- 28.Tagashira, M., T. Kitagawa, S. Isonishi, A. Okamoto, K. Ochiai, and Y. Ohtake. 2000. Mithramycin represses MDR1 gene expression in vitro, modulating multidrug resistance. Biol. Pharm. Bull. 23:926-929. [DOI] [PubMed] [Google Scholar]
- 29.Trefzer, A., C. Fischer, S. Stockert, L. Westrich, E. Künzel, U. Girreser, J. Rohr, and A. Bechthold. 2001. Elucidation of the function of two glycosyltransferase genes (lanGT1 and lanGT4) involved in landomycin biosynthesis and generation of new oligosaccharide antibiotics. Chem. Biol. 8:1239-1252. [DOI] [PubMed] [Google Scholar]
- 30.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100:189-194. [DOI] [PubMed] [Google Scholar]
- 31.Vilches, C., C. Méndez, C. Hardisson, and J. A. Salas. 1990. Biosynthesis of oleandomycin by Streptomyces antibioticus: influence of nutritional conditions and development of resistance. J. Gen. Microbiol. 136:1447-1454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.