Abstract
Maintenance of antimicrobial drug resistance in bacteria can be influenced by factors unrelated to direct selection pressure such as close linkage to other selectively advantageous genes and secondary advantage conveyed by antimicrobial resistance genes in the absence of drug selection. Our previous trials at a dairy showed that the maintenance of the antimicrobial resistance genes is not influenced by specific antimicrobial selection and that the most prevalent antimicrobial resistance phenotype of Escherichia coli is specifically selected for in young calves. In this paper we examine the role of secondary advantages conveyed by antimicrobial resistance genes. We tested antimicrobial-susceptible null mutant strains for their ability to compete with their progenitor strains in vitro and in vivo. The null mutant strains were generated by selection for spontaneous loss of resistance genes in broth supplemented with fusaric acid or nickel chloride. On average, the null mutant strains were as competitive as the progenitor strains in vitro and in newborn calves (in vivo). Inoculation of newborn calves at the dairy with antimicrobial-susceptible strains of E. coli did not impact the prevalence of antimicrobial-resistant E. coli. Our results demonstrate that the antimicrobial resistance genes are not responsible for the greater fitness advantage of antimicrobial-resistant E. coli in calves, but the farm environment and the diet clearly exert critical selective pressures responsible for the maintenance of antimicrobial resistance genes. Our current hypothesis is that the antimicrobial resistance genes are linked to other genes responsible for differential fitness in dairy calves.
Antimicrobial drug-resistant bacteria present a significant risk to public health, and consequently there is great interest in reducing the prevalence of antimicrobial resistance (AR) genes in both commensal and pathogenic bacteria. One strategy to decrease the prevalence of AR bacteria is to discontinue using antimicrobial drugs for growth promotion and prophylaxis in food animals (4, 11, 22, 43, 44). Unfortunately, this strategy has produced mixed results that are dependent on the duration and amount of antimicrobial drug use (10, 16, 24, 25, 37, 38, 41). While pathogenic bacteria are the primary concern, commensal bacteria are an important reservoir for AR genes. It is hypothesized that, because AR genes can easily transfer among diverse bacterial species (6, 29, 32, 35, 39, 45), then they can also move easily from commensal to pathogenic bacteria. Consequently abundance and ease of acquisition of the antimicrobial drug resistance genes ultimately complicate the orthodox treatment of diverse bacterial diseases (13, 18).
It is generally accepted that the prevalence of AR bacteria is directly correlated with antimicrobial drug use (19, 20, 40), but mechanisms unrelated to specific antimicrobial drug selection have also been implicated in the wide distribution and maintenance of AR genes. Some of these mechanisms include plasmid addiction (14, 17) and close linkage to other selectively advantageous genes (2, 9, 16, 21), and in some instances a secondary advantage can be conveyed by the AR genes in the absence of specific antimicrobial drug selection (15, 27, 36).
Previously we have shown that a young cohort of dairy cattle harbor a higher prevalence of antimicrobial drug-resistant Escherichia coli than do older cattle at the Washington State University (WSU) dairy. In that study the predominant AR phenotype was SSuT (resistant to streptomycin, sulfadiazine, and tetracycline and susceptible to ampicillin, chloramphenicol, and nalidixic acid) with these strains comprising ∼60% of all E. coli strains isolated from calves (<3 months old) (23). We also demonstrated that the absence of antimicrobial drug selection did not impact the level in antimicrobial drug-resistant E. coli, at least in the short term. Furthermore, compared to antimicrobial-susceptible strains, the SSuT E. coli strains had a significant fitness advantage in dairy calves but not in older cows. Thus, younger calves represented a clear reservoir for antimicrobial drug resistance genes on the dairy farm, but the mechanism(s) responsible for maintenance of these strains was not immediately apparent.
Clearly, determining mechanisms responsible for the selection and maintenance of AR E. coli may allow formulation of novel strategies to decrease the prevalence of AR bacteria. Our previous study showed that antimicrobial drug selection pressure is not needed to maintain AR strains of E. coli. In the present study we test the hypothesis that the AR genes harbored by the SSuT strains convey a secondary fitness advantage(s) that permits maintenance of these strains in the dairy population. To test this hypothesis, we generated null mutants and tested their ability to compete with other strains both in vitro and in vivo.
MATERIALS AND METHODS
Bacterial strains.
The strains used for the inoculation experiments were isolated from calves (1 to 45 days old) and cows (>1 year old). The strains represented two main AR patterns: “susceptible” (susceptible to ampicillin [16 μg/ml], chloramphenicol [16 μg/ml], nalidixic acid [18 μg/ml], streptomycin [12 μg/ml], sulfadiazine [512 μg/ml], and tetracycline [10 μg/ml]) and SSuT (resistant to streptomycin, sulfadiazine, and tetracycline in the same concentrations). Resistance phenotypes were determined as described below. In some cases strains with spontaneous mutations for nalidixic acid resistance (18 μg/ml) were used because the Nalr phenotype is a useful marker for these experiments. Selection of Nalr strains followed procedures described by Khachatryan et al. (23). Table 1 shows the distribution of these strains among three different experiments.
TABLE 1.
Strains used in vitro and in vivo
| Strain | In vitro competition | Calf expt in isolation | Farm expt | Source |
|---|---|---|---|---|
| SSuT 22 | X, Xc | Xb,c Xc | Xa | Calf |
| SSuT 25 | X, Xc | X,b,c Xc | Calf | |
| SSuT 32 | X | Calf | ||
| SSuT 35 | X | Xa | Calf | |
| SSuT 45 | X | Xa | Calf | |
| SSuT 79 | X | Xa | Calf | |
| SSuT 84 | X | Xa | Calf | |
| SSuT 98 | X | Xa | Calf | |
| Susceptible 66 | X, Xc | Xc | Calf | |
| Susceptible 68 | X, Xc | Xc | Calf | |
| Susceptible 69 | X, Xc | Xc | Calf | |
| Susceptible 71 | X, Xc | Xc | Calf | |
| Susceptible 562 | X | X | Calf | |
| Susceptible 559 | X | X | Calf | |
| Susceptible 561 | X | X | Calf | |
| Susceptible 572 | X | X | Cow | |
| Susceptible 579 | X | X | Cow | |
| Susceptible 580 | X | X | Cow |
Spontaneous susceptible null mutants of SSuT strains (cured SSuT) selected in medium supplemented with nickel chloride.
Cured-SSuT strains selected in medium supplemented with fusaric acid.
Nalidixic acid-resistant spontaneous mutant.
Selection for null mutants.
Because medium supplemented with fusaric acid and heat-inactivated chlortetracycline hydrochloride is toxic to E. coli when a functional tetracycline efflux pump is expressed (5, 28), we used this medium to select for strains of E. coli that spontaneously lose tetracycline and other antimicrobial drug resistance genes. Specifically two distinct SSuT Nalr (nalidixic acid-resistant) E. coli strains were separately incubated in the above-mentioned broth medium at 37.0°C on a shaker (200 rpm). Overnight culture (100 μl) was transferred into fresh medium (5 ml) for 14 consecutive days. On days 3, 6, 9, and 14, a dilution of overnight culture from each serial passage experiment was plated on a nonselective medium and grown overnight. The following day 96 colonies from each passage experiment were picked and tested for antimicrobial drug susceptibility using agar dilution at breakpoint concentrations (23). Antimicrobial drug (Sigma) susceptibilities were tested using IsoSensitest agar medium (Oxoid) supplemented with tetracycline (10 μg/ml), streptomycin (12 μg/ml), and sulfadiazine (512 μg/ml). Replicated test plates included a final plate of antimicrobial drug-free medium to confirm inoculum delivery. Three isolates from each SSuT strain that became susceptible to the above-mentioned antimicrobial drugs (called “cured SSuTs”) were chosen for further analysis. The analysis included validation of antimicrobial drug susceptibility by disk diffusion assay conforming to CLSI (formerly NCCLS) guidelines (26), screening for absence of AR genes by PCR [tet(B) (8), strA and strB (16), and sul-2 (16)] and pulsed-field gel electrophoresis (PFGE) using the XbaI enzyme (12) to confirm similarity to the progenitor resistant strain. Quality control organisms used for the disk diffusion assay included E. coli ATCC 25922, Staphylococcus aureus ATCC 25923, Pseudomonas aeruginosa ATCC 27853, and E. coli ATCC 10536.
Medium supplemented with nickel chloride and heat-inactivated chlortetracycline hydrochloride has also been shown to be effective for selecting tetracycline-resistant null mutants (34, 42). Therefore, we also used this medium to select for susceptible strains of E. coli that had spontaneously lost tetracycline and other antimicrobial drug resistance genes. Ten distinct SSuT E. coli strains were separately incubated in LB (Luria-Bertani) broth supplemented with 0.6 mM nickel chloride and 50-μg/ml heat-inactivated (121°C for 20 min) chlortetracycline hydrochloride at 37.0°C on a shaker (200 rpm). Overnight culture (100 μl) was transferred into fresh medium (5 ml) for six consecutive days. Three antimicrobial drug-susceptible isolates (cured SSuTs) generated from each strain were selected and confirmed as described for the fusaric acid selection method.
In vitro competition experiments.
Each cured-SSuT isolate that was generated from SSuT strains was subjected to in vitro competition (23) (i) with a mixture of five wild-type susceptible E. coli strains isolated from calves, (ii) with its progenitor resistant SSuT strain, and (c) with a progenitor resistant SSuT strain that was acquired from the final passage in corresponding selective medium (“medium-adapted progenitor”). After overnight incubation at 37.0°C on a shaker (200 rpm), equal mixtures of cured-SSuT and competitor strains were combined and serially passaged (10 μl) into fresh LB broth (3 ml) for eight consecutive days. On day 8, a competition index (CI) was calculated by estimating the CFU/ml for the resistant and susceptible strains. The number of resistant colonies was counted on fresh LB agar supplemented with streptomycin (12 μg/ml), whereas the number of susceptible colonies was determined by subtracting the number of resistant colonies from the same dilution grown on Luria-Bertani agar medium without antimicrobial drugs. Three replicate counts were made and averaged. The CI was calculated as (X − Y)/(X + Y), where X was the number of resistant colonies and Y was the number of susceptible colonies. CI values approaching +1 indicated dominance by resistant strains, whereas CI values approaching −1 indicated dominance by susceptible strains.
In vivo competition experiments between SSuT and cured-SSuT strains.
In vivo competition experiments were conducted in biosafety level 2 isolation rooms and included six neonatal calves in the experimental group (24 to 48 h old) and three neonatal calves in the control group (24 to 48 h old). Neonatal calves in the experimental group were inoculated per os with two strains of cured-SSuT (antimicrobial drug-susceptible null mutant) Nalr E. coli and their progenitor SSuT Nalr E. coli strains (109 CFU of each). Neonatal calves in the control group were inoculated per os with two wild-type antimicrobial drug-susceptible (Nalr) strains and two SSuT (Nalr) E. coli strains (109 CFU of each). Neonatal calves were sampled every other day until shedding of inoculum bacteria was not detectable (up to 21 days postinoculation). To determine CFU/ml for SSuT and susceptible E. coli in in vivo experiments, 1 g of freshly collected fecal samples was serially diluted in peptone-buffered saline and 10-fold dilutions were plated on specific selective medium. CI values were calculated as described above where colony counts were averaged within animal (in vivo) across time points, and the null hypothesis (CI = 0) was tested using Student's t test. All in vivo inoculation studies were approved by the WSU Institutional Animal Care and Use Committee.
In vivo inoculation of antimicrobial drug-susceptible strains at a dairy farm.
In vivo inoculation experiments were carried out at the Holstein dairy farm (WSU, Pullman, Wash.). The farm is a closed dairy herd (n = 167), where all the replacement heifers are raised on-site. Calves are separated from their mothers 24 to 48 h after birth and moved to a separate building, where they are housed in individual pens. Calves are weaned at 4 to 6 weeks of age but stay in the calf pens for 90 days, after which they are moved to a separate heifer barn. The dairy follows accepted biosecurity standards in order to prevent introduction of pathogens from outside. All the food is raised off-site and purchased commercially. No antimicrobial drugs were used for chemotherapeutics throughout the duration of the experiment in calves.
The in vivo inoculation experiment at the farm involved 30 neonatal calves randomly assigned to one of three groups. Group 1 received six cured-SSuT strains per os (2 × 109 CFU of each) once within the first 2 days of life (24 to 36 h) and the second time 7 days later. Group 2 received six antimicrobial drug-susceptible wild-type strains in a way similar to group 1. Group 3 received a volumetric equivalent of sterile medium similar to the other two groups. Each calf was sampled every 5 days for 3 months. Fecal samples were collected with sterile tongue depressors and placed into sterile bags. Collected samples were streaked for isolation on three separate plates of violet red bile agar (Remel, Kansas) with 4-methylumbelliferyl-β-d-glucuronide (MUG; Biosynth Ag, Switzerland) within 4 h after collection and were incubated overnight at 37°C. Twenty-one presumptive E. coli colonies (pink coloration and fluorescence under UV light) per animal, seven per sample plate, were used to inoculate E. coli medium (Remel) with MUG broth (200 μl) in a 96-well plate format, leaving 8 to 24 noninoculated, negative-control wells. Each 96-well plate also included two positive-control isolates, Q-89 and Q-90, resistant and susceptible to all tested antimicrobial drugs, respectively. The 96-well plates were then incubated at 44.5°C overnight, and the MUG reaction was confirmed under UV light.
Presumptive E. coli strains were tested for antimicrobial drug susceptibility using agar dilution at breakpoint concentrations (23). Antimicrobial drug (Sigma) susceptibilities were tested using Mueller-Hinton agar medium (Hardy Diagnostics) supplemented with ampicillin (16 μg/ml), tetracycline (10 μg/ml), chloramphenicol (16 μg/ml), streptomycin (12 μg/ml), sulfadiazine (512 μg/ml), and nalidixic acid (18 μg/ml). Replicated test plate series included a final plate of antimicrobial drug-free medium to confirm inoculum delivery. The results of replicator assays were recorded after overnight incubation at 37°C. Results for antimicrobial drug plates were coded as a dichotomous variable, 0 for no growth and 1 for growth. These results were used to calculate the frequencies for different resistance patterns.
Data were entered and analyzed with Microsoft Excel and NCSS 2001 (NCSS Statistical Software, Kaysville, UT). The Student-Newman-Keuls multiple comparison test was used to test for statistical differences in the prevalence of resistant bacteria between groups.
RESULTS
Spontaneous loss of antimicrobial drug resistance genes in strains of E. coli.
Medium supplemented with either fusaric acid or nickel chloride allowed selection for loss of tetracycline and associated streptomycin and sulfadiazine resistance genes. Ten SSuT strains were passaged, and on days 6, 10, and 14, 96 isolates from each tube were assessed for antimicrobial resistance. Medium supplemented with 0.6 mM nickel chloride was more efficient (90%; 6 days) in selecting detectable levels of antimicrobial drug-susceptible strains (cured SSuTs) than medium supplemented with fusaric acid (21%; 14 days). These results were confirmed by PCR for tet(B), sul-2, strA, and strB resistance genes. Medium supplemented with fusaric acid also selected for isolates missing only the tetracycline resistance gene (72%; 14 days), isolates missing tetracycline and streptomycin resistance genes (2%; 14 days), and isolates missing tetracycline and sulfadiazine resistance genes only (1%: 14 days). These results were confirmed using a disk diffusion assay. The close correspondence between loss of tetracycline resistance genes and the loss of other AR genes suggests close physical linkage between these genes on a genetic unit (plasmid, transposon, or phage).
In vitro competition experiments with cured-SSuT strains.
In vitro competition experiments tested the cured-SSuT strains that were selected in two different selective media. From each SSuT strain three susceptible isolates were tested to control for spontaneous changes unique to individual bacteria. The three cured-SSuT isolates were tested in competition experiments individually and as a mixture.
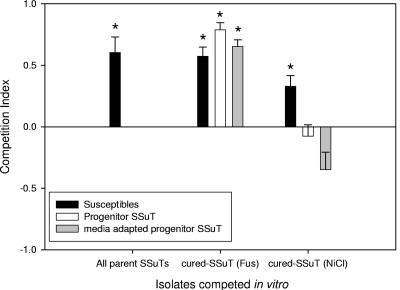
Cured-SSuT isolates (n = 2) selected in medium supplemented with fusaric acid [cured-SSuT (Fus)] consistently outcompeted progenitor SSuT strains, progenitor SSuT strains that retained their AR genes after a 14-day passage in broth culture with fusaric acid (“medium-adapted progenitors”), and wild-type susceptible strains (Student's one-sample t test; P < 0.001) (Fig. 1). The cured-SSuT isolates (n = 8) selected in medium supplemented with nickel chloride [cured-SSuT (NiCl)] outcompeted wild-type susceptible strains (Student's one-sample t test; P < 0.001) but were equal in competitive ability with progenitor AR SSuT strains (Student's one-sample t test; P = 0.413) and with medium-adapted progenitors (6-day passage in broth with nickel chloride) (Student's one-sample t test; P = 0.021) (Fig. 1). At the individual strain level, competition results were variable (Table 2). This variation is consistent with a diverse background of E. coli strains harboring the SSuT genes.
FIG. 1.
In vitro competition experiments with SSuT and cured SSuT strains with mixtures of susceptible (black bars), progenitor SSuT (white bars), and passaged parent SSuT (gray bars) strains. CI = (X − Y)/(X + Y); when values approach +1, strains listed on the x axis dominate (*, dominant strains significantly >0; one-sample t test, P < 0.05). When CI is 0, there is an equal proportion of the competing resistance patterns.
TABLE 2.
Results of the in vitro competition experiments between cured-SSuT and wild-type susceptible, progenitor SSuT, or “medium-adapted progenitor” SSuT strains (passaged in NiCl2)a
| Strain from which cured-SSuT isolate derived | Competitiveness vs:
|
||
|---|---|---|---|
| Wild-type susceptible strains | Progenitor SSuT strains | “Medium-adapted progenitor” SSuT strains (NiCl2) | |
| SSuT 22 | +,+,+ | −,+/−,+ | +,−,+ |
| SSuT 25 | −,−,− | −,−,− | −,−,+ |
| SSuT 32 | +,+,+ | −,−,− | −,−,+ |
| SSuT 35 | +,+,+ | +,+,+ | −,+,− |
| SSuT 45 | +,−,+ | −,−,− | −,−,− |
| SSuT 79 | +/−,+,+ | +,+,+ | +,−,− |
| SSuT 84 | −,−,− | +,+,+ | −,−,− |
| SSuT 98 | +,+,+ | −,−,− | −,−,− |
Results are shown for the three cured SSuT isolates derived from each strain. +, cured-SSuT strains were more competitive (competition index < 0.5); −, cured-SSuT strains were less competitive (competition index > −0.5); +/−, cured-SSuT strains were equally competitive (competition index between 0.5 and −0.5).
In vivo competition between cured-SSuT and SSuT strains in isolation.
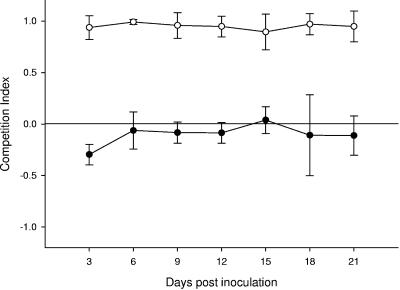
We used a calf challenge model to determine if cured-SSuT strains maintained their competitive advantage in vivo. Newborn calves (24 to 48 h) were inoculated with Nalr strains of cured-SSuT E. coli and Nalr strains of SSuT E. coli (109 CFU of each) (Table 1). Consistent with previously published findings (23), SSuT Nalr E. coli in the control group outcompeted wild-type susceptible Nalr strains in neonatal calves (Student's t test; P < 0.001) (Fig. 2). Cured-SSuT Nalr E. coli strains were generally more competitive than parent SSuT strains in the neonatal intestinal environment (Student's t test; P = 0.002) (Fig. 2). A direct comparison of cured-SSuT and susceptible strains would be possible only if we introduced an additional antimicrobial selection marker. Nevertheless, because cured-SSuT strains were at least as competitive as progenitor strains, we inferred that the cured-SSuT strains would also outcompete susceptible strains in this calf model. Shedding of Nalr E. coli from the experimental animals was not detectable 21 days after inoculation. None of the animals in the in vivo studies shed detectable fecal Nalr E. coli prior to inoculation (0 to 3 days).
FIG. 2.
Competition experiments in neonatal calves: for the control group (n = 3), competition between SSuT Nalr and wild-type susceptible Nalr strains (open circles), and for the experimental group (n = 6), competition between SSuT Nalr and cured-SSuT (susceptible) Nalr strains (closed circles). Each circle represents the mean with the 95% confidence interval bars for the time point shown on the x axis. CI = (X − Y)/(X + Y), where X is the number of resistant colonies and Y is the number of susceptible colonies. When CI is 0, there is an equal proportion of the competing resistance patterns.
Inoculation of antimicrobial drug-susceptible strains at a dairy farm.
Because the select cured-SSuT strains from the isolation experiment maintained competitive fitness in vivo, we tested the hypothesis that the cured-SSuT strains (selected in broth supplemented with nickel chloride) could displace naturally occurring SSuT strains in a farm environment. New cured-SSuT strains were used because (i) nalidixic acid-susceptible strains were to be used in the field environment and (ii) since nickel chloride-supplemented medium selects for cured-SSuT strains more efficiently, there is less time for adaptation of the strains to the in vitro medium environment. Cured-SSuT strains that were generally more competitive than their progenitor strains in vitro were used in vivo.
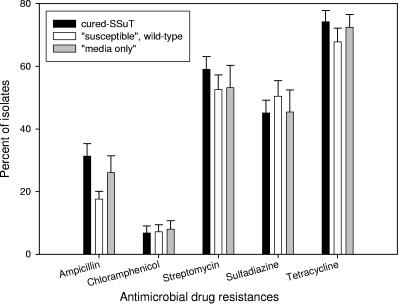
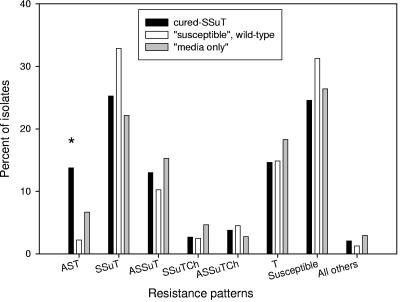
Neonatal calves were inoculated with either six cured-SSuT strains (Nals) (Table 1) or six wild-type susceptible strains (Nals) (Table 1) or “medium only” within 24 to 36 h of birth and 7 days later. We assessed percentages of resistant bacteria two times weekly for 3 months under the hypothesis that competition exclusion would lead to a significant decrease in SSuT strains within the cured-SSuT treatment group. Among the three inoculation groups, however, there was no statistically significant difference in the level of antimicrobial drug resistance to ampicillin, chloramphenicol, streptomycin, sulfadiazine, and tetracycline (Fig. 3). There was no statistically significant difference among the three inoculation groups for main resistance patterns (<2%), except that the group inoculated with cured SSuTs had significantly more E. coli strains resistant to ampicillin, streptomycin, and tetracycline (Fig. 4). Thus, it appears that the cured-SSuT strains were not effective in displacing native SSuT strains at the dairy. One possible explanation for the absence of treatment effect was a selective sweep where our cured-SSuT strains could have contaminated and dominated all treatment groups. To test this hypothesis, we compared 30 susceptible isolates (acquired at 2 to 4 weeks postinoculation from the three groups [three animals per group]) to the inoculum ex-SSuT isolates by PFGE. All “susceptible” isolates (n = 10) from the group inoculated with cured-SSuT E. coli were indistinguishable from the cured-SSuT inoculum strain SSuT-98. None of the “susceptible” isolates from the other two groups shared a distinct fingerprint with any of the cured-SSuT inoculum strains. Also there were no identical fingerprints between the groups receiving wild-type susceptible E. coli and “medium only” (data not shown).
FIG. 3.
Frequency of antimicrobial drug resistance for all E. coli strains shed from calves for the cured-SSuT group (n = 3,415), “susceptible” wild-type group (n = 3,430), and “medium-only” group (n = 3,448).
FIG. 4.
Frequency of antimicrobial drug resistance patterns for all E. coli strains shed from calves for the cured-SSuT group (n = 3,415), “susceptible” wild-type group (n = 3,430), and “medium-only” group (n = 3,448). *, statistically significant (P < 0.05 with Student-Newman-Keuls multiple comparison test for seven tests). Resistance patterns are denoted by letters, where A is ampicillin, Ch is chloramphenicol, S is streptomycin, Su is sulfadiazine, and T is tetracycline. Susceptible isolates were susceptible to all above-mentioned antimicrobial drugs tested.
DISCUSSION
Antimicrobial drug resistance is a significant public health concern that calls for reducing the factors that contribute to the emergence and maintenance of antimicrobial drug resistance. Food animal production consumes large amounts of antimicrobial drugs and thus can serve as an important arena for emergence and maintenance of AR (1, 3, 46). Nevertheless, it is not clear that eliminating all nontherapeutic antimicrobial use in animal production will result in a significant reduction in the prevalence of AR bacteria (7, 10, 24, 25, 37, 41). This is because many factors unrelated to antimicrobial use are also implicated in the increase in prevalence of AR, including animal stress (30, 31), close linkage to other selectively advantageous genes (2, 9, 16, 21), plasmid addiction (14, 17), and secondary advantage conveyed by the AR genes (15, 27, 36).
Our previous work at the WSU dairy showed that calves shed more antimicrobial drug-resistant E. coli isolates than did older cattle, making them a clear reservoir of AR genes on a farm. In that study E. coli strains with the SSuT resistance pattern were shed at a much higher level from calves than from older cattle (∼60% versus ∼5%). A short-term removal of oxytetracycline from a dietary supplement did not affect the high prevalence of AR E. coli in calves, suggesting that other mechanisms were responsible for maintaining the SSuT strains at the dairy. That is, it is possible that the AR genes were being selected in the presence of naturally occurring antimicrobial analogs or that these genes conferred a secondary, but unrecognized, selective advantage in this environment.
Secondary advantages might include cases where the tet(B) proton pump interacts with substrates other than tetracycline and its analogs, or there might be a need for increased folate synthesis conveyed by the sul-2 gene. There might also be nonspecific but important phosphotransferase activity of the strA/strB genes. These potential benefits must be balanced with potential costs. For example, pioneering experiments by Lenski's group showed a fitness reduction for E. coli when the tet(B) gene is expressed and very little cost associated with the gene when it is not expressed (33). We determined that the SSuT strains from our study do not express the tet(B) gene in vitro except in the presence of a tetracycline analog (data not shown). This does not exclude the possibility that the gene will be expressed in the farm environment. There are very few data in the literature that specifically examine the effect of sul-2 (15) and strA/strB resistance genes on the fitness of bacteria.
To test the hypothesis that resistance genes convey secondary fitness advantages, we developed null mutant strains of SSuT E. coli (cured SSuT) and tested these in competition experiments (in vitro) and in isolation (in vivo). There was variation in the competition outcome for individual strains (Table 2), but on average the cured SSuT strains were at least as competitive as parental SSuT strains for in vitro and in vivo competition experiments. These data suggested that there are no significant secondary advantages attributable to the SSuT genes in calves. The study tested only the interaction between the calf and E. coli in isolation and did not account for possible environmental selective pressures that are probably present in a farm environment. It is also worth noting that the inoculum strains were observed for only 21 days postinoculation.
Based on the finding that the AR genes do not influence the competitive outcome in vitro and in vivo, we tested the hypothesis that inoculation of ex-SSuT and other susceptible strains would decrease the level (via competition exclusion) of AR E. coli shed from calves in the farm environment. Contrary to our prediction, the results of the study did not show any treatment effect. Interestingly, compared to the prevalence of AR E. coli in the year 2001 (23), there was a nearly 50% decrease in the number of SSuT E. coli strains in all inoculation groups for the present study and this may have significantly reduced the statistical power of our experiment. PFGE fingerprinting of susceptible isolates from all three groups (n = 30) showed that the reduced level of SSuT E. coli was not due to a selective sweep by any of the cured-SSuT or “susceptible” strains.
In conclusion, we have now dismissed two of the three hypotheses for the maintenance of SSuT strains at the WSU dairy. Neither direct selection from antimicrobial drugs nor secondary benefits from the AR genes are responsible for maintenance of SSuT strains at our study facility. The remaining hypothesis is that the gene(s) conferring a selective advantage to SSuT strains in dairy calves is located proximally to the resistance genes, probably associated with a horizontally transmissible element.
Acknowledgments
This project was funded in part by the Agricultural Animal Health Program (Animal Health Formula Funds), College of Veterinary Medicine, Washington State University, Pullman, WA, and by USDA NRI 2004-35201-14112 and USDA NRI 2001-35212-10844.
Special thanks go to John Swain, Melissa Krug, Stacey LaFrentz, Edward Kuhn, Melissa Oatley, Russell McClanahan, Marilyn Soule, the WSU Field Disease Investigation Unit, the WSU Dairy, and the Animal Resource Unit, Pullman, WA.
REFERENCES
- 1.Aarestrup, F. M. 1999. Association between the consumption of antimicrobial agents in animal husbandry and the occurrence of resistant bacteria among food animals. Int. J. Antimicrob. Agents 12:279-285. [DOI] [PubMed] [Google Scholar]
- 2.Aarestrup, F. M. 2000. Characterization of glycopeptide-resistant Enterococcus faecium (GRE) from broilers and pigs in Denmark: genetic evidence that persistence of GRE in pig herds is associated with coselection by resistance to macrolides. J. Clin. Microbiol. 38:2774-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aarestrup, F. M., H. Kruse, E. Tast, A. M. Hammerum, and L. B. Jensen. 2000. Associations between the use of antimicrobial agents for growth promotion and the occurrence of resistance among Enterococcus faecium from broilers and pigs in Denmark, Finland, and Norway. Microb. Drug Resist. 6:63-70. [DOI] [PubMed] [Google Scholar]
- 4.Aarestrup, F. M., A. M. Seyfarth, H. D. Emborg, K. Pedersen, R. S. Hendriksen, and F. Bager. 2001. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 45:2054-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochner, B. R., H. C. Huang, G. L. Schieven, and B. N. Ames. 1980. Positive selection for loss of tetracycline resistance. J. Bacteriol. 143:926-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, E. F., and D. L. Hartl. 1997. Recent horizontal transmission of plasmids between natural populations of Escherichia coli and Salmonella enterica. J. Bacteriol. 179:1622-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bywater, R. J. 2004. Veterinary use of antimicrobials and emergence of resistance in zoonotic and sentinel bacteria in the EU. J. Vet. Med. B Infect. Dis. Vet. Public Health 51:361-363. [DOI] [PubMed] [Google Scholar]
- 8.Call, D. R., M. K. Bakko, M. J. Krug, and M. C. Roberts. 2003. Identifying antimicrobial resistance genes with DNA microarrays. Antimicrob. Agents Chemother. 47:3290-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calomiris, J. J., J. L. Armstrong, and R. J. Seidler. 1984. Association of metal tolerance with multiple antibiotic resistance of bacteria isolated from drinking water. Appl. Environ. Microbiol. 47:1238-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaslus-Dancla, E., G. Gerbaud, M. Lagorce, J. P. Lafont, and P. Courvalin. 1987. Persistence of an antibiotic resistance plasmid in intestinal Escherichia coli of chickens in the absence of selective pressure. Antimicrob. Agents Chemother. 31:784-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook, R. 1999. EU ban on four antibiotic growth promoters. Vet. Rec. 144:158. [PubMed] [Google Scholar]
- 12.Davis, M. A., D. D. Hancock, T. E. Besser, and D. R. Call. 2003. Evaluation of pulsed-field gel electrophoresis as a tool for determining the degree of genetic relatedness between strains of Escherichia coli O157:H7. J. Clin. Microbiol. 41:1843-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desenclos, J. C., and D. Guillemot. 2004. Consequences of bacterial resistance to antimicrobial agents. Emerg. Infect. Dis. 10:759-760. [DOI] [PubMed] [Google Scholar]
- 14.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53:43-70. [DOI] [PubMed] [Google Scholar]
- 15.Enne, V. I., P. M. Bennett, D. M. Livermore, and L. M. Hall. 2004. Enhancement of host fitness by the sul2-coding plasmid p9123 in the absence of selective pressure. J. Antimicrob. Chemother. 53:958-963. [DOI] [PubMed] [Google Scholar]
- 16.Enne, V. I., D. M. Livermore, P. Stephens, and L. M. Hall. 2001. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357:1325-1328. [DOI] [PubMed] [Google Scholar]
- 17.Gerdes, K., P. B. Rasmussen, and S. Molin. 1986. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 83:3116-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helms, M., P. Vastrup, P. Gerner-Smidt, and K. Molbak. 2002. Excess mortality associated with antimicrobial drug-resistant Salmonella typhimurium. Emerg. Infect. Dis. 8:490-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinton, M., D. J. Hampson, E. Hampson, and A. H. Linton. 1985. The effects of oxytetracycline on the intestinal Escherichia coli flora of newly weaned pigs. J. Hyg. (London) 95:77-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson, C. R., P. J. Fedorka-Cray, J. B. Barrett, and S. R. Ladely. 2004. Effects of tylosin use on erythromycin resistance in enterococci isolated from swine. Appl. Environ. Microbiol. 70:4205-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kehrenberg, C., and S. Schwarz. 2001. Occurrence and linkage of genes coding for resistance to sulfonamides, streptomycin and chloramphenicol in bacteria of the genera Pasteurella and Mannheimia. FEMS Microbiol. Lett. 205:283-290. [DOI] [PubMed] [Google Scholar]
- 22.Kelly, L., D. L. Smith, E. L. Snary, J. A. Johnson, A. D. Harris, M. Wooldridge, and J. G. Morris, Jr. 2004. Animal growth promoters: to ban or not to ban? A risk assessment approach. Int. J. Antimicrob. Agents 24:205-212. [DOI] [PubMed] [Google Scholar]
- 23.Khachatryan, A. R., D. D. Hancock, T. E. Besser, and D. R. Call. 2004. Role of calf-adapted Escherichia coli in maintenance of antimicrobial drug resistance in dairy calves. Appl. Environ. Microbiol. 70:752-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klare, I., D. Badstubner, C. Konstabel, G. Bohme, H. Claus, and W. Witte. 1999. Decreased incidence of vanA-type vancomycin-resistant enterococci isolated from poultry meat and from fecal samples of humans in the community after discontinuation of avoparcin usage in animal husbandry. Microb. Drug Resist. 5:45-52. [DOI] [PubMed] [Google Scholar]
- 25.Langlois, B. E., K. A. Dawson, T. S. Stahly, and G. L. Cromwell. 1984. Antibiotic resistance of fecal coliforms from swine fed subtherapeutic and therapeutic levels of chlortetracycline. J. Anim. Sci. 58:666-674. [DOI] [PubMed] [Google Scholar]
- 26.Lorian, V. 1996. Antibiotics in laboratory medicine, 4th ed. Williams & Wilkins, Baltimore, Md.
- 27.Luo, N., S. Pereira, O. Sahin, J. Lin, S. Huang, L. Michel, and Q. Zhang. 2005. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc. Natl. Acad. Sci. USA 102:541-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maloy, S. R., and W. D. Nunn. 1981. Selection for loss of tetracycline resistance by Escherichia coli. J. Bacteriol. 145:1110-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazodier, P., and J. Davies. 1991. Gene transfer between distantly related bacteria. Annu. Rev. Genet. 25:147-171. [DOI] [PubMed] [Google Scholar]
- 30.Moro, M. H., G. W. Beran, R. W. Griffith, and L. J. Hoffman. 2000. Effects of heat stress on the antimicrobial drug resistance of Escherichia coli of the intestinal flora of swine. J. Appl. Microbiol. 88:836-844. [DOI] [PubMed] [Google Scholar]
- 31.Moro, M. H., G. W. Beran, L. J. Hoffman, and R. W. Griffith. 1998. Effects of cold stress on the antimicrobial drug resistance of Escherichia coli of the intestinal flora of swine. Lett. Appl. Microbiol. 27:251-254. [DOI] [PubMed] [Google Scholar]
- 32.Netherwood, T., R. Bowden, P. Harrison, A. G. O'Donnell, D. S. Parker, and H. J. Gilbert. 1999. Gene transfer in the gastrointestinal tract. Appl. Environ. Microbiol. 65:5139-5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen, T. N., Q. G. Phan, L. P. Duong, K. P. Bertrand, and R. E. Lenski. 1989. Effects of carriage and expression of the Tn10 tetracycline-resistance operon on the fitness of Escherichia coli K12. Mol. Biol. Evol. 6:213-225. [DOI] [PubMed] [Google Scholar]
- 34.Podolsky, T., S. T. Fong, and B. T. Lee. 1996. Direct selection of tetracycline-sensitive Escherichia coli cells using nickel salts. Plasmid 36:112-115. [DOI] [PubMed] [Google Scholar]
- 35.Poppe, C., L. C. Martin, C. L. Gyles, R. Reid-Smith, P. Boerlin, S. A. McEwen, J. F. Prescott, and K. R. Forward. 2005. Acquisition of resistance to extended-spectrum cephalosporins by Salmonella enterica subsp. enterica serovar Newport and Escherichia coli in the turkey poult intestinal tract. Appl. Environ. Microbiol. 71:1184-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pruss, G. J., and K. Drlica. 1986. Topoisomerase I mutants: the gene on pBR322 that encodes resistance to tetracycline affects plasmid DNA supercoiling. Proc. Natl. Acad. Sci. USA 83:8952-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rollins, L. D., S. A. Gaines, D. W. Pocurull, H. D. Mercer, and L. T. Frobish. 1976. Persistence of transferable drug resistance in the lactose-fermenting enteric flora of swine following antimicrobial feeding. Can. J. Comp. Med. 40:175-183. [PMC free article] [PubMed] [Google Scholar]
- 38.Seppala, H., T. Klaukka, J. Vuopio-Varkila, A. Muotiala, H. Helenius, K. Lager, P. Huovinen, et al. 1997. The effect of changes in the consumption of macrolide antibiotics on erythromycin resistance in group A streptococci in Finland. N. Engl. J. Med. 337:441-446. [DOI] [PubMed] [Google Scholar]
- 39.Shoemaker, N. B., H. Vlamakis, K. Hayes, and A. A. Salyers. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singer, R. S., R. Finch, H. C. Wegener, R. Bywater, J. Walters, and M. Lipsitch. 2003. Antibiotic resistance—the interplay between antibiotic use in animals and human beings. Lancet Infect. Dis. 3:47-51. [DOI] [PubMed] [Google Scholar]
- 41.Smith, H. W. 1975. Persistence of tetracycline resistance in pig E. coli. Nature 258:628-630. [DOI] [PubMed] [Google Scholar]
- 42.Stavropoulos, T. A., and C. A. Strathdee. 2000. Expression of the tetA(C) tetracycline efflux pump in Escherichia coli confers osmotic sensitivity. FEMS Microbiol. Lett. 190:147-150. [DOI] [PubMed] [Google Scholar]
- 43.Taylor, D. J. 1999. EU ban on four antibiotic growth promoters. Vet. Rec. 144:158. [PubMed] [Google Scholar]
- 44.Wierup, M. 2001. The Swedish experience of the 1986 year ban of antimicrobial growth promoters, with special reference to animal health, disease prevention, productivity, and usage of antimicrobials. Microb. Drug Resist. 7:183-190. [DOI] [PubMed] [Google Scholar]
- 45.Winokur, P. L., D. L. Vonstein, L. J. Hoffman, E. K. Uhlenhopp, and G. V. Doern. 2001. Evidence for transfer of CMY-2 AmpC β-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 5:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Witte, W. 2000. Selective pressure by antibiotic use in livestock. Int. J. Antimicrob. Agents 16:S19-S24. [DOI] [PubMed] [Google Scholar]