Abstract
Various xenobiotic-degrading genes on many catabolic plasmids are often flanked by two copies of an insertion sequence, IS1071. This 3.2-kb IS element has long (110-bp) terminal inverted repeats (IRs) and a transposase gene that are phylogenetically related to those of the class II transposons. However, the transposition mechanism of IS1071 has remained unclear. Our study revealed that IS1071 was only able to transpose at high frequencies in two environmental β-proteobacterial strains, Comamonas testosteroni and Delftia acidovorans, and not in any of the bacteria examined which belong to the α- and γ-proteobacteria. IS1071 was found to have the functional features of the class II transposons in that (i) the final product of the IS1071 transposition was a cointegrate of its donor and target DNA molecules connected by two directly repeated copies of IS1071, one at each junction; (ii) a 5-bp duplication of the target sequence was observed at the insertion site; and (iii) a tnpA mutation of IS1071 was efficiently complemented by supplying the wild-type tnpA gene in trans. Deletion analysis of the IS1071 IR sequences indicated that nearly the entire region of the IRs was required for its transposition, suggesting that the interaction between the transposase and IRs of IS1071 might be different from that of the other well-characterized class II transposons.
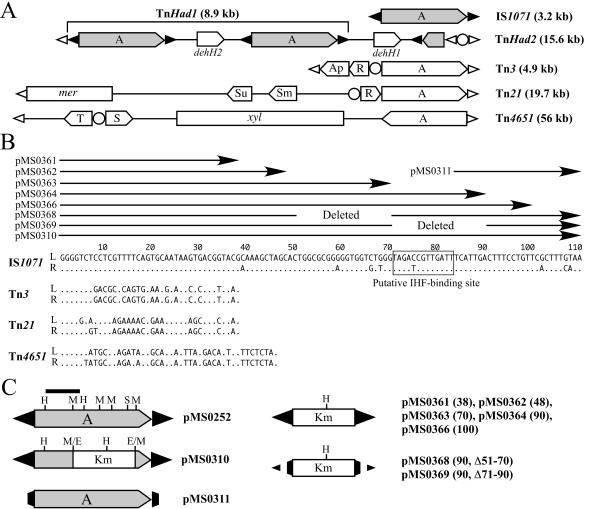
Bacterial class II (Tn3-like) transposons generally carry the genes for their transposition (tnpA, tnpR, and res) and one or more phenotypic traits between their terminal inverted repeats (IRs), which have sizes of less than 50 bp (Fig. 1A and B) (23). These transposons move by a two-step and replicative mechanism (6, 23). In the first step, the tnpA product (transposase) acts at the IRs to generate a cointegrate of the donor and target molecules connected by two directly repeated copies of the transposon, one at each junction. In the second step, the cointegrate resolves at the resolution (res) sites by means of the tnpR product (resolvase). A 5-bp duplication of the target sequence is generated upon transposition. The transposases of the class II transposons are able to catalyze their transposition even when the tnpA gene and cognate IRs are located on separate molecules (6, 23). Several class II transposons have been reported to play an important role in the wide dissemination of various catabolic gene clusters, such as toluene-xylenes, naphthalene, and carbazole (17, 29-31). To date, three major groups (Tn3, Tn21, and Tn4651) of class II transposons have been characterized in detail with respect to their structural and functional aspects (Fig. 1A).
FIG. 1.
Structures of class II transposons, their IR sequences, and IS1071 derivatives. (A) Schematic structures of the class II transposons. The sizes are arbitrary. The black and white arrowheads indicate the terminal IR sequences of IS1071 and those of the representative transposons, respectively, and the circle represents the res site. The pentagon shows the orientation of the gene. Abbreviations: A, tnpA gene; R, S, and T, genes for cointegrate resolution; Ap, Sm, Su, and mer, genes for resistance to ampicillin, streptomycin, sulfonamide, and mercury, respectively; xyl, genes for degradation of toluene and xylenes. (B) Comparison of IR sequences. The left (L) and right (R) ends of IS1071 are defined as those located upstream and downstream, respectively, of the tnpA gene, and those of other transposons are defined as the distal and proximal ends, respectively, to the tnpA gene. A dot indicates a nucleotide identical to that in the left IR of IS1071. The arrows indicate the IR sequences that are carried in the IS1071 derivatives on the plasmids depicted. The boxed sequence represents the putative IHF-binding site. (C) Structures of the IS1071 derivatives on the pMS plasmids. The black bar indicates the DNA fragment used as a probe for the Southern analysis (Fig. 3). E, H, M, and S represent EcoRI, HindIII, MunI, and SalI sites, respectively. The values in parentheses are the lengths of the mutant IRs in the pMS plasmids.
IS1071 is a 3.2-kb insertion sequence (IS) that was originally identified in a chlorobenzoate-catabolic transposon, Tn5271, from Comamonas testosteroni BR60 (20). On the basis of structural features of its 110-bp IRs and 2,913-bp tnpA gene, IS1071 has been considered to belong to the class II transposons (7, 20). However, this IS element shows the uniqueness in its long (110-bp) IRs and its lack of the resolution function. The identification of many IS1071 sequences in close proximity to various xenobiotic-degrading genes on self-transmissible plasmids from environmental bacteria, e.g., Pseudomonas (18), Comamonas (2, 13), and Wautersia (3), indicates that IS1071 must have been involved in the recruitment of catabolic genes to these plasmids and in the dissemination of these genes among various host strains. We have also identified a haloacetate-catabolic IS1071-composite transposon, TnHad1, on an IncP-1β plasmid, pUO1, from Delftia acidovorans strain B (Fig. 1A) (24, 25). TnHad1 is located within a defective class II transposon, TnHad2, which is a Tn21-related transposon that lacks the tnpA and tnpR genes (Fig. 1A) (24). We have previously reported that the two intact copies and one truncated copy of IS1071 in TnHad2 might have been incorporated into an ancestor of TnHad2 (24). However, no clear transposition events of the TnHad1-specified IS1071 element were observed.
No functional analysis of IS1071 has been carried out since its discovery more than a decade ago. Our functional analysis of IS1071 in this study has indicated that (i) efficient transposition of IS1071 occurred in two specific host strains, (ii) IS1071 had the functional features of the class II transposons, and (iii) almost the entire region of the 110-bp IR was required for transposition.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains and plasmids used are listed in Table 1. Luria broth (LB) and LB agar (1) were used throughout this study. Escherichia coli cells were cultivated at 37°C and the others at 30°C. The agents added to the media were as follows: ampicillin, 100 μg/ml; chloramphenicol, 50 μg/ml; kanamycin, 50 μg/ml; nalidixic acid, 30 μg/ml; tetracycline, 10 μg/ml; sulfathiazole, 350 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| Agrobacterium tumefaciens C58 | Free of IS1071 | 35 |
| Comamonas testosteroni JCM5832 | Free of IS1071; type strain | JCMa |
| Delftia acidovorans B | Harboring pUO1 | 14 |
| D. acidovorans B123 | pUO1-cured derivative of strain B, more than one copy of IS1071 in the chromosome | 14 |
| Escherichia coli DH5α | recA lacZΔM15; free of IS1071 | 1 |
| E. coli HB101 | recA; free of IS1071 | 1 |
| E. coli JM109 | NalrrecA hsdR17; free of IS1071 | 1 |
| Pseudomonas alcaligenes JCM5967 | Free of IS1071; type strain | JCMa |
| P. putida PpN1 | Free of IS1071 | 4 |
| Plasmids | ||
| pUO1 | Hgr, haloacetate degradation, TnHad1, TnHad2 (Fig. 1A) | 14 |
| pBBR1MCS | Cmr, broad-host-range cloning vector | 16 |
| pBBR1MCS-3 | Tcr, broad-host-range cloning vector | 15 |
| pCB182 | Apr, promoter-probe vector, lacZ gene without its promoter sequence (Fig. 2B) | 22 |
| pME6012 | Tcr, broad-host-range vector, pVS1 and p15A replicons | 8 |
| pNIT6012 | Tcr, pME6012 derivative carrying the oriT sequence of RP4 at BamHI site | Laboratory collection |
| pMS0252 | Tcr, pBBR1MCS-3 derivative carrying IS1071 (Fig. 1C) | This study |
| pMS0310 | Tcr Kmr, pNIT6012 derivative carrying a Kmr-IS1071 derivative with 110-bp IRs (Fig. 1B and C) | This study |
| pMS0311 | Cmr, pBBR1MCS derivative carrying an IS1071 derivative that lacked the outermost 83-bp sequence in both IRs (Fig. 1B and C) | This study |
| pMS0321 | Apr, pCB182 derivative carrying the 145-bp sequence upstream of the tnpA gene in IS1071 between BglII and SalI sites (Fig. 2A and B) | This study |
| pMS0324 | Cmr, pBBR1MCS derivative carrying the BamHI-PstI fragment of pMS0321 (Fig. 2B) | This study |
| pMS0326 | Cmr, pBBR1MCS derivative carrying the BglII-PstI fragment of pCB182 (Fig. 2B) | This study |
| pMS0361 | Tcr Kmr, pNIT6012 derivative carrying a Kmr-IS1071 derivative with the outermost 38-bp sequences of the IRs (Fig. 1B and C) | This study |
| pMS0362 | Tcr Kmr, pNIT6012 derivative carrying a Kmr-IS1071 derivative with the outermost 48-bp sequences of the IRs (Fig. 1B and C) | This study |
| pMS0363 | Tcr Kmr, pNIT6012 derivative carrying a Kmr-IS1071 derivative with the outermost 70-bp sequences of the IRs (Fig. 1B and C) | This study |
| pMS0364 | Tcr Kmr, pNIT6012 derivative carrying a Kmr-IS1071 derivative with the outermost 90-bp sequences of the IRs (Fig. 1B and C) | This study |
| pMS0366 | Tcr Kmr, pNIT6012 derivative carrying a Kmr-IS1071 derivative with the outermost 100-bp sequences of the IRs (Fig. 1B and C) | This study |
| pMS0368 | Tcr Kmr, pNIT6012 derivative carrying a Kmr-IS1071 derivative whose IRs have a 20-bp deletion at positions 51 to 70 (Fig. 1B and C) | This study |
| pMS0369 | Tcr Kmr, pNIT6012 derivative carrying a Kmr-IS1071 derivative whose IRs have a 20-bp deletion at positions 71 to 90 (Fig. 1B and C) | This study |
| pUC4K | Apr Kmr, cloning vector, Kmr gene is flanked by the EcoRI, BamHI, SalI, and PstI sites | 26 |
| R388 | Tpr Sur, conjugal plasmid free of transposons, IncW replicon | 32 |
JCM, Japan Collection of Microorganisms.
DNA methodology.
Standard methods were used for extracting plasmid DNA, DNA digestion with restriction endonucleases, ligation, gel electrophoresis, and transformation of bacterial cells (1). The PCR was carried out with ExTaq DNA polymerase (TAKARA BIO). Purification of PCR-amplified DNA fragments was done with a GFX PCR DNA and Gel Band Purification Kit (Amersham Biosciences) according to the manufacturer's protocol. Nucleotide sequencing was performed with an ABI PRISM model 310 sequencer (Applied Biosystems). Southern hybridization (1) was done with an ECL Random-Prime Labeling and Detection System (Amersham Biosciences) according to the manufacturer's protocol.
PCR amplification and construction of plasmids.
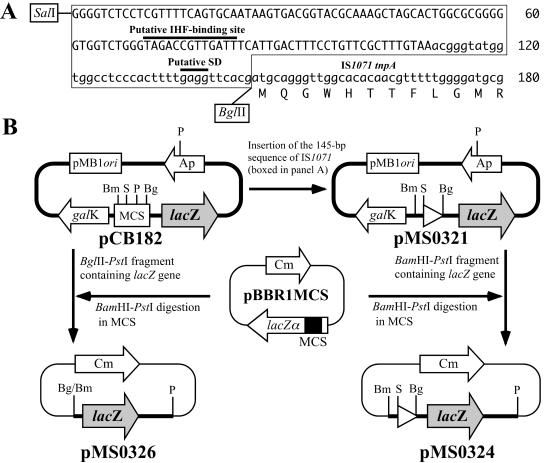
Primer 1071Kpn (5′-ACGTGGTACCGGGGTCTCCTCGTTTTCAGT-3′) contained a KpnI site (underlined) and the outermost 20-base sequence of the 110-bp IR of IS1071 (bold letters). This primer was used for PCR amplification of the entire IS1071 sequence with pUO1 as the template. The product was cloned into the KpnI site of pBBR1MCS-3 (15) to generate pMS0252 (Fig. 1C). The MunI fragment in the IS1071 element on pMS0252 was replaced by the 1.3-kb pUC4K-derived EcoRI fragment carrying a Kmr determinant (26), and the resulting Kmr-IS1071 derivative was inserted into the KpnI site on broad-host-range plasmid pNIT6012 (Table 1) to construct pMS0310 (Fig. 1C). Seven pNIT6012-based plasmids, pMS0361, pMS0362, pMS0363, pMS0364, pMS0366, pMS0368, and pMS0369, carried the Kmr-IS1071 derivatives with mutant IRs (38-, 48-, 70-, 90-, 100-, 90-, and 90-bp IRs, respectively), and the IRs in the last two plasmids lacked internal 20-bp sequences at different positions (Fig. 1B and C). These plasmids were constructed by PCR with appropriate primers. Primer 1071-84 (5′-GGCCGCTAGCTCATTGACTTTCCTGTTC-3′) had an NheI site (underlined) and the 18-bp sequence (bold letters) that annealed to the internal nucleotides (positions 84 to 101, Fig. 1B) of the 110-bp IRs of IS1071. This primer was used to amplify an IS1071 derivative lacking the outermost 83-bp sequences at both ends. Cloning of the amplified product into the XbaI site of pBBR1MCS (16) generated pMS0311 (Fig. 1C). Primers 1071pro1 (5′-TTTTGTCGACGGGGTCTCCTCGTTTTCAGT-3′) and 1071pro3 (5′-TTTTAGATCTCGTGAACCTCAAAAGTGGGA-3′) were used to amplify the 145-bp sequence upstream of the putative tnpA gene of IS1071 (Fig. 2A). The product flanked by SalI (GTCGAC, underlined in 1071pro1) and BglII (AGATCT, underlined in 1071pro3) sites was inserted between the corresponding sites in promoter-probe vector pCB182 (22) to construct pMS0321 (Fig. 2B). The BamHI-PstI fragment of pMS0321 containing the 145-bp fragment and a promoterless lacZ gene was cloned to the corresponding sites in pBBR1MCS to construct pMS0324 (Fig. 2B). The BglII-PstI fragment of pCB182 that carried the promoterless lacZ gene was cloned between the BamHI and PstI sites in pBBR1MCS to generate pMS0326 (Fig. 2B). Primers 1071-287 (5′-GGTCTGGCGCTCCATATTGGTTTCCTGCGC-3′) and 1071-1358 (5′-GCAGCTTGGCAAGGTACTCGATGGCAGGAT-3′) were used to amplify the 1.1-kb portion of IS1071 (Fig. 1C). The product was used as a probe for Southern hybridization.
FIG. 2.
Construction of plasmids used for tnpA promoter analysis. (A) Nucleotide sequence upstream of the tnpA gene of IS1071. Capital letters indicate the 110-bp IR sequence of IS1071. The boxed region (145 bp) was amplified by PCR with primers 1071pro1 and 1071pro3 and used to construct pMS0321. (B) Construction of plasmids. Abbreviations for restriction endonucleases are as follows: B, BamHI; Bg, BglII; P, PstI; S, SalI. Plasmid pCB182 is a promoter-probe vector and carries a multiple-cloning site (MCS) between the galK and lacZ genes, each of which lacks its promoter sequence (22). The BglII-PstI fragment of pCB182 that carried the promoterless lacZ gene was cloned between the BamHI and PstI sites in pBBR1MCS (16) to generate pMS0326. The 145-bp sequence of IS1071 (white arrowhead) flanked by SalI and BglII sites (see panel A) was inserted between the corresponding sites in pCB182 to generate pMS0321. Subsequent cloning of the BamHI-PstI fragment of pMS0321 containing the lacZ gene between the corresponding sites in pBBR1MCS led to the construction of pMS0324.
Transposition assays.
Transposition of the IS1071 derivatives was assayed by the “mating-out” experiments described previously (24). For this purpose, we introduced pMS0252 and R388 (32) into the bacterial strains listed in Table 2. The resulting strains were employed as donors to mate with E. coli JM109 on a membrane filter, and Tcr Nalr transconjugants were selected. The transposition frequency was expressed as the number of Tcr Nalr transconjugants per number of Sur Nalr transconjugants. The Tcr Nalr transconjugants were analyzed for their plasmid profiles. Complementation of the IS1071 tnpA mutation and transposition of the Kmr-IS1071 derivatives with mutant ends were also examined by the mating-out experiment. The donor strain was C. testosteroni JCM5832 harboring the following three plasmids: (i) R388; (ii) pMS0311, a pBBR1MCS-based plasmid carrying the tnpA gene of IS1071; and (iii) pMS0310 or one of the other pNIT6012-based plasmids carrying a Kmr-IS1071 derivative with mutant ends (Fig. 1B and C). Such a JCM5832 derivative was mated with JM109, and Kmr Nalr transconjugants were selected and analyzed for their plasmid profiles.
TABLE 2.
Transposition of IS1071
| Host straina | Proteobacterial division | Transposition frequencyb
|
|
|---|---|---|---|
| pBBR1MCS-3 | pMS0252 | ||
| A. tumefaciens C58 | α | <1.2 × 10−7 | <8.7 × 10−8 |
| C. testosteroni JCM5832 | β | <3.3 × 10−7 | 1.5 × 10−3 |
| D. acidovorans B123c | β | <1.4 × 10−5 | 4.0 × 10−4 |
| E. coli HB101 | γ | <9.8 × 10−9 | <1.6 × 10−8 |
| P. alcaligenes JCM5967 | γ | <9.7 × 10−8 | <2.1 × 10−8 |
| P. putida PpN1 | γ | <1.9 × 10−8 | <1.4 × 10−8 |
D. acidovorans B123 carried more than one copy of IS1071 in its chromosome, and the other strains were free of IS1071 (data not shown).
Transposition of IS1071 was detected by the cointegration between pMS0252 and R388 (32). Plasmid pMS0252 is a pBBR1MCS-3 derivative carrying IS1071 (Fig. 1C). The transposition frequency is expressed as the number of Tcr Nalr transconjugants per number of Sur Nalr transconjugants. A value with the symbol< means that no Tcr Nalr transconjugants were obtained in the experiment. All values are averages from at least three independent experiments.
The transfer frequency of R388 was low (2 × 10−4 per donor cell) for an unknown reason(s) when D. acidovorans B123 was used as the donor host.
β-Gal assays.
The bacterial strains harboring pMS0324 or pMS0326 (Fig. 2B) were grown to the early stationary phase in LB containing chloramphenicol and used for β-galactosidase (β-Gal) assays according to the method of Miller (19).
Nucleotide sequence accession number.
Our partial sequencing and the previously deposited sequences of IncW plasmid R388 revealed its complete sequence (33,913 bp), and the sequence has been deposited in the DDBJ/EMBL/GenBank databases under accession number BR000038.
RESULTS
Transposition of IS1071.
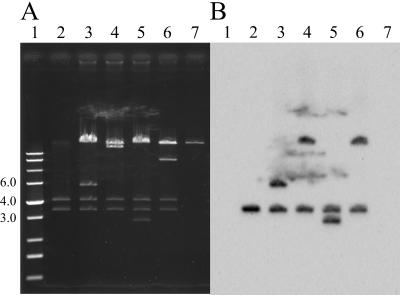
Although we previously detected a very low frequency of transposition (1.3 × 10−7) of the pUO1-derived IS1071 element in E. coli (24), our subsequent and repeated attempts to detect such a transposition event in any E. coli strains were unsuccessful. Therefore, in this study we investigated IS1071 transposition in several environmental bacterial strains listed in Table 2. These strains, harboring pMS0252 (=pBBR1MCS-3::IS1071) and R388, were employed as the donor host strains in the mating-out experiments. Tcr Nalr transconjugants were obtained at high frequencies when C. testosteroni JCM5832 and D. acidovorans B123 were employed as the donor host strains (Table 2) but not with any of the other donor strains. Restriction and Southern analyses of the plasmids from several Tcr Nalr transconjugants revealed that these plasmids were cointegrates of pMS0252 and R388 connected by two directly repeated copies of IS1071, one at each junction, since each plasmid had (i) two (4.2- and 3.5-kb) fragments identical to those from pMS0252 (Fig. 3A) and (ii) two copies of IS1071 (Fig. 3B). It was theoretically possible that these cointegrates were generated by nonreplicative transposition of an IS1071-composite transposon from dimers of pMS0252. This possibility was unlikely since agarose gel analysis did not reveal the preferential presence of the pMS0252 dimers in the two host strains (data not shown). Sequence analysis of the insertion sites indicated that IS1071 transposed to various sites in R388 with concomitant generation of a 5-bp duplication of the target sequence. These results demonstrated that (i) IS1071 is highly mobile, but only in specific bacterial strains; (ii) IS1071 is duplicated upon transposition; and (iii) IS1071 generates a 5-bp duplication of the target sequence. Subsequent transposition experiments in this study were carried out with C. testosteroni JCM5832 since this strain was free of IS1071.
FIG. 3.
Analysis of IS1071-mediated cointegrates by agarose gel electrophoresis (A) and Southern hybridization patterns with an internal 1.1-kb fragment of IS1071 (Fig. 1C) as a probe (B). All samples were digested with SalI, which cleaved IS1071 at one site (Fig. 1C). Lane 1, marker DNA; lane 2, pMS0252; lanes 3 to 6, cointegrates of pMS0252 and R388 obtained in the independent experiments; lane 7, R388. The sizes (in kilobases) of three fragments in the marker DNA are shown on the left.
Complementation of cointegration function.
The tnpA mutations of the class II transposons are usually complemented efficiently by supplying their cognate wild-type tnpA genes in trans (23). To investigate whether this was the case with IS1071, we conducted a mating-out experiment with E. coli JM109 as a recipient strain and a C. testosteroni JCM5832 derivative harboring pMS0311 (the supplier of tnpA), pMS0310 (the Kmr-IS1071 carrier) (Fig. 1B and C), and R388 as a donor strain. Kmr Nalr transconjugants were obtained at a frequency of 3.7 × 10−3 (Table 3, experiment 1). All of the 100 transconjugants examined showed resistance to tetracycline, which was encoded by the vector portion (pNIT6012) of pMS0310. This suggested that all the transconjugants carried the fusion plasmids that have portions of pMS0310 and R388. Detailed analysis of the plasmids from several transconjugants with restriction endonucleases further revealed that they were cointegrates of the two plasmids connected by two directly repeated copies of the Kmr-IS1071 derivative, one at each junction (data not shown). The absence of the R388::Kmr-IS1071 plasmids in the 100 transconjugants indicated that the C. testosteroni JCM5832-encoded DNA recombination systems (e.g., RecA system) did not function efficiently to resolve the cointegrates rapidly. These results indicated that (i) the wild-type tnpA gene of IS1071 is able to complement its mutation in trans and (ii) the final product of the IS1071 transposition is a cointegrate of its donor and target DNA molecules. The Kmr Nalr transconjugants were also obtained at very low frequencies when the tnpA gene was not supplied (Table 3, experiment 1). However, all the plasmids residing in these transconjugants had an identical structure in which pMS0310 and R388 were fused but not connected by the two copies of Kmr-IS1071 (data not shown). Such a structure also supported the idea that the IS1071 derivative was not involved in the cointegration. We did not further examine the mechanism by which such a fusion product was formed because IS1071 transposition was not involved.
TABLE 3.
Transposition of Kmr-IS1071 derivatives with mutant IRsa
| Expt | Donor of Kmr-IS1071b (IR length, bp) | Transposition frequencyc
|
|
|---|---|---|---|
| pBBR1MCS | pMS0311 | ||
| 1 | pMS0310 (110) | 8.5 × 10−7 | 3.7 × 10−3 |
| 2 | PMS0366 (100) | 1.5 × 10−6 | 8.7 × 10−6 |
| 3 | pMS0364 (90) | 2.4 × 10−6 | 6.0 × 10−6 |
| 4 | pMS0363 (70) | 8.0 × 10−7 | 4.2 × 10−7 |
| 5 | pMS0362 (48) | 8.7 × 10−7 | 3.8 × 10−6 |
| 6 | pMS0361 (38) | 4.7 × 10−7 | 2.9 × 10−6 |
| 7 | pMS0368 (90, Δ51-70) | 3.7 × 10−6 | 8.7 × 10−6 |
| 8 | pMS0369 (90, Δ71-90) | 3.1 × 10−6 | 2.8 × 10−6 |
The donor strain was a C. testosteroni JCM5832 derivative harboring the three following plasmids: (i) R388, (ii) a pNIT6012 derivative carrying a Kmr-IS1071 derivative with the mutant IRs, and (iii) either pBBR1MCS or pMS0311, a pBBR1MCS derivative that carried the tnpA gene of IS1071 (Fig. 1B and C).
The IR sequences and structure, respectively, of each Kmr-IS1071 derivative are shown in Fig. 1B and C.
The transposition frequency is expressed as the number of Kmr Nalr transconjugants per number of Sur Nalr transconjugants. All values are averages from at least three independent experiments.
Transposition of IS1071 derivatives with mutant ends.
IS1071 has 110-bp IRs, the outermost 38-bp sequences of which are similar to the IRs of Tn3 and Tn21 (Fig. 1B). Considering that Tn3 and Tn21 are able to transpose by using their 38-bp IRs (23), we constructed several Kmr-IS1071 derivatives with shorter IRs and investigated their transposition. A C. testosteroni JCM5832 derivative harboring pMS0311, R388, and a pNIT6012-based plasmid carrying a Kmr-IS1071 derivative with mutant ends (Fig. 1B and C) was mated with JM109. Use of the seven pNIT6012 derivatives generated the Kmr Nalr transconjugants at frequencies of 10−6 to 10−7, and these frequencies were very similar regardless of the presence or absence of the tnpA gene (Table 3, experiments 2 to 8). Furthermore, the transconjugants in each experiment had a fusion product of R388 and the pNIT6012-based plasmid that was not generated by Kmr-IS1071 (data not shown). These results strongly suggest that IS1071-mediated cointegration required almost the entire region of its 110-bp IRs.
Transcriptional activity of the tnpA gene.
To know why IS1071 was transposable in the Comamonas and Delftia cells but not in the Agrobacterium, E. coli, and Pseudomonas cells (Table 2), we investigated the promoter activity of the IS1071-specified 145-bp sequence that was located just upstream of its tnpA gene (Fig. 2A). Plasmid pMS0324, which carried this sequence in front of a promoterless lacZ gene (Fig. 2B), was introduced into the strains listed in Table 2, and the β-Gal activities of the resulting strains were assayed. As shown in Table 4, C. testosteroni JCM5832, Agrobacterium tumefaciens C58, and Pseudomonas putida PpN1 cells harboring pMS0324 showed low β-Gal activities but the activities were not detected in E. coli DH5α, D. acidovorans B123, and Pseudomonas alcaligenes JCM5967. These results were inconsistent with the transposition experiments since transposition of IS1071 in A. tumefaciens C58 and P. putida PpN1 was not detected despite the positive transcriptional activity of the tnpA promoter in these hosts. Thus, differences in promoter activity cannot be the cause of the observed differences in transposition activity among the strains examined.
TABLE 4.
Promoter analysis of the tnpA genea
| Host strain | IS1071-mediated cointegration | β-Gal activity (Miller units)
|
|
|---|---|---|---|
| pMS0324 | pMS0326 | ||
| A. tumefaciens C58 | No | 142 ± 31 | 7.6 ± 2.1 |
| C. testosteroni JCM5832 | Yes | 9.2 ± 0.6 | NDb |
| D. acidovorans B123 | Yes | ND | ND |
| E. coli DH5α | No | ND | ND |
| P. alcaligenes JCM5967 | No | ND | ND |
| P. putida PpN1 | No | 33 ± 4.0 | ND |
β-Gal activity was assayed by the method of Miller (19). Plasmid pMS0324 carries a tnpA promoter-lacZ transcriptional fusion, whereas pMS0326 lacks the tnpA promoter region and was used as a negative control. The nucleotide sequence upstream of the tnpA gene is shown in Fig. 2A, and the structures of the plasmids used in this analysis are shown in Fig. 2B. The values are averages from three independent experiments and are shown with the standard deviations.
ND, not detected in a 60-min reaction.
DISCUSSION
In this study, we showed that (i) IS1071 transposed by a replicative mode to generate a cointegrate of its donor and target molecules as the final product, (ii) a 5-bp duplication of the target sequence was generated upon transposition, and (iii) a tnpA mutation of IS1071 was efficiently complemented by the supply of the wild-type tnpA gene in trans. The experimental data obtained in this study confirm that IS1071 is classified as a class II transposon. A remarkable difference between IS1071 and other typical class II transposons is the absence of the resolution function. Such a function is not absolutely required for completion of the transposition reaction of IS1071 since the host-specified RecA system is also able to resolve the cointegrate by homologous recombination between two directly repeated copies of a transposon (6). It has been proposed that the class II transposons would have evolved from an IS1071 or IS1071-like element by acquisition of the resolution systems (7, 20). This is consistent with the fact that the phylogenetic tree of the enzymes for the resolution systems does not agree with that of the transposases from various class II transposons (7); for example, the transposases of Tn5403 (accession no. X75779) and Tn5393 (M95402) have 62% amino acid identity but their resolvases share only 35% identity. It would be interesting to examine by what special molecular mechanisms the resolution function was acquired.
The length (110 bp) of the IRs is another remarkable characteristic of IS1071 since the IRs of other prokaryotic class II transposons are less than 50 bp long (Fig. 1B). Our deletion analysis in this study revealed that the outermost 100-bp part of the IRs was not sufficient for the transposition of IS1071 (Table 3). It is known that transposases of several class II transposons cooperatively bind to their IRs with the integration host factor (IHF) (12, 34). A well-studied example is Tn4652, a deletion derivative of Tn4651 from P. putida KT2440 (12, 34). Tn4652 requires the IHF for its efficient transposition (10, 12). It has been considered that an unidentified Pseudomonas-specific host factor(s) activates the transcription of the tnpA gene promoter and that binding of the Pseudomonas IHF to the ends of Tn4652 is also required for its transposition (10, 12). Since the IHF generally binds to the flanking region of its target sequence (12) and IS1071 carries a putative IHF-binding site in its both IRs (Fig. 1B), it is likely that lack of the IHF-binding sites in the mutant IRs of IS1071 might be a reason why our IS1071 mutants were unable to transpose. The apparent requirement of the long IR sequences for IS1071 transposition also indicates that the specific recognition and binding of the IS1071 transposase to its IRs might be different from those of other typical class II transposons such as Tn3 and Tn21. In vivo binding experiments of the IS1071 transposase and the IHF with its cognate IRs will provide some clues to clarify the unique interaction between the transposase and IRs.
In this study, high-frequency transposition of IS1071 was detected only in two β-proteobacteria and not in any of the other bacteria tested (Table 2), which belong to the α- and γ-proteobacteria. Since E. coli JM109, used as the recipient cell in the transposition experiments, has an hsdR (restriction-defective) mutation, the host-specific detection of the IS1071 transposition was not due to the restriction of the transferred cointegrates in JM109. It should be noted that our experiment with pMS0252 and R388 cannot detect the IS1071 transposition if (i) IS1071 transposes into R388 by a simple insertion mechanism or (ii) the cointegrate of pMS0252 and R388 resolves rapidly by the host-encoded DNA recombination systems (6). However, this idea is not valid because (i) we never obtained the R388::Kmr-IS1071 plasmid and (ii) the cointegrate of pMS0252 and R388 was stably maintained in A. tumefaciens, P. putida, and P. alcaligenes (data not shown). The strain-specific transposition of IS1071 is consistent with our database searches, which showed that IS1071 and its remnants are, in addition to their preferential localization on the broad-host-range IncP-1β plasmids (18, 21, 27, 28), mainly distributed on the chromosomes of several β-proteobacterial strains such as D. acidovorans P4a (9), Wautersia metallidurans CH34 (accession no. X90708), and Burkholderia xenovorans LB400 (5). Moreover, the nucleotide sequence of IS1071 is highly conserved (>99%) in many bacterial strains, indicating that this element might have been maintained only in a limited number of closely related bacterial strains. One possible explanation for the host-specific transposition of IS1071 is that its transposition requires or is inhibited by some host-specific factor(s). The other possibility is the defect in posttranscriptional steps of the tnpA transcript (translation, holding, and maturation, etc.). It could also be envisioned that the host specificity is involved in the codon usages of the host cells, because (i) taxonomically closely related organisms have similar codon usages (11) and (ii) both C. testosteroni and D. acidovorans are members of the family Comamonadaceae (33). These ideas are consistent with our preliminary experiment in that IS1071 transposed only at a very low frequency (9.6 × 10−7) in E. coli cells, even when its tnpA gene was expressed under the control of a tac promoter. We do not know which one of the three ideas is the most plausible. It is of great interest to uncover the specific host factor(s) of Comamonas and Delftia cells that plays a crucial role in IS1071 transposition.
Acknowledgments
We are grateful to Eva M. Top (University of Idaho) for critically reviewing the manuscript and Masaru Nagai (Institute for Environmental Sciences) for experimental assistance.
This work was carried out under contract with the Ministry of Education, Culture, Sports, Science and Technology. This work was also supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology and the Ministry of Agriculture, Forestry, and Fisheries (HC-05-2323-5), Japan.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. Wiley, New York, N.Y.
- 2.Boon, N., J. Goris, P. De Vos, W. Verstraete, and E. M. Top. 2001. Genetic diversity among 3-chloroaniline- and aniline-degrading strains of the Comamonadaceae. Appl. Environ. Microbiol. 67:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clement, P., D. H. Pieper, and B. Gonzalez. 2001. Molecular characterization of a deletion/duplication rearrangement in tfd genes from Ralstonia eutropha JMP134(pJP4) that improves growth on 3-chlorobenzoic acid but abolishes growth on 2,4-dichlorophenoxyacetic acid. Microbiology 147:2141-2148. [DOI] [PubMed] [Google Scholar]
- 4.Dean, H. F., and A. F. Morgan. 1983. Integration of R91-5::Tn501 into the Pseudomonas putida PPN chromosome and genetic circularity of the chromosomal map. J. Bacteriol. 153:485-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goris, J., P. De Vos, J. Caballero-Mellado, J. Park, E. Falsen, J. F. Quensen III, J. M. Tiedje, and P. Vandamme. 2004. Classification of the biphenyl- and polychlorinated biphenyl-degrading strain LB400T and relatives as Burkholderia xenovorans sp. nov. Int. J. Syst. Evol. Microbiol. 54:1677-1681. [DOI] [PubMed] [Google Scholar]
- 6.Grindley, N. D., and R. R. Reed. 1985. Transpositional recombination in prokaryotes. Annu. Rev. Biochem. 54:863-896. [DOI] [PubMed] [Google Scholar]
- 7.Grindley, N. D. F. 2002. The movement of Tn3-like elements: transposition and cointegrate resolution, p. 272-302. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D. C.
- 8.Heeb, S., Y. Itoh, T. Nishijyo, U. Schnider, C. Keel, J. Wade, U. Walsh, F. O'Gara, and D. Haas. 2000. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol. Plant-Microbe. Interact. 13:232-237. [DOI] [PubMed] [Google Scholar]
- 9.Hoffmann, D., S. Kleinsteuber, R. H. Muller, and W. Babel. 2003. A transposon encoding the complete 2,4-dichlorophenoxyacetic acid degradation pathway in the alkalitolerant strain Delftia acidovorans P4a. Microbiology 149:2545-2556. [DOI] [PubMed] [Google Scholar]
- 10.Horak, R., and M. Kivisaar. 1998. Expression of the transposase gene tnpA of Tn4652 is positively affected by integration host factor. J. Bacteriol. 180:2822-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikemura, T. 1985. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 2:13-34. [DOI] [PubMed] [Google Scholar]
- 12.Ilves, H., R. Horak, R. Teras, and M. Kivisaar. 2004. IHF is the limiting host factor in transposition of Pseudomonas putida transposon Tn4652 in stationary phase. Mol. Microbiol. 51:1773-1785. [DOI] [PubMed] [Google Scholar]
- 13.Junker, F., and A. M. Cook. 1997. Conjugative plasmids and the degradation of arylsulfonates in Comamonas testosteroni. Appl. Environ. Microbiol. 63:2403-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawasaki, H., N. Tone, and K. Tonomura. 1981. Plasmid-determined dehalogenation of haloacetates in Moraxella species. Agric. Biol. Chem. 45:29-34. [Google Scholar]
- 15.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II., and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 16.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II., and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 17.Maeda, K., H. Nojiri, M. Shintani, T. Yoshida, H. Habe, and T. Omori. 2003. Complete nucleotide sequence of carbazole/dioxin-degrading plasmid pCAR1 in Pseudomonas resinovorans strain CA10 indicates its mosaicity and the presence of large catabolic transposon Tn4676. J. Mol. Biol. 326:21-33. [DOI] [PubMed] [Google Scholar]
- 18.Martinez, B., J. Tomkins, L. P. Wackett, R. Wing, and M. J. Sadowsky. 2001. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J. Bacteriol. 183:5684-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, J. H. 1992. Short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Nakatsu, C., J. Ng, R. Singh, N. Straus, and C. Wyndham. 1991. Chlorobenzoate catabolic transposon Tn5271 is a composite class I element with flanking class II insertion sequences. Proc. Natl. Acad. Sci. USA 88:8312-8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlüter, A., H. Heuer, R. Szczepanowski, L. J. Forney, C. M. Thomas, A. Pühler, and E. M. Top. 2003. The 64,508 bp IncP-1β antibiotic multiresistance plasmid pB10 isolated from a waste-water treatment plant provides evidence for recombination between members of different branches of the IncP-1β group. Microbiology 149:3139-3153. [DOI] [PubMed] [Google Scholar]
- 22.Schneider, K., and C. F. Beck. 1986. Promoter-probe vectors for the analysis of divergently arranged promoters. Gene 42:37-48. [DOI] [PubMed] [Google Scholar]
- 23.Sherratt, D. 1989. Tn3 and related transposable elements: site-specific recombination and transposition, p. 163-184. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 24.Sota, M., M. Endo, K. Nitta, H. Kawasaki, and M. Tsuda. 2002. Characterization of a class II defective transposon carrying two haloacetate dehalogenase genes from Delftia acidovorans plasmid pUO1. Appl. Environ. Microbiol. 68:2307-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sota, M., H. Kawasaki, and M. Tsuda. 2003. Structure of haloacetate-catabolic IncP-1β plasmid pUO1 and genetic mobility of its residing haloacetate-catabolic transposon. J. Bacteriol. 185:6741-6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor, L. A., and R. E. Rose. 1988. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 16:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tralau, T., A. M. Cook, and J. Ruff. 2001. Map of the IncP1β plasmid pTSA encoding the widespread genes (tsa) for p-toluenesulfonate degradation in Comamonas testosteroni T-2. Appl. Environ. Microbiol. 67:1508-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trefault, N., R. De la Iglesia, A. M. Molina, M. Manzano, T. Ledger, D. Perez-Pantoja, M. A. Sanchez, M. Stuardo, and B. Gonzalez. 2004. Genetic organization of the catabolic plasmid pJP4 from Ralstonia eutropha JMP134 (pJP4) reveals mechanisms of adaptation to chloroaromatic pollutants and evolution of specialized chloroaromatic degradation pathways. Environ. Microbiol. 6:655-668. [DOI] [PubMed] [Google Scholar]
- 29.Tsuda, M., and H. Genka. 2001. Identification and characterization of Tn4656, a novel class II transposon carrying a set of toluene-degrading genes from TOL plasmid pWW53. J. Bacteriol. 183:6215-6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuda, M., and T. Iino. 1990. Naphthalene degrading genes on plasmid NAH7 are on a defective transposon. Mol. Gen. Genet. 223:33-39. [DOI] [PubMed] [Google Scholar]
- 31.Tsuda, M., K. Minegishi, and T. Iino. 1989. Toluene transposons Tn4651 and Tn4653 are class II transposons. J. Bacteriol. 171:1386-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward, J. M., and J. Grinsted. 1982. Physical and genetic analysis of the Inc-W group plasmids R388, Sa, and R7K. Plasmid 7:239-250. [DOI] [PubMed] [Google Scholar]
- 33.Wen, A., M. Fegan, C. Hayward, S. Chakraborty, and L. I. Sly. 1999. Phylogenetic relationships among members of the Comamonadaceae, and description of Delftia acidovorans (den Dooren de Jong 1926 and Tamaoka et al. 1987) gen. nov., comb. nov. Int. J. Syst. Bacteriol. 49:567-576. [DOI] [PubMed] [Google Scholar]
- 34.Wiater, L. A., and N. D. Grindley. 1988. γδ transposase and integration host factor bind cooperatively at both ends of γδ. EMBO J. 7:1907-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]