Abstract
The human gastrointestinal pathogen Campylobacter jejuni is a microaerophilic bacterium with a respiratory metabolism. The genome sequence of C. jejuni strain 11168 reveals the presence of genes that encode terminal reductases that are predicted to allow the use of a wide range of alternative electron acceptors to oxygen, including fumarate, nitrate, nitrite, and N- or S-oxides. All of these reductase activities were present in cells of strain 11168, and the molybdoenzyme encoded by Cj0264c was shown by mutagenesis to be responsible for both trimethylamine-N-oxide (TMAO) and dimethyl sulfoxide (DMSO) reduction. Nevertheless, growth of C. jejuni under strictly anaerobic conditions (with hydrogen or formate as electron donor) in the presence of any of the electron acceptors tested was insignificant. However, when fumarate, nitrate, nitrite, TMAO, or DMSO was added to microaerobic cultures in which the rate of oxygen transfer was severely restricted, clear increases in both the growth rate and final cell density compared to what was seen with the control were obtained, indicative of electron acceptor-dependent energy conservation. The C. jejuni genome encodes a single class I-type ribonucleotide reductase (RNR) which requires oxygen to generate a tyrosyl radical for catalysis. Electron microscopy of cells that had been incubated under strictly anaerobic conditions with an electron acceptor showed filamentation due to an inhibition of cell division similar to that induced by the RNR inhibitor hydroxyurea. An oxygen requirement for DNA synthesis can thus explain the lack of anaerobic growth of C. jejuni. The results indicate that strict anaerobiosis is a stress condition for C. jejuni but that alternative respiratory pathways can contribute significantly to energy conservation under oxygen-limited conditions, as might be found in vivo.
The genus Campylobacter comprises a number of gastrointestinal pathogens which have assumed increasing prominence in recent years as the causative agents of major animal and human diseases. They are gram-negative, spiral-shaped bacteria which are characterized as being microaerophilic and which also require elevated levels of carbon dioxide for growth (27). Campylobacter jejuni is now recognized as the leading cause of acute bacterial gastroenteritis in both the Western world and developing countries (7), where it is acquired by handling or ingesting contaminated food, milk, or water. Poultry is a particularly common source of contamination (7), and C. jejuni is a commensal of the gastrointestinal tract of many species of birds. Acute symptoms of C. jejuni infection in humans include diarrhea, fever, and abdominal pain, but the sequelae can include colitis, reactive arthritis, and Guillain-Barré syndrome (25). The pathogenic mechanisms of C. jejuni are mediated by a number of virulence factors, including motility, adhesion, and the ability to invade host cells, as well as the production of several toxins (at present not well characterized) which are likely to be responsible for many of the acute manifestations of infection (20). Despite the increasing incidence of C. jejuni as a food-borne pathogen, there are many aspects of the physiology of this bacterium that remain poorly defined. However, physiological studies are crucial in understanding both the mechanisms of pathogenicity of C. jejuni and its ability to survive in the environment, the host, and the food chain. Such studies may also lead to the identification of potential targets for controlling or preventing the growth of these bacteria.
Currently, C. jejuni is regarded as an obligate microaerophile, with a respiratory metabolism based on the use of oxygen as the terminal electron acceptor. The genome sequence of C. jejuni strain 11168 (19) reveals the presence of two terminal oxidases, a cb-type cytochrome c oxidase, and a bd-like quinol oxidase, confirming the results of early work on the respiratory chain composition (4, 10). However, it is clear that C. jejuni 11168 encodes a number of reductases which are predicted to allow the bacterium to carry out respiration with several alternative electron acceptors. These include fumarate reductase, a nitrate reductase of the periplasmic Nap type; a putative S- or N-oxide reductase; and an NrfA homologue, encoding a periplasmic nitrite reductase (12, 19). Thus, C. jejuni appears to have considerable potential to carry out anaerobic respiration, which, in principle, could allow growth in the absence of oxygen. This has not been thoroughly investigated previously, despite the obvious possibility of growth under anaerobiosis or severe oxygen limitation in vivo.
In this paper, we demonstrate that under strictly anaerobic conditions with hydrogen or formate as the major electron donors, none of the predicted alternative electron acceptors were able to support significant growth. However, when the growth rate was restricted by the further limitation of oxygen diffusion into microaerobically grown cultures, a clear stimulation of both growth rate and cell yields was observed in the presence of all of the alternative electron acceptors tested. Examination of the genome sequence (19) for the oxygen-requiring processes which may explain these observations suggested the obligatory reliance of C. jejuni on a class I (NrdAB-type) ribonucleotide reductase (RNR), which utilizes oxygen in the reaction mechanism (11). Consequently, DNA synthesis cannot occur in the absence of oxygen.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
C. jejuni strain 11168 was routinely grown at 37°C under microaerobic conditions (10% [vol/vol] O2, 5% [vol/vol] CO2, and 85% [vol/vol] N2) in a MACS-VA500 incubator (Don Whitley Scientific Ltd., Shipley, United Kingdom) on Columbia agar containing 5% (vol/vol) lysed horse blood and 10 μg each of amphotericin B and vancomycin per ml. For investigation of the capacity of C. jejuni to utilize alternative electron acceptors for growth, liquid cultures of C. jejuni were grown in brain heart infusion broth supplemented with 5% (vol/vol) fetal calf serum (BHI-FCS) and the above-mentioned antibiotics at 37°C under three distinct conditions: (i) standard microaerobic conditions (gas concentrations as described above), with 250 ml of BHI-FCS medium contained in a 500-ml conical flask and mixed by continuous orbital shaking at 120 rpm; (ii) oxygen-limited conditions, where the gas atmosphere and mixing conditions were the same as those described for the standard microaerobic conditions but where the diffusion of oxygen was restricted by using 500 ml of medium contained in a 500-ml conical flask; and (iii) anaerobic conditions, with 250 ml of medium contained in 500-ml conical flasks incubated in an atmosphere of 5% (vol/vol) CO2, 10% (vol/vol) H2, and 85% (vol/vol) N2, achieved by use of an anaerobic cabinet containing a palladium catalyst to remove traces of oxygen (MKIII anaerobic workstation; Don Whitley Scientific Ltd.). Medium for all growth experiments was preincubated in the appropriate gas atmosphere for 24 h at 37°C before inoculation with a microaerobically grown BHI-FCS starter culture. Electron acceptors were added from filter-sterilized stock solutions to a final concentration of 20 mM, except where stated otherwise below. In some cases, sodium formate was added as an additional electron donor (final concentration, 20 mM). Growth was monitored by determining the optical density at 600 nm (OD600) in an Ultrospec 2000 spectrophotometer (Amersham Pharmacia Biotech, Cambridge, United Kingdom). The data presented here are from representative single experiments, but all growth experiments were repeated at least once with similar results.
For selection of C. jejuni mutants, chloramphenicol (CHL) at a final concentration of 30 μg ml−1 was added to the medium. Escherichia coli DH5α was cultured in Luria-Bertani medium supplemented with appropriate antibiotics at 37°C.
Assay of reductase activities.
Methyl or benzyl viologen-linked reductase assays were carried out with intact cells in a 1-ml assay volume by use of a Shimadzu UV-240 recording spectrophotometer. The assay mixture contained 10 mM Tris-HCl (pH 7.5), 0.1 mM methyl or benzyl viologen, and a 5 mM concentration of electron acceptor (potassium nitrate, sodium nitrite, sodium fumarate, trimethylamine-N-oxide [TMAO], or dimethyl sulfoxide [DMSO]) in a screw-topped glass cuvette fitted with a silicone seal. After addition of cells to the buffer-plus-viologen mixture, aliquots of a freshly prepared sodium dithionite solution were injected into the cuvette until the absorbance at 585 nm was 0.8 to 0.9. The assay was started by the injection of an anaerobic solution of the appropriate electron acceptor. Cell protein was determined by use of the Lowry assay.
Nitrite assay.
Diluted culture supernatants (50 μl) were added to 850 μl of 1% (wt/vol) sulfanilamide dissolved in 1 M HCl and 100 μl of 0.02% (wt/vol) naphthylethylenediamine. After 15 min, the absorbance at 540 nm was measured and nitrite concentrations were determined by reference to a standard curve.
Determination of TMAO and TMA concentrations by NMR spectroscopy.
Culture samples (1.5 ml) were centrifuged to remove cells (13,800 × g for 5 min), and the supernatants were used for nuclear magnetic resonance (NMR) analysis. Under conditions similar to those described previously (15), 1H NMR was carried out by use of a Bruker AMX500 spectrometer operating at 500 MHz. Spectra were acquired into 8,000 complex points over a spectral width of 12.5 kHz, and the solvent (H2O) signal was reduced by presaturation for 2 s. This is several times the spin-lattice relaxation time for the signals of interest, so it allows for the reliable integration of signal intensity. Samples (0.5 ml) were run in 5-mm-diameter tubes at 30°C. TMAO and trimethylamine (TMA) each contain nine equivalent protons and so give a single spectral peak; chemical shifts and the concentrations of TMAO and TMA were established by reference to a standard (trimethylsilylpropionate; 0 ppm) added to the samples.
DNA isolation and manipulation.
Plasmid DNA was isolated by using the Miniprep kit (Qiagen). C. jejuni genomic DNA was extracted by use of a modified sodium dodecyl sulfate lysis procedure (17). Standard techniques were employed for the cloning, transformation, preparation, and restriction analysis of plasmid DNA from E. coli (23).
Insertional inactivation of Cj0264c.
Primers 264-F (5′-ATGCGGCGCTAAAGGCAGTAAGTTTTGGTA-3′) and 264-R (5′-ATGCTGGAGCGAGCTTAAGTGCTTTACCTC-3′) were designed to amplify a 2.52-kbp fragment containing the entire coding region of Cj0264c from strain 11168 genomic DNA by PCR with Taq polymerase. This fragment was subsequently cloned into pGEM-T Easy (Promega) to generate pSJH1. A CHL acetyltransferase cassette originating from Campylobacter coli (29) was cloned into the unique ClaI site within Cj0264c to generate pSJH2, which was introduced into C. jejuni 11168 by electroporation. Transformants were plated onto Columbia agar plates without CHL and incubated microaerobically at 37°C for 1 day before being replated on media supplemented with CHL (30 μg ml−1) and incubated for 3 days. Single colonies were restreaked, and chromosomal DNA was extracted by the microLYSIS kit (Web Scientific Ltd., Crewe, United Kingdom) for screening by PCR with the above-mentioned primers to ensure that allelic exchange had occurred by double homologous recombination.
Electron microscopy.
A drop of cell suspension was placed onto a Formvar-coated grid for 2 min, excess liquid was removed by blotting, and the grid was allowed to air dry for 5 min. Cells were negatively stained by adding a drop of 1% (vol/vol) phosphotungstic acid, which was immediately removed by blotting. The grids were air dried and then viewed and photographed with a Phillips CM10 electron microscope.
RESULTS
Analysis of the pathways of electron transport in C. jejuni.
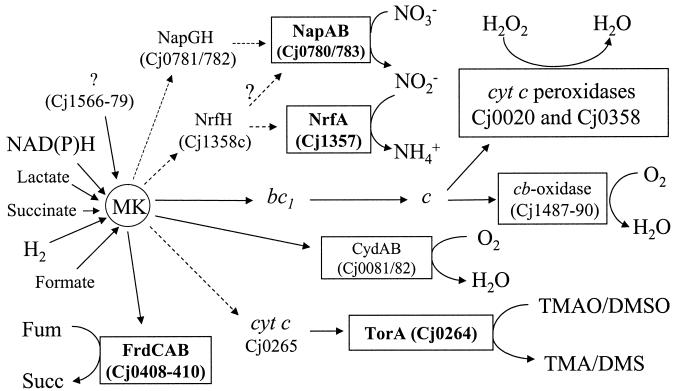
Figure 1 shows a reconstruction of the most likely pathways of electron transport to oxygen and alternative electron acceptors in C. jejuni based on early biochemical studies (4, 10), the annotated genome sequence of strain 11168 (19), additional examination of the sequence, and the known pathways in other bacteria. Electron input to the menaquinone pool is via an Ndh-1 (complex I)-type membrane-bound quinone reductase (Cj1566-1579), a hydrogenase (Cj1266-1267), a succinate dehydrogenase (Cj0437-0439), and a variety of other primary substrate dehydrogenases, including formate dehydrogenase (Cj1508-1511). However, like that of Helicobacter pylori (6, 18), the C. jejuni Ndh-1 lacks the two subunits which in other bacteria bind NADH, so the actual source of electrons for this enzyme may not be reduced nicotinamide nucleotides (18). In terms of oxygen-dependent electron transport, the four-subunit cb-type cytochrome c oxidase likely receives electrons from the proton-translocating cytochrome bc1 complex via a high-potential periplasmic c-type cytochrome and would thus terminate a highly coupled proton-translocating respiratory chain. The alternative oxidase (CydAB homologues) is predicted to oxidize menaquinol directly and provide a less-coupled route for oxygen respiration. Two unlinked genes encoding periplasmic cytochrome c peroxidases may be important in detoxifying hydrogen peroxide in the periplasm. Based on an analogy with peroxide-linked electron transport in a related microaerophile, Campylobacter mucosalis (9), soluble cytochrome c is the most likely electron donor to these enzymes.
FIG. 1.
Predicted electron transport pathways in C. jejuni. A variety of potential electron donors (not all shown here) feed electrons to the menaquinone pool (MK). Cj1566-79 encodes an Nuo/NdhI-type enzyme which probably does not use NAD(P)H (6, 12, 18). Oxygen-linked respiration occurs via two membrane-bound terminal oxidases, a cytochrome c oxidase and a quinol oxidase. Two unlinked periplasmic cytochrome c peroxidases are also predicted to be reduced by cytochrome c. With the exception of fumarate reductase (FrdCAB), all of the other oxygen-independent terminal reductases are predicted to be periplasmic. Electron transport from quinol to nitrate is more likely to be via the NapGH proteins than the NapC/NrfH homologue (question marks). The molybdoenzyme encoded by Cj0264c is shown in this study to be a TMAO and DMSO reductase with greater activity with TMAO, so it is designated TorA. It probably receives electrons from quinol via a small monoheme c-type cytochrome encoded in the same operon. Fum, fumarate; succ, succinate; bc1, cytochrome bc1 complex. Dashed arrows indicate uncertainty regarding the exact electron transport pathway or the likely participation of additional redox proteins.
Several well-defined oxygen-independent electron transport chains can be identified in C. jejuni (Fig. 1). First, a membrane-bound fumarate reductase encoded by Cj0408-0410 (FrdCAB homologues) is present. A periplasmic nitrate reductase of the Nap type is likely to be encoded within an operon comprising Cj0780-0786. The catalytic NapA subunit gene (Cj0780) and the gene for NapB, a diheme cytochrome c (Cj0783), are separated by napG (Cj0781) and napH (Cj0782) genes, encoding iron-sulfur proteins which may mediate electron transport between quinol and NapAB, as this operon does not contain a napC gene which in other bacteria encodes a membrane-bound tetraheme cytochrome c that fulfills this function (3, 21). However, Cj1358c encodes a NapC/NirT/NrfH homologue, which is directly upstream of a gene (Cj1357c) encoding a pentaheme cytochrome c homologous to the periplasmic nitrite reductase (NrfA) of E. coli and other bacteria. Thus, C. jejuni appears to be equipped with both nitrate and nitrite reductases located in the periplasm, each predicted to receive electrons from menaquinol via distinct redox proteins (Fig. 1). An electron transport chain from quinol to N- or S-oxides like TMAO or DMSO may be formed by Cj0264, a DorA/TorA homologue, and Cj0265, a monoheme c-type cytochrome. Both Cj0264 and NapA (Cj0780) are molybdoproteins containing conserved residues for the binding of a molybdenum guanosine dinucleoside cofactor, and they possess a motif in their N termini (RRXFL/IK) which is similar to the core “twin-arginine” translocase recognition motif (S/TRRXFLK), characteristic of extracytoplasmic proteins containing complex redox cofactors (2).
The cellular location of the terminal reductases has consequences for energy conservation. Menaquinol-dependent fumarate reduction may be electrogenic, resulting in the generation of a proton motive force, as the active site of the membrane-bound fumarate reductase faces the cytoplasm (4, 13, 26). However, as all of the other reductases identified here are predicted to be located in the periplasm, the oxidation of quinol and the reduction of the terminal electron acceptor should release and consume protons, respectively, in the same cellular compartment. Thus, electron transport from menaquinol to nitrate, nitrite, TMAO, or DMSO should not lead to the generation of a proton motive force, which can therefore be generated only at the level of the primary dehydrogenases.
Growth with fumarate as a terminal electron acceptor.
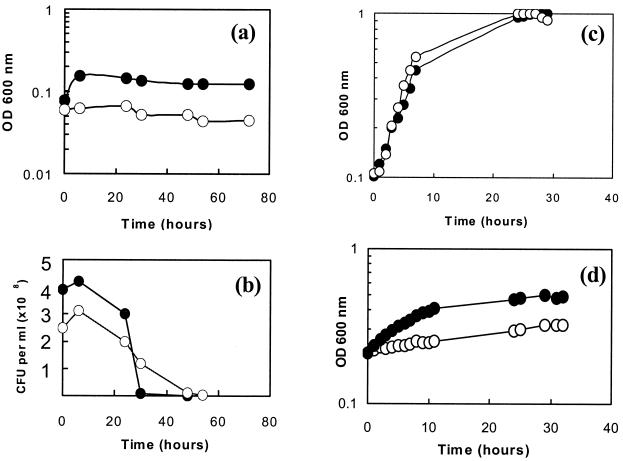
It has previously been reported that C. jejuni is incapable of anaerobic growth with fumarate as the electron acceptor (28). However, the presence of fumarate reductase activity (4, 26) encoded by typical frd genes distinct from those encoding succinate dehydrogenase (12, 19) suggests that anaerobic growth supported by fumarate respiration might be possible. Figure 2a shows the growth of strain 11168 in the presence and absence of fumarate under strictly anaerobic conditions in an atmosphere of 85% (vol/vol) nitrogen, 10% (vol/vol) carbon dioxide, and 5% (vol/vol) hydrogen, with the latter acting as the major electron donor under these conditions. In the presence of fumarate, there was only a slight initial increase in OD compared with what was observed with the control (presumably as a result of the utilization of residual oxygen introduced with the inoculum), with growth stopping after a few hours. Viable counts showed only a slight increase in the first few hours after inoculation, before viability declined in both cultures (Fig. 2b). Similar results were obtained with anaerobic experiments where 20 mM sodium formate was added to the cultures as an additional electron donor (data not shown). Thus, in agreement with the results of Véron et al. (28), there is no evidence that fumarate supports either the anaerobic growth or survival of C. jejuni. As shown in Fig. 2c, there was also no effect on either the growth rate or final cell yield when fumarate was added to cultures growing under standard microaerobic growth conditions.
FIG. 2.
Growth of C. jejuni on fumarate under anaerobic (a and b), microaerobic (c), and oxygen-limited (d) conditions. Shown are the growth curves for C. jejuni in BHI-FCS medium with no added fumarate (open circles) and with 20 mM sodium fumarate (closed circles).
Fumarate reductase enzyme activity was constitutive and was easily detectable in intact C. jejuni cells harvested from blood-agar plates with reduced benzyl viologen as electron donor (>50 nmol of benzyl viologen oxidized/min/mg of cell protein), so we considered the possibility that some essential metabolic reaction(s) in C. jejuni has an obligatory requirement for oxygen and that this prevents fumarate-dependent growth under strictly anaerobic conditions. In order to test this possibility, the rate of diffusion of oxygen into liquid cultures incubated in a standard microaerobic atmosphere of 10% (vol/vol) oxygen, 5% (vol/vol) carbon dioxide, and 85% (vol/vol) nitrogen was limited by increasing the volume of medium in the flasks and thus decreasing the surface area/volume ratio (see Materials and Methods). Under these conditions, the growth rate of C. jejuni was significantly reduced (compare Fig. 2c and d). The effect of the addition of fumarate to such cultures was dramatic (Fig. 2d). Clear increases in both growth rate (doubling times of >40 h without fumarate compared to ∼7 h with fumarate) and final cell density were observed, which was fully consistent with energy conservation by the utilization of fumarate as a terminal electron acceptor under these conditions.
Growth with nitrate and nitrite as electron acceptors.
Figure 3a, c, and e show the results of a series of growth experiments conducted under anaerobic, standard microaerobic, and oxygen-limited conditions, respectively, in the presence or absence of 20 or 100 mM nitrate. The accumulation of nitrite in these cultures is shown in Fig. 3b, d, and f. Under strictly anaerobic culture conditions (Fig. 3a), no growth was observed in the control and, as in the presence of fumarate, only a very small initial increase in turbidity (OD600 of <0.1) occurred in the presence of nitrate. Nitrate reductase activity was clearly evidenced by the accumulation of nitrite, but conversion was nonquantitative (the maximum conversions of 20 and 100 mM nitrate to nitrite were 65 and 17%, respectively). However, under standard microaerobic conditions, 20 mM nitrate was quantitatively converted to nitrite after 6 h of growth, while 100 mM nitrate was converted to 70 mM nitrite (Fig. 3d). Nevertheless, there was no nitrate-dependent growth rate advantage under these conditions (Fig. 3c) and the nitrate-containing cultures reached a slightly lower final OD compared to that of the control without added nitrate. In contrast, under oxygen-limited conditions (produced in the same way as for the fumarate growth experiments) there were clear nitrate-dependent increases in the growth rate and final cell density (Fig. 3e), indicating the use of nitrate for energy conservation. These cultures showed nitrate reduction characteristics very similar to those of standard microaerobic cultures (Fig. 3f), with quantitative conversion of 20 mM nitrate to nitrite after 10 h. Overall, the patterns of growth observed with nitrate as the electron acceptor were very similar to those seen with fumarate.
FIG. 3.
(a, c, and e) Growth of C. jejuni under anaerobic (a), microaerobic (c), and oxygen-limited (e) conditions in BHI-FCS medium with no added nitrate (open circles), with 20 mM sodium nitrate (open squares), and with 100 mM sodium nitrate (closed squares). (b, d, and f) Nitrite accumulation in culture supernatants under anaerobic (b), microaerobic (d), and oxygen-limited (f) conditions after addition of 20 mM (open squares) and 100 mM (closed squares) sodium nitrate.
Following the exhaustion of nitrate in microaerobic and oxygen-limited cultures with 20 mM nitrate (Fig. 3d and f), there was a decrease in nitrite concentration, which suggested the operation of nitrite reductase. Methyl viologen-linked nitrite reductase activity was readily detectable (>500 nmol of methyl viologen oxidized/min/mg of cell protein) in intact cells grown with or without nitrite. The accumulation of high concentrations of nitrite did not appear to severely inhibit growth in the experiment for which results are shown in Fig. 3, but unlike with the other electron acceptors tested, experiments under oxygen-limited conditions with 20 or 5 mM sodium nitrite did not reveal any stimulation of growth rate or cell yield (data not shown). In order to avoid potential nitrite toxicity problems, the experiment for which results are shown in Fig. 4 was performed. Pulses of 1 mM sodium nitrite were added at intervals to an oxygen-limited culture, and growth in this culture was compared to that in a similar culture without nitrite. After each addition of nitrite, the cells consumed nitrite rapidly (Fig. 4b) and over the course of the experiment higher growth rate and cell density were clearly apparent compared to what was observed with the control (Fig. 4a). These data show that C. jejuni possesses an energy-conserving nitrite reduction system.
FIG. 4.
Nitrite-dependent growth of C. jejuni under oxygen-limited conditions. (a) Growth curves for C. jejuni in BHI-FCS medium with no added nitrite (open squares) and with sodium nitrite added to a final concentration of 1 mM at time zero and at the times indicated by the arrows in panel b (closed squares). (b) Nitrite concentrations in culture supernatants from the nitrite-containing culture in panel a.
Growth with TMAO or DMSO as electron acceptor.
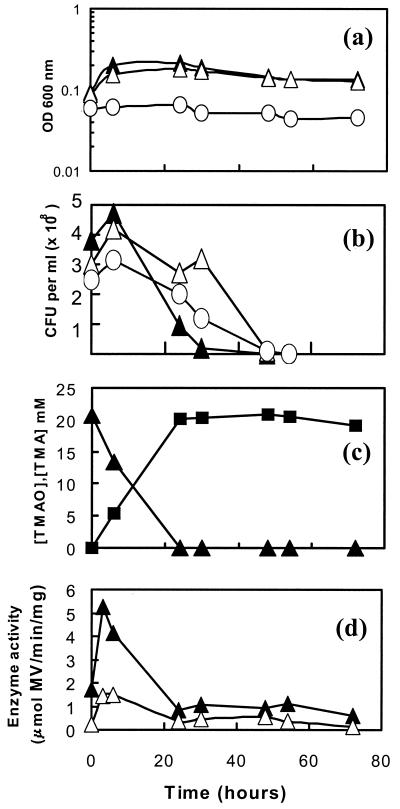
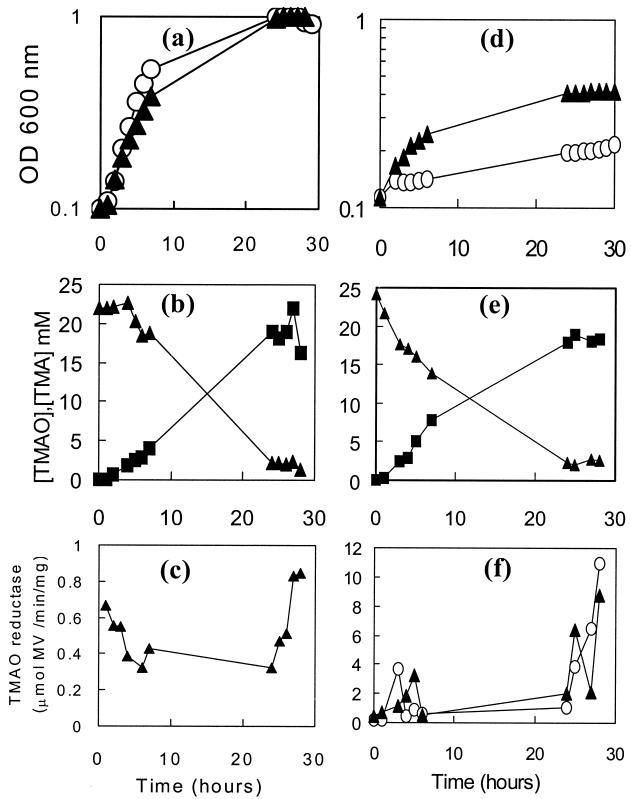
Under strictly anaerobic conditions, C. jejuni 11168 did not grow significantly with either TMAO or DMSO as the electron acceptor (Fig. 5a). Viable counts (Fig. 5b) also indicated that survival was not maintained in the presence of either electron acceptor compared to what was observed with the control. However, 1H NMR measurements of culture supernatants showed that TMAO was quantitatively converted into TMA (Fig. 5c) and that DMSO was reduced to dimethyl sulfide (DMS; data not shown) (DMSO and DMS are volatile and difficult to quantify). Methyl viologen-dependent TMAO and DMSO reductase activities were readily detectable in intact cells, and the specific activities were relatively constant after an initial rise in the first few hours of anaerobic incubation (Fig. 5d). Under standard microaerobic conditions, cultures with or without added TMAO grew at the same rate (doubling times of ∼2 h) and reached the same final OD (Fig. 6a). Almost all of the TMAO was converted to TMA by the end of the experiment (Fig. 6b). Low-to-moderate methyl viologen-dependent TMAO reductase activities were maintained during the exponential growth phase and were increased slightly in the late stationary phase in the TMAO-containing culture (Fig. 6c). In cultures which were oxygen limited (Fig. 6d), the addition of TMAO significantly increased both the growth rate (doubling time of ∼4 h with TMAO and >40 h without TMAO) and the final stationary-phase cell density. These cultures also converted TMAO into TMA almost quantitatively (Fig. 6e) and displayed moderate levels of methyl viologen-dependent TMAO reductase activity, which increased significantly in the late stationary phase (Fig. 6f). However, there was no evidence that the presence of TMAO in the growth medium induced an increase of reductase activity (Fig. 6f). DMSO was also found to stimulate growth under oxygen-limited conditions but not in standard microaerobic cultures (data not shown).
FIG. 5.
Anaerobic growth of C. jejuni on TMAO or DMSO. (a) Growth curves for C. jejuni in BHI-FCS medium with no added electron acceptor (open circles), with 20 mM DMSO (open triangles), and with 20 mM TMAO (closed triangles). (b) Viable counts, with the same symbols as those in panel a. (c) Reduction of TMAO (closed triangles) to TMA (closed squares) in BHI-FCS plus 20 mM TMAO. Culture supernatants were prepared at the time points shown and subjected to 1H NMR spectroscopy as described in Materials and Methods. (d) Methyl viologen-linked reductase activity in intact cells. Closed triangles, TMAO reductase activity in washed cells from BHI-FCS plus 20 mM TMAO; open triangles, DMSO reductase activity in washed cells from BHI-FCS plus 20 mM DMSO.
FIG. 6.
Growth of C. jejuni on TMAO under microaerobic (a to c) or oxygen-limited (d to f) conditions. (a and d) Growth curves for C. jejuni in BHI-FCS medium in the absence (open circles) or presence (closed triangles) of 20 mM TMAO. (b and e) Reduction of TMAO (closed triangles) to TMA (closed squares) in BHI-FCS plus 20 mM TMAO. Culture supernatants were prepared at the time points shown and subjected to 1H NMR spectroscopy as described in Materials and Methods. (c and f) Methyl viologen-linked TMAO reductase activity in intact cells. Closed triangles, TMAO reductase activity in washed cells from BHI-FCS plus 20 mM TMAO; open circles, TMAO reductase activity in washed cells from BHI-FCS.
Cj0264c encodes the sole TMAO and DMSO reductase in C. jejuni.
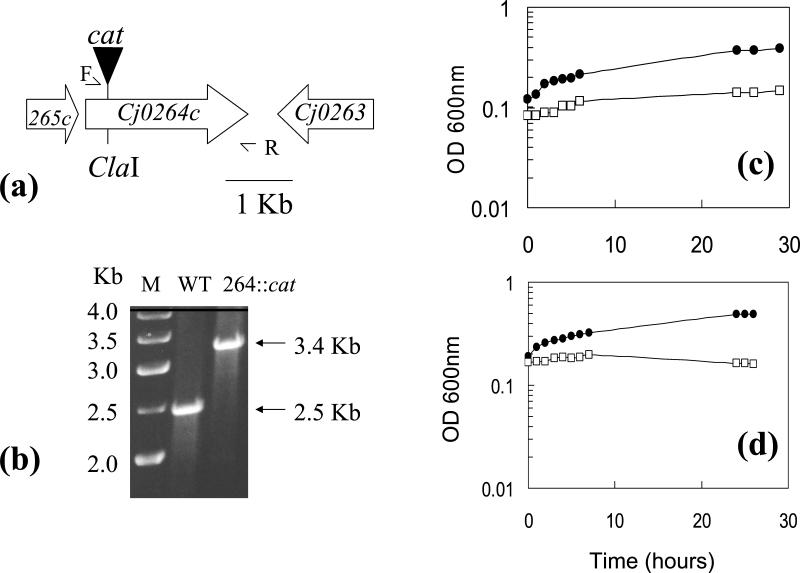
In order to determine whether the molybdoprotein encoded by Cj0264c is a TMAO and/or DMSO reductase responsible for the ability of C. jejuni to respire these electron acceptors, a Cj0264c-null mutant was constructed (Fig. 7a). By use of gene-specific primers, the correct construction of the mutant was shown by the amplification from genomic DNA of a single PCR product, which was 0.9 kb larger than that obtained with wild-type DNA (Fig. 7b). Wild-type parent and mutant cells were grown for 24 h under oxygen-limited conditions in BHI-FCS medium plus either 20 mM TMAO or 20 mM DMSO, and the cells were assayed for reductase activity with reduced methyl viologen as the electron donor. The mutant possessed no detectable methyl viologen-linked TMAO or DMSO reductase activity, whereas high activities were measurable in the wild type (in one typical experiment, TMAO reductase activity of 4,703 nmol of methyl viologen oxidized/min/mg of protein in TMAO-grown cells and DMSO reductase activity of 980 nmol of methyl viologen oxidized/min/mg of protein in DMSO-grown cells were measured). Under oxygen-limited conditions, no TMAO- or DMSO-dependent growth was observed with the mutant compared to what was observed with the parent strain (Fig. 7c and d). These data demonstrate that Cj0264c encodes the sole TMAO and DMSO reductase in C. jejuni, and this enzyme is responsible for the TMAO- and DMSO-dependent growth enhancement seen in wild-type cells under oxygen-limited conditions.
FIG. 7.
TMAO- or DMSO-stimulated growth under oxygen-limited conditions depends on the reductase encoded by Cj0264c. (a) Strategy for insertional inactivation of Cj0264c. Primers 264-F (F) and 264-R (R) were used to PCR amplify a 2.52-kb product containing the gene, which was cloned into pGEM and insertionally inactivated at a unique ClaI site, by use of a cat cassette containing its own promoter and transcriptional terminators. As the downstream gene (Cj0263) is transcribed in the opposite orientation to that of Cj0264c, there is no polarity effect for this insertion. Plasmid pSJH2 was then electroporated into C. jejuni 11168, with selection for CHL resistance. (b) Verification of mutant construction. Primers F and R were used in a PCR with genomic DNA from the wild-type (WT) parent strain 11168 or from a CHL-resistant transformant (264::cat). Agarose gel electrophoresis showed a single product in each case, which was 0.9-kb larger in the mutant strain due to the cat insertion. Lane M contained a 1-kb DNA ladder. (c and d) Growth of strain 11168 (closed circles) and the Cj0264c::cat mutant (open squares) under oxygen-limited conditions in BHI-FCS plus 20 mM TMAO (c) or 20 mM DMSO (d).
Use of an oxygen-dependent class I RNR for DNA synthesis can explain the lack of anaerobic growth of C. jejuni.
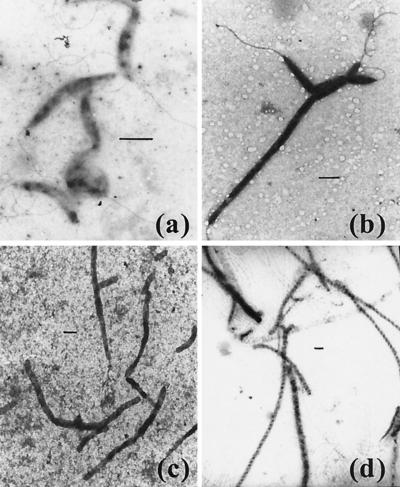
The results described above clearly show that some oxygen is required for the growth of C. jejuni on a range of structurally distinct electron acceptors. A likely explanation for this requirement is the existence of one or more essential oxygen-requiring metabolic reactions in this bacterium. There are two obvious possibilities. (i) The oxidative decarboxylation of coproporphyrinogen III to protoporphyrin IX is an essential step in heme biosynthesis, which can be catalyzed by HemF, an oxygen-requiring oxidase, or HemN, an oxygen-independent dehydrogenase (1). Scrutiny of the C. jejuni 11168 genome sequence reveals the presence of genes encoding several HemN homologues rather than HemF, so the inability to synthesize heme anaerobically is unlikely to be the explanation for the lack of anaerobic growth. (ii) Reduction of ribonucleotides by RNR to provide 2′-deoxyribonucleotides for DNA synthesis and repair is an essential process in all cells that can be catalyzed by several distinct classes of enzymes (11). Most bacteria possess class II RNRs, which are adenosylcobalamin cofactored and active under both aerobic and anaerobic conditions. Class III RNRs function only anaerobically due to the participation of an oxygen-sensitive glycyl radical in catalysis. In contrast, class I enzymes contain a relatively stable tyrosyl radical which is generated by the formation of an oxygen-linked di-iron center in a reaction which is strictly dependent on molecular oxygen (11). C. jejuni 11168 contains genes for the oxygen-dependent class I type of RNR only (NrdA and NrdB homologues; Cj0024 and Cj0231c genes, respectively) and is thus obligatorily dependent on oxygen for DNA synthesis. This could explain its inability to grow under strictly anaerobic conditions. To obtain further evidence of this possibility, we investigated the morphological effects of the RNR inhibitor hydroxyurea on microaerobically growing cells and compared these effects with those produced by the incubation of cells under strictly anaerobic conditions in the presence of an electron acceptor (Fig. 8). Electron microscopy showed that, as expected, hydroxyurea caused cellular filamentation due to the prevention of cell division (compare Fig. 8a and b). Very similar filamentation effects were observed under anaerobic conditions in the presence of nitrate (Fig. 8c) or fumarate (Fig. 8d), which is consistent with the prevention of DNA synthesis and the inability of cells to complete division due to the absence of oxygen in these cultures.
FIG. 8.
Filamentation induced by the DNA synthesis inhibitor hydroxyurea or by incubation of cells anaerobically with nitrate or fumarate. (a) C. jejuni 11168 grown for 24 h under standard microaerobic conditions in BHI-FCS medium, showing normal morphology. (b) Filamentation induced after microaerobic growth in BHI-FCS for 24 h in the presence of 20 mM hydroxyurea. (c and d) Filamentous cells produced under strictly anaerobic conditions after 48 h in BHI-FCS plus either 20 mM nitrate (c) or 20 mM fumarate (d). Bars, 1 μm.
DISCUSSION
Early research on respiration in campylobacters led to the realization that some species, e.g., Campylobacter fetus (28), could grow anaerobically by using fumarate as an electron acceptor, but this capacity was reported as being absent from C. jejuni (28). In this study, we have obtained clear evidence that C. jejuni is capable of growth that is supported by respiration of a range of electron acceptors, including fumarate, nitrate, nitrite, and TMAO or DMSO. However, this is apparent only in cultures in which growth is limited by a reduced oxygen supply but which are not anaerobic. This requirement for oxygen accounts for the inability of previous investigators to obtain the growth of C. jejuni under strictly anaerobic conditions (28). Although other oxygen-requiring processes could contribute to the prevention of anaerobic growth, the most likely essential role of oxygen is to satisfy the requirement for deoxyribonucleotide in DNA synthesis due to the use of an oxygen-utilizing class I-type RNR. This is consistent with the lack of alternative types of RNR encoded in the genome and the cellular filamentation observed during anaerobic growth experiments, which was similar to that induced by hydroxyurea due to the inhibition of cell division. Although strict anaerobiosis can thus be viewed as a stress-inducing condition for C. jejuni, the discovery of the organism's ability to use alternative electron acceptors in energy-conserving reactions in the presence of oxygen raises the possibility that growth in the oxygen-limited avian or mammalian gut may be partially dependent on the use of fumarate or nitrate, for example, which might be readily available from host or dietary sources.
In Mycobacterium bovis, membrane-bound nitrate reductase (Nar) activity has been shown to contribute to the organism's virulence in mice (30). In gram-negative bacterial pathogens, periplasmic nitrate reductases (Nap) are more common than Nar-type enzymes (21). What is the role of Nap in C. jejuni? We have shown that electron transport to Nap can enhance growth in vitro at high nitrate concentrations, but nitrate concentrations in human body fluids are only in the range of 10 to 50 μM (16). However, the E. coli Nap enzyme has a much higher affinity for nitrate compared to that of the membrane-bound Nar (22), making Nap ideally suited to a role in scavenging the low nitrate concentrations encountered in vivo. In many bacteria, the catalytic NapAB subunits are supplied with electrons via a membrane-bound tetraheme cytochrome c (NapC homologue) that acts as a quinol oxidase (3, 21). As noted above, a NapC homologue is encoded by a gene (Cj1358c) directly upstream of nrfA in C. jejuni, rather than being in the nap operon itself, and is thus likely to be the electron donor to the nitrite reductase. While it is possible that NapAB may also receive electrons from the Cj1358 protein, a role for the NapG- and NapH-like Fe-S proteins encoded in the nap operon is perhaps more likely (Fig. 1), but this has not yet been tested experimentally.
Nitrite from 20 mM nitrate added to oxygen-limited cultures accumulated to stoichiometric concentrations, but the buildup of nitrite did not inhibit growth and nitrite appeared to be utilized to some extent after the exhaustion of nitrate. However, we could only demonstrate nitrite-dependent growth under oxygen-limited conditions when the concentration of nitrite in the culture was kept low by the addition of pulses of the electron acceptor at intervals, suggesting some degree of nitrite toxicity. Unphysiological nitrate and nitrite concentrations had to be employed in these experiments; there is evidence of the formation of multimeric Nap-Nrf complexes in E. coli (21), and at environmental (i.e., low) nitrate concentrations, C. jejuni is likely to be able to efficiently reduce nitrate to ammonia by using such a complex, preventing nitrite from diffusing away, thus allowing maximal energy conservation and avoiding nitrite toxicity. The NrfA nitrite reductase of C. jejuni is a pentaheme protein of particular interest, as the ligation of the active site heme is unusual; a conventional CXXCH motif is present in the N-terminal region of NrfA instead of CXXCK, which is found in all other periplasmic NrfA proteins presently known (5). This presumably explains the absence in C. jejuni of nrfEFGIJKLM genes encoding heme ligases and assembly proteins which are found in other bacteria (5, 24), but the consequences for the activity or function of this enzyme are unknown at present.
C. jejuni is also clearly equipped with the capacity for TMAO and DMSO reduction, and based on the phenotype of the Cj0264 mutant, it is clear that this gene encodes the sole reductase for these electron acceptors in the bacterium. TMAO is an enzyme-protective osmolyte found in some fish and aquatic invertebrates (31), and DMSO is a cryoprotectant produced by some algae (14). These electron acceptors would thus be available to C. jejuni in aquatic environments; their reduction by an energy-conserving electron transport chain may specifically aid survival outside the host. The electron transport chain to TMAO or DMSO is likely to involve the transfer of electrons to the molybdenum guanosine dinucleoside cofactor of Cj0264 from the 22-kDa monoheme c-type cytochrome encoded by Cj0265c (Fig. 1). Interestingly, Cj0265 has greatest sequence similarity to the monoheme C-terminal domain of the pentaheme TorC protein in E. coli, which has been shown to directly donate electrons to the TorA reductase (8). Additional redox proteins are likely to be involved in electron transfer from quinol to Cj0265.
The regulation of alternative respiratory pathways in C. jejuni is also of interest. We detected several terminal reductase enzyme activities irrespective of the presence or absence of electron acceptors in growth medium, suggesting constitutive synthesis. In the case of TMAO reductase, which was studied in most detail, there was evidence of an increase of enzyme-specific activity in late-stationary-phase oxygen-limited cultures. One explanation for this increase could be the involvement of a low-oxygen-sensing transcription factor. Interestingly, C. jejuni does encode a protein (Cj0466) related to the CRP/FNR family which might regulate reductase expression in response to oxygen limitation, although the deduced sequence lacks the N-terminal cysteine residues essential for the sensing of oxygen in FNR-type proteins.
In conclusion, we have demonstrated that C. jejuni has the ability to utilize a range of structurally distinct electron acceptors of differing redox potentials in energy-conserving reactions that support growth, provided sufficient oxygen is available for DNA synthesis to continue. We are currently constructing additional mutants in several of the electron transport genes identified in this study in order to determine the nature of the electron transport pathways and their importance in vivo.
Acknowledgments
This work was supported by a research grant to D.J.K. from the U.K. Biotechnology and Biological Sciences Research Council (BBSRC) and by a BBSRC studentship to M.J.S.
We thank Gavin Thomas for useful discussions.
REFERENCES
- 1.Beale, S. I. 1996. Biosynthesis of hemes, p. 731-748. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 2.Berks, B. C. 1996. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22:393-404. [DOI] [PubMed] [Google Scholar]
- 3.Berks, B. C., S. J. Ferguson, J. W. B. Moir, and D. J. Richardson. 1995. Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim. Biophys. Acta 1232:97-173. [DOI] [PubMed] [Google Scholar]
- 4.Carlone, G. M., and J. Lascelles. 1982. Aerobic and anaerobic respiratory systems in Campylobacter fetus subsp. jejuni grown in atmospheres containing hydrogen. J. Bacteriol. 152:306-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Einsle, O., P. Stach, A. Messerschmidt, J. Simon, A. Kroger, R. Huber, and P. M. H. Kroneck. 2000. Cytochrome c nitrite reductase from Wolinella succinogenes. J. Biol. Chem. 275:39608-39616. [DOI] [PubMed] [Google Scholar]
- 6.Finel, M. 1998. Does NADH play a central role in energy metabolism in Helicobacter pylori? Trends Biochem. Sci. 23:412-414. [DOI] [PubMed] [Google Scholar]
- 7.Friedman, C. R., J. Neiman, H. C. Wegener, and R. V. Tauxe. 2000. Epidemiology of Campylobacter jejuni in the United States and other industrialized nations, p. 121-138. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 8.Gon, S., M. T. Giudici-Orticoni, V. Mejean, and C. Iobbi-Nivol. 2001. Electron transfer and binding of the c-type cytochrome TorC to the trimethylamine N-oxide reductase in Escherichia coli. J. Biol. Chem. 276:11545-11551. [DOI] [PubMed] [Google Scholar]
- 9.Goodhew, C. F., A. B. elKurdi, and G. W. Pettigrew. 1988. The microaerophilic respiration of Campylobacter mucosalis. Biochim. Biophys. Acta 933:114-123. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman, P. S., and T. G. Goodman. 1982. Respiratory physiology and energy conservation efficiency of Campylobacter jejuni. J. Bacteriol. 150:319-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jordan, A., and P. Reichard. 1998. Ribonucleotide reductases. Annu. Rev. Biochem. 67:71-98. [DOI] [PubMed] [Google Scholar]
- 12.Kelly, D. J. 2001. The physiology and metabolism of Campylobacter jejuni and Helicobacter pylori. J. Appl. Microbiol. 90:16S-24S. [DOI] [PubMed] [Google Scholar]
- 13.Lancaster, C. R. D., A. Kroger, M. Auer, and H. Michel. 1999. Structure of fumarate reductase from Wolinella succinogenes at 2.2 Å resolution. Nature 402:377-385. [DOI] [PubMed] [Google Scholar]
- 14.Lee, P. A., and S. J. de Mora. 1999. Intracellular dimethylsulfoxide in unicellular marine algae: speculations on its origin and possible biological role. J. Phycol. 35:8-18. [Google Scholar]
- 15.Leighton, M. P., D. J. Kelly, M. P. Williamson, and J. G. Shaw. 2001. An NMR and enzyme study of carbon metabolism in Neisseria meningitidis. Microbiology 147:1473-1482. [DOI] [PubMed] [Google Scholar]
- 16.Lentner, C. (ed.). 1984. Composition of blood, p. 81. In Geigy scientific tables, vol. 3. Ciba-Geigy Ltd., Basel, Switzerland.
- 17.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 18.Mendz, G. L., M. A. Smith, M. Finel, and V. Korolik. 2000. Characteristics of the aerobic respiratory chains of the microaerophiles Campylobacter jejuni and Helicobacter pylori. Arch. Microbiol. 174:1-10. [DOI] [PubMed] [Google Scholar]
- 19.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M.-A. Rajandream, K. M. Rutherford, A. H. M. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 20.Pickett, C. L. 2000. Campylobacter toxins and their role in pathogenesis, p. 179-190. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 21.Potter, L., H. Angrove, D. Richardson, and J. Cole. 2001. Nitrate reduction in the periplasm of gram-negative bacteria. Adv. Microb. Physiol. 45:51-112. [DOI] [PubMed] [Google Scholar]
- 22.Potter, L. C., P. Millington, L. Griffiths, G. H. Thomas, and J. A. Cole. 1999. Competition between Escherichia coli strains expressing either a periplasmic or a membrane bound nitrate reductase: does Nap confer a selective advantage during nitrate-limited growth? Biochem. J. 344:77-84. [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Simon, J., R. Gross, O. Einsle, P. M. H. Kroneck, A. Kroger, and O. Klimmek. 2000. A NapC/NirT-type cytochrome c (NrfH) is the mediator between the quinone pool and the cytochrome c nitrite reductase of Wolinella succinogenes. Mol. Microbiol. 35:686-696. [DOI] [PubMed] [Google Scholar]
- 25.Skirrow, M. B., and M. J. Blaser. 2000. Clinical aspects of Campylobacter infection, p. 69-88. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 26.Smith, M. A., G. L. Mendz, M. A. Jorgensen, and S. L. Hazell. 1999. Fumarate metabolism and the microaerophily of Campylobacter jejuni. Int. J. Biochem. Cell Biol. 31:961-975. [DOI] [PubMed] [Google Scholar]
- 27.Vandamme, P. 2000. Taxonomy of the family Campylobacteriaceae, p. 3-26. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 28.Véron, M., A. Lenvoisé-Furet, and P. Beaune. 1981. Anaerobic respiration of fumarate as a differential test between Campylobacter fetus and Campylobacter jejuni. Curr. Microbiol. 6:349-354. [Google Scholar]
- 29.Wang, Y., and D. E. Taylor. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94:23-28. [DOI] [PubMed] [Google Scholar]
- 30.Weber, I., C. Fritz, S. Ruttkowski, A. Kreft, and F.-C. Bang. 2000. Anaerobic nitrate reductase (narGHJI) activity of Mycobacterium bovis BCG in vitro and its contribution to virulence in immunodeficient mice. Mol. Microbiol. 35:1017-1025. [DOI] [PubMed] [Google Scholar]
- 31.Yancey, P. H., and J. F. Siebenaller. 1999. Trimethylamine-N-oxide stabilizes teleost and mammalian lactate dehydrogenases against inactivation by hydrostatic pressure and trypsinolysis. J. Exp. Biol. 202:3597-3603. [DOI] [PubMed] [Google Scholar]