Abstract
The abundant and widespread coccolithophore Emiliania huxleyi plays an important role in mediating CO2 exchange between the ocean and the atmosphere through its impact on marine photosynthesis and calcification. Here, we use long serial analysis of gene expression (SAGE) to identify E. huxleyi genes responsive to nitrogen (N) or phosphorus (P) starvation. Long SAGE is an elegant approach for examining quantitative and comprehensive gene expression patterns without a priori knowledge of gene sequences via the detection of 21-bp nucleotide sequence tags. E. huxleyi appears to have a robust transcriptional-level response to macronutrient deficiency, with 42 tags uniquely present or up-regulated twofold or greater in the N-starved library and 128 tags uniquely present or up-regulated twofold or greater in the P-starved library. The expression patterns of several tags were validated with reverse transcriptase PCR. Roughly 48% of these differentially expressed tags could be mapped to publicly available genomic or expressed sequence tag (EST) sequence data. For example, in the P-starved library a number of the tags mapped to genes with a role in P scavenging, including a putative phosphate-repressible permease and a putative polyphosphate synthetase. In short, the long SAGE analyses have (i) identified many new differentially regulated gene sequences, (ii) assigned regulation data to EST sequences with no database homology and unknown function, and (iii) highlighted previously uncharacterized aspects of E. huxleyi N and P physiology. To this end, our long SAGE libraries provide a new public resource for gene discovery and transcriptional analysis in this biogeochemically important marine organism.
Coccolithophores are an abundant and widespread phytoplankton functional group responsible for significant amounts of calcification in the ocean. This group is intensively studied for its roles in the marine carbon and sulfur cycles, the production of alkenones, and marine calcification. The coccolithophore Emiliania huxleyi is the most abundant species of this functional group in the modern ocean, and it blooms in both coastal and open ocean regions (24). E. huxleyi both fixes CO2 through photosynthesis and generates CO2 through the biomineralization of calcium carbonate (calcification). Photosynthesis and calcification are important components of the global carbon (C) cycle. Ultimately, both the presence of E. huxleyi blooms and the ratio of photosynthesis to calcification within the population mediate exchange between atmospheric and oceanic CO2. As such, coccolithophores are being intensively studied for their role in the C cycle and their potential influence on global climate.
Nitrogen (N) and phosphorus (P) are two critical macronutrients for E. huxleyi growth, and their availability can impact when and where E. huxleyi blooms are able to occur (20). Further, N and P starvation can influence CO2 exchange by changing rates of photosynthesis and calcification (24). For example, P starvation typically increases calcification rates relative to photosynthesis (25). In short, N and P availability in the field may influence bloom dynamics, calcification, and their concomitant impact on C cycling and on the ocean's ability to buffer changing CO2 concentrations in the atmosphere.
To cope with low macronutrient availability in nature, marine phytoplankton have evolved inducible systems that enable them to efficiently scavenge dissolved inorganic N (DIN) and dissolved inorganic P (DIP), the concentrations of which are often growth limiting in marine systems. Phytoplankton also have the ability to utilize N and P from a diverse suite of dissolved organic N (DON) and P (DOP) compounds (1, 5). The concentrations of DON and DOP often exceed those of DIN and DIP in surface waters, so these organic compounds can be an important nutrient source in DIN- or DIP-depleted environments, such as the oligotrophic oceans. Understanding the complexity of phytoplankton nutrient scavenging systems and how they are expressed in response to depletion of N or P in the ocean is an ongoing area of study for biological oceanographers. Previous work with E. huxleyi cultures suggests that this coccolithophore has the ability to scavenge nitrogen from diverse sources. For example, it is able to grow on several DON substrates as a sole N source, including formamide, hypoxanthine, and urea (28). E. huxleyi is also able to scavenge P from diverse sources, expressing the enzyme alkaline phosphatase under low-DIP conditions allowing for the hydrolysis of certain DOP compounds (13, 32). In fact, E. huxleyi is known for being a good competitor relative to other algae in low-DIP systems and elevating phosphate uptake at growth-limiting DIP concentrations (32). Although some N and P starvation-inducible proteins have been identified for E. huxleyi (13, 29), our transcriptional understanding of E. huxleyi biology and particularly nutrient scavenging and nutrient starvation responses is limited.
While genomic research with marine cyanobacteria is rapidly advancing our understanding of their role in the sea (12, 27), there are few genome sequences (3), differential gene expression studies (2, 22, 37), and transcriptome analyses with eukaryotic marine algae. In the case of coccolithophores, fundamental gaps in our molecular-level understanding of calcification and even basic N and P scavenging mechanisms remain. Gene expression analyses are one way to work towards closing these gaps, providing a dynamic link between the E. huxleyi genome and its cellular functioning in the ocean.
Methods for assessing gene expression on a genomic scale include DNA microarrays, massively parallel signature sequencing, differential display reverse transcriptase PCR (RT-PCR), subtraction hybridization, and serial analysis of gene expression (SAGE) (38). In long SAGE (33), a short, 21-bp sequence tag from the most poly(A) proximal NlaIII restriction site of an mRNA molecule is used to uniquely identify the source gene from within the genome. Short sequence tags are sampled from all NlaIII-positive transcripts in an mRNA sample and are linked together to form long concatenated molecules that are cloned and sequenced. Quantification of all tags provides a relative measure of gene expression (i.e., mRNA abundance). SAGE thus provides both the identities of expressed genes and the levels of their expression. Detection level and sensitivity in SAGE are a function of sampling depth, i.e., the more tags sampled, the more likely detection of rare transcripts and the stronger statistical resolution of differential abundance of transcripts among mRNA samples. Of the aforementioned approaches, SAGE is advantageous in that it requires no a priori knowledge of the genome, responsive genes, or gene sequences and directly samples transcript abundance. The material benefits of SAGE are its simplicity, low cost, and efficiency. Perhaps the most common approach to expression studies with eukaryotic marine algae involves expressed sequence tag (EST) sequencing (34, 39, 40). In contrast to EST sequencing, SAGE can provide a much deeper sampling of the transcriptome, is not subject to the difficulties involved in normalization, and is quantitative in nature.
Despite its clear value, SAGE is typically applied to model systems with available genome sequences (e.g., mouse, Arabidopsis thaliana) and its utility in nontraditional models has only recently been addressed (11). Here we used long SAGE to examine genes responsive to N or P starvation by sampling 21-bp sequence tags from three E. huxleyi libraries: replete, nitrogen starved (−N), and phosphorus starved (−P). Because P starvation resulted in increased calcification, we were also able to screen the −P library for genes potentially responsive to changes in calcification. To our knowledge, these are the first transcriptome analyses of N and P starvation for a coccolithophore.
MATERIALS AND METHODS
Cell culture.
Emiliania huxleyi CCMP 1516 was obtained from the Provasoli-Guillard Center for the Culture of Marine Phytoplankton, Bigelow Laboratories. Cultures were grown at 18°C on a 14 h:10 h light:dark cycle (140 μmol quanta m−2 s−1). Nitrogen- and phosphate-replete (35 μM NO3− and 1.5 μM PO43−), −N (10 μM NO3−), and −P (0 μM PO43−) cells were grown in f/50 medium without Si (16). Locally collected seawater was filtered (pore size, 0.2 μm) and autoclaved. Filter-sterilized inorganic nutrients, trace metals, and vitamins (thiamine, biotin, and B12) were added after autoclaving. The cells were grown in 8-liter batch cultures. The growth of cultures was monitored daily by cell number counted with a hemacytometer and by relative fluorescence with a Turner Designs AU fluorometer. Replete cells were harvested in mid-log phase, while −N and −P cells were harvested at the onset of stationary phase. At the onset of stationary phase, inorganic nitrogen (35 μM NO3−) and inorganic phosphate (1.5 μM PO43−) were added to a 20-ml aliquot of −N cells and −P cells, respectively, and compared to a 20-ml aliquot of the cultures with no amendment. Nutrient samples were collected aseptically, filtered through a GF/F filter to remove cells, and collected into an acid-cleaned tube. All samples were stored frozen at −20°C until analysis and assayed throughout the experiment. Soluble reactive phosphate was assayed via the molybdate blue method (19), and nitrate was determined with a nutrient autoanalyzer by use of standard protocols. Calcification was assayed with a 10-ml aliquot of cells removed at the point of harvest from control, −N, and −P cultures as described elsewhere (26).
Total RNA extraction.
Approximately 2 × 107 cells were harvested (8,000 × g for 10 min at 22°C), and the RNA was extracted using TRI reagent (Sigma, St. Louis, MO) according to the manufacturer's instructions. Briefly, cell pellets were resuspended and lysed in 0.75 ml TRI reagent. After complete lysis of the cells, 0.15 ml chloroform was added. Total RNA was precipitated with 0.35 ml of isopropanol and washed with 75% (vol/vol) ethanol. The RNA pellets were solubilized in 50 μl diethyl pyrocarbonate-treated water. RNA concentrations were obtained with a UV spectrophotometer. Integrity of the total RNA was assessed by 1% (wt/vol) agarose gel electrophoresis.
SAGE.
SAGE libraries were constructed using ∼30 μg RNA isolated from extractions of replete, −P, and −N E. huxleyi cells following the I-SAGE Long kit protocol (Invitrogen, Carlsbad, CA). Recombinant pZEro1 clones produced by SAGE were purified using GeneMachines RevPrep Orbit (Genomic Solutions, Ann Arbor, MI) and were sequenced on an ABI 3730xl DNA sequencer (Applied Biosystems, Foster City, CA). Sequences collected were analyzed with software created at the Marine Biological Laboratory specifically for E. huxleyi SAGE analysis. The SAGE software extracts ditag sequences from the ABI 3730xl results according to the SAGE sequence grammar, parses out individual SAGE tags, excludes tags with sequence ambiguities, and reduces all SAGE tags to a look-up table of unique SAGE tag sequences and their observed frequencies among all of the E. huxleyi SAGE libraries. SAGE tags not found more than once in at least one SAGE library were excluded from analyses as putative sequencing errors, unless the tag sequence had a perfect match to available genomic and EST data. The unique tag sequences were mapped to all available E. huxleyi DNA sequences to determine the identities of expressed genes. For this, 11,880 genome and EST sequences were obtained from NCBI on 1 April 2005 and assembled with the computer program PHRAP (15). The 21-bp SAGE tags were mapped to assembled contigs and singletons based on exact sequence matches. Tags were then visualized with a generic model organism database using the Gbrowse interface (35) in the context of EST sequences and predicted open reading frames (ORFs). As SAGE tags are generated from the most 3′ NlaIII restriction site of transcripts, tags with significant differential expression among the SAGE libraries were manually assigned to predicted genes based on their positions relative to ESTs and predicted ORFs. These genes were annotated by BLAST and other homology detection methods. SAGE tags were scored for differential expression among the three libraries by using the R statistic of Stekel et al. (36), a log likelihood ratio statistic which scores tags by their deviation from the null hypothesis of equal frequencies given the tag sampling depth for each SAGE library. Higher scores represent a greater deviation from the null hypothesis, while scores close to zero represent near constitutive expression. To reduce the effects of sampling error in highlighting differential expression, only tags with an R value of 2 or greater were considered for RT-PCR and other post-SAGE analysis.
RT-PCR.
RNA was extracted from cultures as described above. Genomic DNA contamination was removed from the replete, −N, and −P RNA samples with a Turbo DNA-free kit (Ambion, Austin, TX) according to the directions provided. Equivalent concentrations of total RNA from each condition were reverse transcribed to single-stranded cDNA by using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) and amplified using gene-specific primers (Table 1). All genes and conditions were tested for genomic contamination using no RT controls. PCR reaction conditions were as follows: 15-μl total volume of 1× PCR buffer, 0.25 mM deoxynucleoside triphosphates (Bio-Rad), 0.3 μM primers, 1 U Taq DNA polymerase (Bio-Rad) or Optimase high-fidelity polymerase (Transgenomic, Cambridge, MA), and 1 ng cDNA. PCR conditions were as follows: 1 cycle at 95°C for 5 min; 30 cycles at 95°C for 1 min, x°C for 1 min, and 72°C for 1 min; and 1 cycle at 72°C for 7 min. See Table 1 for annealing temperatures and amplicon sizes. PCR products were resolved on a 2% agarose gel, stained with ethidium bromide, and imaged with a ChemiImaging system (Alpha Innotech, San Leandro, CA).
TABLE 1.
PCR primers and amplification conditions for the genes used in the RT-PCR validation of the long SAGE analysisa
| Tag no./gene | Primer sequence | Annealing temp (°C) | Amplicon size (bp) |
|---|---|---|---|
| 45 | 5′GCATCACGTTGACGTAGTCG | 56.2 | 267 |
| 3′GGAGTTCGCCTCACTGATTC | |||
| 285 | 5′AGCTTGAAGGTCTTGGTGGA | 63 | 115 |
| 3′CGGGCTACTTCATCTTCGAG | |||
| 12112 | 5′GATCATCGAGAGCACACCAA | 59.5 | 286 |
| 3′GATTCCGAAGATGACCCTCA | |||
| Type 1 actin (Q41205) | 5′GATCTGGCACCACACCTTCT | 55.5 | 116 |
| 3′TGATCTGCGTCATCTTCTCG |
PCR conditions are given in the text.
RESULTS
Culture physiology.
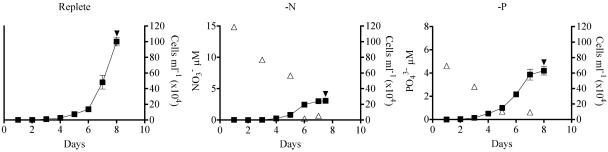
Any transcriptome analysis is highly dependent on the experimental culture conditions. Here we manipulated the dissolved inorganic N and P concentrations in batch cultures of E. huxleyi CCMP 1516 to induce N or P starvation and potentially capture the up-regulation of genes involved in N and P stress responses. Growth curves are presented for each condition (Fig. 1). At the onset of stationary phase, nitrate was drawn down in the −N culture and phosphate was drawn down in the −P culture (Fig. 1). The −N and −P cell cultures were split at the onset of stationary phase and re-fed with the appropriate nutrient to f/50 levels. Both of the re-fed cultures resumed growth relative to a no-addition control, confirming N and P starvation conditions (data not shown). Calcification was detectable under all conditions but enhanced nearly threefold in the −P culture relative to the −N and replete cultures (Table 2).
FIG. 1.
Growth curves of E. huxleyi under replete, −N, and −P conditions. Graphs are plotted as cell number (filled symbols) and nutrient concentration (open symbols) versus day in culture, noting the difference in scale of the y axes. Error bars denote standard errors of the means (n = 4). Filled arrowhead indicates point of harvest.
TABLE 2.
Summary of library characteristics, including calcification data and long SAGE analyses
| Library condition | % Calcification | Total no. of tags sampled | Total no. of tag sequences | No. of tagsa
|
|||
|---|---|---|---|---|---|---|---|
| Up-regulated twofold or greater | Down-regulated twofold or greater | Uniquely present | Uniquely absent | ||||
| Replete | 3.09 | 14,089 | 4,306 | 5 | 8 | ||
| −N | 3.57 | 15,055 | 4,219 | 38 | 31 | 4 | 37 |
| −P | 9.84 | 11,578 | 3,817 | 74 | 45 | 54 | 30 |
Numbers include only those tags with R values of 2 or greater.
SAGE analyses.
Correcting for sequencing error, we have sampled 11,000 to 15,000 long SAGE tags from each library and detected 3,817 to 4,306 unique tags per library (Table 2). With the use of log likelihood statistics (R ≥ 2) to reduce the effects of sampling error, we identified a total of 38 unique tags that were up-regulated twofold or greater in the −N library and 74 unique tags that were up-regulated twofold or greater in the −P library (Table 2). In addition, a total of 4 tags were uniquely present in the −N library, and 54 tags were uniquely present in the −P library (Table 2). Approximately 48% of all differentially expressed tags could be mapped to publicly available sequence data. Expression data for tags that map to genes with a putative function are presented in Table 3. The remaining tags either map to E. huxleyi sequence data with no database homology, match more than one EST or genome sequence (unresolved tags), or do not have a sequence match (orphan tags) and apparently represent nascent E. huxleyi sequences (Table 4) (also see the supplemental material). The five most highly expressed (i.e., greatest change [n-fold]) tags encompass these tag categories (Table 4). The complete data set including all of the differentially expressed genes is available in Table S1 in the supplemental material. Additional information on all the gene tags, including those that are not differentially expressed, is available on our website (http://gmod.mbl.edu/emiliania_huxleyi).
TABLE 3.
Successfully annotated differentially expressed (R ≥ 2) SAGE tags
| Tag IDa | Tag sequence | No. of copiesb in:
|
R value | Change (n-fold)c or result for:
|
Annotation | |||
|---|---|---|---|---|---|---|---|---|
| −N library | Replete library | −P library | −N library | −P library | ||||
| 1064 | CATGGGATAATGAAATAGGAC | 21 | 41 | 137 | 32.03 | −2.09 | 4.07 | Probable 18S rRNA transcript |
| 24856 | CATGGCGTTAACGGGTAACGG | 28 | 50 | 137 | 26.78 | −1.91 | 3.33 | Probable 18S rRNA transcript |
| 11533 | CATGATTTGTCGACCGAAGAT | 0 | 2 | 40 | 19.30 | Uniquely absent | 24.34 | Putative fructose-1,6-bisphosphate aldolase |
| 11736 | CATGCTAATTTTTAAAAGAAA | 0 | 1 | 38 | 19.22 | Uniquely absent | 46.24 | Mitochondrial-encoded large-subunit rRNA |
| 24531 | CATGCAAGTCGAACGAGAGTT | 0 | 6 | 37 | 15.45 | Uniquely absent | 7.50 | Plastid-encoded small-subunit rRNA |
| 289 | CATGCCGACTAGGGATTGGAG | 20 | 29 | 75 | 12.49 | −1.55 | 3.15 | Probable 18S rRNA transcript |
| 11766 | CATGGTTGTCGTCAGTTCGTG | 0 | 0 | 21 | 11.48 | NA | Uniquely present | Mitochondrial-encoded large-subunit rRNA |
| 25306 | CATGGCCGTTCTTAGTTGGTG | 40 | 71 | 104 | 10.68 | −1.90 | 1.78 | Probable 18S rRNA transcript |
| 2053 | CATGTTTTCCGTGCCGGTCTG | 9 | 7 | 39 | 9.28 | 1.20 | 6.78 | Putative chloroplast ferredoxin |
| 25385 | CATGTTGGCTAGTGTGATCTT | 8 | 14 | 43 | 9.09 | −1.87 | 3.74 | Putative light-harvesting complex protein |
| 285 | CATGCGCCGATTCGTGCGCAC | 12 | 47 | 41 | 6.92 | −4.19 | −0.94 | Putative fucoxanthin chlorophyll a/c binding protein |
| 24557 | CATGGATAGTATAGAGAGTCC | 1 | 2 | 16 | 5.68 | −2.14 | 9.74 | Putative polyphosphate synthetase |
| 39 | CATGATGGCCAAGTAGGCTCC | 34 | 40 | 5 | 5.58 | −1.26 | −6.57 | Putative calmodulin |
| 30 | CATGGCTGCGATGTACGACCC | 35 | 16 | 3 | 5.32 | 2.05 | −4.38 | Putative polyphosphoinositide-binding protein |
| 196 | CATGGGAGCGCAGGACGGCGC | 3 | 5 | 20 | 4.96 | −1.78 | 4.87 | Putative fucoxanthin chlorophyll a/c binding protein |
| 12215 | CATGTGGAGTGTCCTCCTTTT | 0 | 0 | 9 | 4.92 | NA | Uniquely present | Putative enolase (2-phosphoglycerate dehydratase) |
| 12088 | CATGAATTGGGTTTAAAACGA | 0 | 0 | 9 | 4.92 | NA | Uniquely present | Mitochondrial-encoded large-subunit rRNA |
| 25078 | CATGTAGAGGTATTCTACACC | 11 | 20 | 36 | 4.81 | −1.94 | 2.19 | Putative fucoxanthin chlorophyl a/c protein |
| 25102 | CATGCCCCTTATGCCCTGGGC | 1 | 4 | 15 | 4.51 | −4.27 | 4.56 | Plastid-encoded small-subunit rRNA |
| 3320 | CATGTGTTGTTGTTTTGGATA | 3 | 14 | 21 | 4.44 | −4.99 | 1.83 | Putative fucoxanthin chlorophyll a/c binding protein |
| 25010 | CATGGCTGTCGTCAGCTCGTG | 1 | 8 | 16 | 4.41 | −8.55 | 2.43 | Plastid-encoded small-subunit rRNA |
| 12892 | CATGCAACATTTGTAAAAATG | 0 | 0 | 8 | 4.38 | NA | Uniquely present | Mitochondrial-encoded large-subunit rRNA |
| 25026 | CATGCTGTGTGAAGGGGCACA | 4 | 10 | 22 | 4.28 | −2.67 | 2.68 | Putative phosphoglycerate kinase |
| 9469 | CATGCGAGCCTTCTCGTAGGC | 0 | 3 | 11 | 4.24 | Uniquely absent | 4.46 | Phosphate-repressible phosphate permease |
| 9481 | CATGTGTTTTTAATAAACAGT | 0 | 2 | 10 | 4.04 | Uniquely absent | 6.08 | Mitochondrial-encoded large-subunit rRNA |
| 25244 | CATGCAGGAGTTCCCGACTCA | 12 | 16 | 33 | 4.02 | −1.42 | 2.51 | Probable 18S rRNA transcript |
| 8393 | CATGCCGATATGTTGTCTGCC | 1 | 0 | 9 | 3.94 | Uniquely absent in replete | Uniquely absent in replete | Putative 30S ribosomal protein S1 |
| 24482 | CATGGGAGCTGGTCATACCCA | 0 | 3 | 10 | 3.80 | Uniquely absent | 4.06 | Plastid-encoded small-subunit rRNA |
| 1484 | CATGGGCTTCGTCTCGGAGGC | 4 | 12 | 20 | 3.55 | −3.21 | 2.03 | Putative fucoxanthin chlorophyll a/c binding protein |
| 5002 | CATGTATGTATAAATTACTCG | 1 | 0 | 8 | 3.44 | Uniquely absent in replete | Uniquely absent in replete | Putative sulfate adenylyltransferase (ATP-sulfurylase) |
| 4313 | CATGTCGTGTGTTTGTGTCCT | 2 | 0 | 8 | 3.07 | Uniquely absent in replete | Uniquely absent in replete | Putative chlorophyll a/b binding protein |
| 37 | CATGTCGTGGCAGGCCTTTGT | 45 | 24 | 12 | 2.93 | 1.75 | −1.64 | Putative calcium-dependent protein kinase |
| 191 | CATGGGTGCCGTGCGCCGGGG | 17 | 40 | 15 | 2.89 | −2.51 | −2.19 | Putative ribosomal protein S15 |
| 2203 | CATGGCCTTCGTCTCCGAGTC | 6 | 11 | 0 | 2.87 | −1.96 | Uniquely absent | Putative fucoxanthin chlorophyll a/c binding protein |
| 271 | CATGTCGCCCTGCCCAGCGCC | 11 | 8 | 0 | 2.82 | 1.29 | Uniquely absent | Similarity to a number of hypothetical proteins |
| 10410 | CATGATTCTTGATCGTGGCGC | 0 | 0 | 5 | 2.73 | NA | Uniquely present | Putative photosystem II stability/assembly factor HCF136 |
| 11952 | CATGATCAACGAGGTTGACGC | 0 | 0 | 5 | 2.73 | NA | Uniquely present | Putative calmodulin |
| 255 | CATGGGGTTTACATATGCACT | 14 | 4 | 1 | 2.59 | 3.28 | −3.29 | Putative P450 protein |
| 639 | CATGCTCAACGAGGACGAGCT | 7 | 20 | 4 | 2.54 | −3.05 | −4.11 | ADP-ribosylation factor |
| 1714 | CATGGAGTGCGTTTAGCGAGA | 3 | 12 | 1 | 2.49 | −4.27 | −9.86 | Putative acetyl-coenzyme A:acetoacetyl-coenzyme A transferase |
| 18 | CATGATACCCCGTTTGTGCGA | 43 | 19 | 36 | 2.43 | 2.12 | 2.31 | Putative nitrate transporter |
| 5573 | CATGCGATTGTGCGTCAGTGA | 1 | 6 | 10 | 2.42 | −6.41 | 2.03 | Putative RAB11A (member RAS oncogene family) |
| 470 | CATGTGTGCTCCCGCATACGC | 6 | 9 | 0 | 2.35 | −1.60 | Uniquely absent | Putative Myb-related domain |
| 5434 | CATGTAGACGCGTCTGTACAG | 0 | 5 | 0 | 2.30 | Uniquely present in replete | Putative glutaredoxin-related protein | |
| 2558 | CATGTACCGATCGCCGCTACG | 6 | 7 | 17 | 2.30 | −1.25 | 2.96 | Putative light-harvesting chlorophyll a/b protein of photosystem I |
| 90 | CATGGATAGAAGAGGCGCTGC | 38 | 20 | 11 | 2.27 | 1.78 | −1.49 | Possible sterol desaturase-related protein |
| 1848 | CATGCAAGAGCGACTGCCTGC | 0 | 4 | 6 | 2.20 | Uniquely absent | 1.83 | Probable 18S rRNA transcript |
| 12112 | CATGAGGAGAGCGACACTTTG | 0 | 0 | 4 | 2.19 | NA | Uniquely present | Putative inorganic pyrophosphatase |
| 12181 | CATGTCCATCCAGGGCAAGTC | 0 | 3 | 6 | 2.18 | Uniquely absent | 2.43 | Putative photosystem I subunit X |
| 1388 | CATGACCGGGCCGAGAGGGAT | 5 | 0 | 0 | 2.16 | Uniquely present | NA | Putative GDP dissociation inhibitor |
| 4062 | CATGCAGCGCAGCGCGCGTCG | 1 | 4 | 9 | 2.15 | −4.27 | 2.74 | Putative acyl-coenzyme A-desaturase with similarity to 4-methyl-5(B-hydroxyethyl)-thiazole |
| 10890 | CATGCAGGGTGTCGTGCAGTC | 0 | 1 | 5 | 2.02 | Uniquely absent | 6.08 | Monophosphate biosynthesis protein |
| 17129 | CATGCTCGTCTGCCCGACCGA | 0 | 1 | 5 | 2.02 | Uniquely absent | 6.08 | Putative U6 snRNA-associated Sm-like protein LSm7 |
The tag identification (ID) can be used to view the data on a particular tag at http://gmod.mbl.edu/emiliania_huxleyi.
Raw frequencies in each SAGE library are presented for each tag.
Change (n-fold) includes correction for sampling depth, with negative symbols used for down-regulation. NA, tag was not detected.
TABLE 4.
Differentially (R ≥ 2) expressed tags with the greatest change (n-fold) in the −N and −P libraries
| Tag IDa | Tag sequence | No. of copiesb in:
|
R value | Change (n-fold)c or result for:
|
Annotationd | |||
|---|---|---|---|---|---|---|---|---|
| −N library | Replete library | −P library | −N library | −P library | ||||
| 11736 | CATGCTAATTTTTAAAAGAAA | 0 | 1 | 38 | 19.22249452 | Uniquely absent | 46.24131974 | Mitochondrial-encoded large-subunit rRNA |
| 1855 | CATGATTGTTAAGAAAGCGCA | 1 | 1 | 32 | 14.48724537 | −1.068564128 | 38.94005873 | Unresolved SAGE tag |
| 11533 | CATGATTTGTCGACCGAAGAT | 0 | 2 | 40 | 19.30477518 | Uniquely absent | 24.33753671 | Putative fructose-1,6-bisphosphate aldolase |
| 25408 | CATGAACTACTTCTTGAATTC | 0 | 1 | 19 | 9.127231112 | Uniquely absent | 23.12065987 | Orphan SAGE tag |
| 25238 | CATGGCCAGTGTGAAGCTTAG | 1 | 2 | 35 | 15.10682258 | −2.137128256 | 21.29534462 | Orphan SAGE tag |
| 1083 | CATGAGCTACCTTTACGATGA | 11 | 1 | 1 | 2.731229859 | 10.29418798 | 0.821775854 | Orphan SAGE tag |
| 294 | CATGGTTAGGCGAGTGGCAGT | 11 | 1 | 2 | 2.31564119 | 10.29418798 | 2.433753671 | Orphan SAGE tag |
| 827 | CATGCTCGGTGTGGGGTGGCG | 9 | 1 | 0 | 2.934929692 | 8.422517436 | Uniquely absent | Orphan SAGE tag |
| 1041 | CATGGCGCAACCCGGGGGCGC | 9 | 1 | 0 | 2.934929692 | 8.422517436 | Uniquely absent | Orphan SAGE tag |
| 128 | CATGGTGCTCCTCCCGCTGTC | 8 | 1 | 0 | 2.551511149 | 7.486682165 | Uniquely absent | EST has no meaningful BLASTX hit |
The Tag identification (ID) can be used to view the data on a particular tag at http://gmod.mbl.edu/emiliania_huxleyi.
Raw frequencies in each SAGE library are presented for each tag.
Change (n-fold) includes correction for sampling depth, with negative symbols used for down-regulation. The top five tags are presented for each library.
Unresolved SAGE tags are tags matching more than one EST or genome sequence. Orphan SAGE tags are tags not having a sequence match in all available EST and genome sequences.
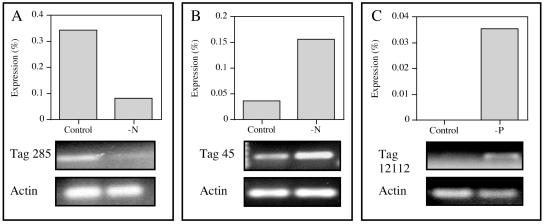
Type 1 actin and three tags (285, 45, and 12112) that mapped to gene sequence data were examined with qualitative RT-PCR to validate the expression patterns observed with the long SAGE analysis (Fig. 2). Under the conditions used in this study, type 1 actin appears to be constitutively expressed (Fig. 2). Tag 285 maps to a putative fucoxanthin chlorophyll a/c binding protein (fcp) of the E. huxleyi light-harvesting complex, and it is down-regulated over fourfold in the −N library (Table 3). This gene also appears to be down-regulated by −N conditions when assayed with RT-PCR (Fig. 2A), as the band intensity is lower in the −N sample than in the replete sample. Additional RT-PCR analysis was performed with tag 45. Tag 45 maps to an EST encoding for the C-terminal portion of a hypothetical protein with no database homology (the complete tag sequence can be found in Table S1 in the supplemental material). This tag is up-regulated over fourfold in the −N library, and this expression pattern was confirmed with qualitative RT-PCR, where the band intensity of the tag 45 amplicon was substantially reduced in the replete sample relative to the band intensity observed for the −N sample (Fig. 2B). Tag 12112 maps to an EST with homology to an inorganic pyrophosphatase (Table 3). This tag is uniquely present in the −P library, with a frequency of 4 (Table 3). Even at this low copy number the expression pattern observed with long SAGE was also observed with RT-PCR, as no band was detected for the replete sample (Fig. 2C). In no case was amplification observed for the no-RT controls (data not shown).
FIG. 2.
(A) Differential expression of long SAGE tag 285. Frequency of tag 285 in the SAGE libraries generated from replete (control) and −N E. huxleyi (top). RT-PCR amplification of the fucoxanthin chlorophyll a/c binding protein gene that maps to tag 285 (115 bp) and type 1 actin (116 bp) from control and −N conditions (bottom). (B) Differential expression of long SAGE tag 45. Frequency of tag 45 in the SAGE libraries generated from control and −N E. huxleyi (top). RT-PCR amplification of the EST sequence which maps to tag 45 (267 bp) and type 1 actin (116 bp) from control and −N E. huxleyi (bottom). (C) Differential expression of long SAGE tag 12112. Frequency of tag 12112 in the SAGE libraries generated from control and −P E. huxleyi (top). RT-PCR amplification of a 286-bp fragment of the putative inorganic pyrophosphatase gene that maps to tag 12112 and a 116-bp fragment of type 1 actin from control and −P samples (bottom). All PCRs were performed with equal template concentrations.
DISCUSSION
Genomic research with marine phytoplankton is rapidly advancing our understanding of how they function and interact with their environment. By comparing three long SAGE libraries (replete, −N, and −P), we have identified hundreds of differentially regulated tags (Table 2). To our knowledge, this is the first time long SAGE has been employed for gene expression profiling over multiple conditions with a photosynthetic marine alga, although a modified approach has been used for gene discovery with a dinoflagellate (11).
Validation.
Nearly half of the long SAGE tags could be mapped to gene sequence data from an E. huxleyi strain. Although this mapping is most straightforward with an available genome sequence, we show herein that it is tractable to map these sequence tags to existing sequence information in the absence of a genome. It is worth noting that the data mapped to multiple strains, suggesting that analysis of these SAGE data are not be restricted to strain 1516. Using RT-PCR, we examined the relative expression patterns of three tags (285, 45, and 12112), which mapped to publicly available sequence data and were either up- or down-regulated in one of the libraries. In all cases, RT-PCR generated an expression pattern similar to that observed with the long SAGE analysis. Although these tags were consistently differentially regulated, type 1 actin was constitutively expressed across the conditions tested in this study. Particularly striking are the results for tag 12112, which amplified from the −P sample but not the replete sample and is uniquely present (four copies) in the −P library. In this case, long SAGE results could be confirmed even for a low copy number tag with a small R value. Overall, these validation results demonstrate the value of SAGE libraries as a tool for transcriptome profiling, where subtle changes in expression, not just presence/absence, can be examined across multiple conditions. With the increasing number of whole-genome sequences or EST collections for diatoms, dinoflagellates, and other groups of eukaryotic marine phytoplankton, long SAGE is a promising new approach for transcriptome profiling with these different taxa. However, long SAGE is equally promising for phytoplankton lacking available EST or genome information, as high-throughput methods exist to identify full transcripts from SAGE tag sequences (6, 7). In some cases, SAGE tag sequences have been used to prime 5′ rapid amplification of cDNA ends for isolation of nearly complete transcripts (A. G. McArthur, unpublished data).
Nitrogen responses.
Little is known about transcriptional responses to N starvation in E. huxleyi. However, many algae become chlorotic, with a decrease in light-harvesting pigments, under N starvation, and the −N library was characterized by the down-regulation of genes mapping to multiple fucoxanthin chlorophyll a/c binding proteins (fcp) of the E. huxleyi light-harvesting complex. RT-PCR validation confirmed the regulation of one fcp (tag 285) (Fig. 2). The expression of this gene is also down-regulated by −N conditions in Pleurochrysis carterae (10), suggesting this may be a common transcriptional response to N starvation. Of the 38 up-regulated tags (e.g., 255, a putative 21-hydroxylase-like P450 protein) in the −N library, only 6 could be assigned a putative function (Table 3). Many of the genes up-regulated in the −N library were either orphan tags or mapped to sequence with no database homology (e.g., Table 4). This observation emphasizes how SAGE can be an important tool for the identification of new genes as well as for the regulation of genes with no putative function. For example, tags 1083, 294, 827, and 1041 (Table 4) are all orphan tags which do not map to available sequence data but are strongly up-regulated eightfold or more in the −N library. Tag 128 maps to an EST sequence without a significant BLASTX hit and thus has an unknown function (Table 4). Although the function of the gene is unclear, the specific N regulation observed with our long SAGE analysis (Table 4) suggests an involvement with N scavenging or N homeostasis. Here long SAGE has identified several new genes or gene targets for more-directed study of N scavenging in this organism.
Phosphorus responses.
E. huxleyi blooms in many oligotrophic, low-DIP areas, such as the North Atlantic (17), and has a strong physiological response to P starvation in culture, inducing several phosphate-regulated surface-associated proteins, as well as increasing alkaline phosphatase activity and phosphate uptake rate (13, 32). As was the case with the −N library, we have identified many orphan tags and tags which map to sequences with no database homology that are up-regulated in the −P library. These tags warrant further study, yet the overall results from the −P library suggest a robust transcriptional response to P starvation, with the up-regulation of several genes likely to be involved in P scavenging or P metabolism. One previously observed response to P deficiency in eukaryotic algae is the up-regulation of genes encoding phosphate-repressible permeases, allowing cells to transport phosphate with high affinity (8). In our analysis, tag 9469 maps to a putative E. huxleyi phosphate-repressible permease (9) that is up-regulated over fourfold in the −P library. Although this putative phosphate-repressible permease was previously identified (9), this is the first data confirming its regulation by phosphate supply. E. huxleyi can increase phosphate uptake under P-deficient conditions (32), and this gene is likely involved with this process.
Tag 12112 is uniquely present in the −P library (Table 3), and its expression was confirmed with RT-PCR. This tag has been successfully mapped to a complete ORF with homology to soluble inorganic pyrophosphatases (sPPase) (data not shown). Transcriptional activation of sPPase genes has been observed for P-starved cyanobacteria (14), and genes for sPPase have been identified for a number of protists (30), but the presence and P regulation of an sPPase gene has not previously been demonstrated with a coccolithophore.
An additional tag (24557) mapped to a gene with a putative role in polyphosphate metabolism. Polyphosphate is ubiquitously found in microorganisms and is either demonstrated to be or thought to be involved in diverse biological functions, including acting as a reservoir for inorganic phosphate (Pi) (18). In Saccharomyces cerevisiae, genes for polyphosphate synthesis and polyphosphate hydrolysis are both up-regulated in response to low extracellular phosphate as part of the PHO pathway involved in the acquisition of phosphate (23). In E. huxleyi, tag 24557, a putative polyphosphate synthetase, was up-regulated over ninefold in the −P library (Table 3). As highlighted above, we observed the up-regulation of a putative sPPase (tag 12112) which releases Pi from pyrophosphate, but we were unable to map any tags to a putative exopolyphosphatase, a processive enzyme that releases the terminal orthophosphate group from linear polyphosphates and would be required for the degradation of polyphosphate. It is important to note that a transcript encoding an exopolyphosphatase could be present in the −P library but not up-regulated sufficiently to be detected in this analysis. The seemingly paradoxical increase in the cell's ability to convert Pi into polyphosphate in response to P starvation might represent a strategy for accumulating and preserving Pi. Studies with yeast suggest that polyphosphate accumulation is required, presumably as a sink, to sustain a high rate of Pi uptake (23), and Escherichia coli has been shown to transiently accumulate polyphosphate in P-deficient medium (31). In E. huxleyi, where P deficiency results in the up-regulation of a phosphate permease (tag 9469) and increased rates of P uptake (32), the production of polyphosphate may be an important short-term storage pool for Pi. Ultimately the ability to both store and mobilize polyphosphate might help explain why E. huxleyi is such a good competitor for P in the oceans, where a given cell may transiently experience fluctuations in DIP concentration.
The annotation of the long SAGE data yielded some unexpected results. Notably, there was up-regulation of a tag (5002) which maps to a putative sulfate adenylyltransferase (ATP-sulfurylase) in the −P library. In plants, this enzyme mediates the first step in the metabolism of sulfate (21), and the up-regulation of this gene would be a potential mechanism for E. huxleyi cells to accumulate S during P deficiency. E. huxleyi is known to produce high levels of dimethylsulfoniopropionate (DMSP), and in diatoms there can be an accumulation of intracellular DMSP in nutrient-limited batch cultures (4). This accumulation of DMSP may be fueled by increased sulfate metabolism. We are in the initial stages of understanding E. huxleyi S metabolism, and additional expression studies of the putative ATP-sulfurylase and the extent to which its expression is linked to DMSP accumulation are certainly warranted. Also, we identified SAGE tags from rRNA in our libraries. This has been observed with other organisms from the genera Giardia and Trypanosoma (McArthur, unpublished), as rRNA contains stretches of poly(A). In most cases, these tags are not differentially expressed, but results from the −P library suggest that mitochondrial and plastid rRNA transcripts are more abundant in the −P condition (Table 3). There was some up-regulation of nuclear rRNA genes in the −P library as well, which is sometimes observed for Trypanosoma spp. at different cell densities (McArthur, unpublished). At this juncture, the mechanisms driving increased abundance of mitochondrial rRNA and plastid rRNA are unclear; however, this may be related to increased calcification in the −P condition. Additional targeted genome expression studies in this organism over a greater range of conditions would address the consistency of this observation.
Calcification.
Calcification was enhanced in the −P condition, and nearly twofold more genes were up-regulated in the −P library than in the −N library. In addition, a total of 54 tags were detected only in the −P library. The high number of unique and up-regulated tags in the −P library suggests that we may be observing the up-regulation of genes involved in calcification or calcium homeostasis (e.g., tag 11952 [calmodulin]) in addition to those involved in P scavenging. Substantial efforts have been made to identify the genes involved in calcification, and the most recent approaches have compared E. huxleyi EST libraries (40) or used suppressive subtractive hybridization with calcifying and noncalcifying cells (22). Of the five tags most highly expressed in our long SAGE −P library (Table 4), none clearly map to the most prevalent ESTs in the calcifying library analyzed by Nguyen et al. (22). Moreover, of the tags up-regulated or unique to the −P library, over 60 are orphan tags, which do not map to any of the genes identified to date in calcifying cells (22, 40). Considering the sensitivity of the transcriptome to culture conditions and the differences between EST, suppressive subtractive hybridization, and long SAGE analyses, these inconsistencies are not surprising; rather, these data underscore the value of using multiple genomic approaches over a range of conditions to identify the genes involved in uncharacterized metabolic pathways such as calcification. Further analysis of the tags up-regulated in our −P library will be helpful for future studies of transcriptional responses associated with increased calcification, and these data emphasize the potential of long SAGE analyses for building our understanding of the genes involved in calcification as well as other poorly understood pathways, such as alkenone biosynthesis.
Conclusions.
Genomic tools such as SAGE hold great promise for phytoplankton ecology and biological oceanography, where the molecular genetics of key marine organisms and the biogeochemical processes they mediate are still poorly understood. With our initial SAGE analyses, we have identified many new E. huxleyi sequences, assigned regulation data to sequences of unknown function, and highlighted previously uncharacterized aspects of E. huxleyi P scavenging, including polyphosphate synthesis. As discussed in the introduction, both N and P bioavailability can have a substantial influence on the timing, magnitude, and C cycling of E. huxleyi blooms. Our data demonstrate the nutrient regulation of a number of potential targets for RT-PCR assays in field populations of E. huxleyi, where the expression patterns of key genes relative to actin could be used to identify the N or P status of a population. As sequence data from the E. huxleyi genome project are released, an increasing number of E. huxleyi long SAGE tags will be resolved to genes in our public database (http://gmod.mbl.edu/emiliania_huxleyi), thus providing a dynamic tool for future gene discovery, transcriptome profiling, and genome annotation efforts. As scientists work toward an understanding of how E. huxleyi influences the marine C cycle and global climate, further analysis of the genes examined herein will help identify the molecular underpinnings that drive the cellular functioning of this organism in the sea.
Supplementary Material
Acknowledgments
We thank Eric Webb for helpful comments on an early draft of the manuscript.
This work was supported by the Woods Hole Oceanographic Institution Ocean Life Institute, the J. Lamar Worzel Assistant Scientist Fund, and the Frank and Lisina Hoch Endowed Fund. A.G.M., S.R.B., and M.J.C. were supported in part by the Marine Biological Laboratory's Program in Global Infectious Diseases, funded by the Ellison Medical Foundation. Computational resources were provided by the Josephine Bay Paul Center for Comparative Molecular Biology and Evolution (Marine Biological Laboratory) through funds provided by the W. M. Keck Foundation and the G. Unger Vetlesen Foundation.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Antia, N. J., P. J. Harrison, and L. Oliveira. 1991. The role of dissolved organic nitrogen in phytoplankton nutrition, cell biology and ecology. Phycologia 30:1-89. [Google Scholar]
- 2.Armbrust, E. V. 1999. Identification of a new gene family expressed during the onset of sexual reproduction in the centric diatom Thalassiosira weissflogii. Appl. Environ. Microbiol. 65:3121-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armbrust, E. V., J. A. Berges, C. Bowler, B. R. Green, D. Martinez, N. H. Putnam, S. Zhou, A. E. Allen, K. E. Apt, M. Bechner, A. A. Brzezinski, B. K. Chaal, A. Chiovitti, A. K. Davis, M. S. Demarest, J. C. Detter, T. Glavina, D. Goodstein, M. Z. Hadi, U. Hellsten, M. Hildebrand, B. D. Jenkins, J. Jurka, V. V. Kapitonov, N. Kröger, W. W. Y. Lau, T. W. Lane, F. W. Larimer, J. C. Lippmeir, S. Lucas, M. Medina, A. Montsant, M. Obornik, M. Schnitzler Parker, B. Palenik, G. J. Pazour, P. M. Richardson, T. A. Rynearson, M. A. Saito, D. C. Scwartz, K. Thamatraklon, K. Valentin, A. Vardi, F. P. Wilkerson, and D. S. Rokhsar. 2004. The genome of the diatom Thalassiosira pseudonana: ecology, evolution and metabolism. Science 306:79-86. [DOI] [PubMed] [Google Scholar]
- 4.Bucciarelli, E., and W. G. Sunda. 2003. Influence of CO2, nitrate, phosphate and silicate limitation on intracellular dimethylsulfoniopropionate in batch cultures of the coastal diatom Thalassiosira pseudonana. Limnol. Oceanogr. 48:2256-2265. [Google Scholar]
- 5.Cembella, A. D., N. J. Antia, and P. J. Harrison. 1984. The utilization of inorganic and organic phosphorous compounds as nutrients by eukaryotic microalgae: a multidisciplinary perspective: part 1. Crit. Rev. Microbiol. 10:317-391. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J., S. Lee, G. Zhou, and S. M. Wang. 2002. High-throughput GLGI procedure for converting a large number of serial analysis of gene expression tag sequences in 3′ complementary DNAs. Genes Chromosomes Cancer 33:252-261. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J. J., J. D. Rowley, and S. M. Wang. 2000. Generation of longer cDNA fragments from serial analysis of gene expression tags for gene identification. Proc. Natl. Acad. Sci. USA 97:349-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung, C.-C., S.-P. L. Hwang, and J. Chang. 2003. Identification of a high-affinity phosphate transporter gene in a prasinophyte alga, Tetraselmis chui, and its expression under nutrient limitation. Appl. Environ. Microbiol. 69:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corstjens, P. L., M. Zuiderwijk, M. Nilsson, H. Feindt, R. Sam Niedbala, and H. J. Tanke. 2003. Lateral-flow and up-converting phosphor reporters to detect single-stranded nucleic acids in a sandwich-hybridization assay. Anal. Biochem. 312:191-200. [DOI] [PubMed] [Google Scholar]
- 10.Corstjens, P. L. A. M., and E. L. González. 2004. Effects of nitrogen and phosphorus availability on the expression of the coccolith-vesicle v-ATPase (subunit C) of Pleurochrysis (Haptophyta). J. Phycol. 40:82-87. [Google Scholar]
- 11.Coyne, K. J., J. M. Burkholder, R. A. Feldman, D. A. Hutchins, and S. C. Cary. 2004. Modified serial analysis of gene expression method for construction of gene expression profiles of microbial eukaryotic species. Appl. Environ. Microbiol. 70:5298-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doney, S. C., M. R. Abbott, J. J. Cullen, D. M. Karl, and L. Rothstein. 2004. From genes to ecosystems: the ocean's new frontier. Front. Ecol. Environ. 2:457-466. [Google Scholar]
- 13.Dyhrman, S. T., and B. Palenik. 2003. A characterization of ectoenzyme activity and phosphate-regulated proteins in the coccolithophorid Emiliania huxleyi. J. Plankton Res. 25:1-11. [Google Scholar]
- 14.Gómez-García, M. R., M. Losada, and A. Serrano. 2003. Concurrent transcriptional activation of ppa and ppx genes by phosphate deprivation in the cyanobacterium Synechocystis sp. strain PCC 6803. Biochem. Biophys. Res. Commun. 302:601-609. [DOI] [PubMed] [Google Scholar]
- 15.Gordon, D., C. Abajian, and P. Green. 1998. CONSED: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 16.Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates, p. 29-60. In W. L. Smith and M. H. Chanley (ed.), Culture of marine invertebrate animals. Plenum, New York, N.Y.
- 17.Holligan, P. M., E. Fernández, J. Aiken, W. M. Balch, P. Boyd, P. H. Burkill, M. Finch, S. B. Groom, G. Malin, K. Muller, D. Purdie, C. Robinson, C. C. Trees, S. M. Turner, and P. van der Wal. 1993. A biogeochemical study of the coccolithophore, Emiliania huxleyi, in the North Atlantic. Global Biogeochem. Cycles 7:879-900. [Google Scholar]
- 18.Kornberg, A., N. N. Rao, and D. Ault-Riché. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89-125. [DOI] [PubMed] [Google Scholar]
- 19.Koroleff, F. 1983. Determination of phosphorus, p. 125-138. In K. Grasshoff, M. Ehrhardt, and K. Kremling (ed.), Methods of seawater analysis. Verlag Chemie, Weinheim, Denmark.
- 20.Lessard, E. J., A. Merico, and T. Tyrrell. 2005. Nitrate: phosphate ratios and Emiliania huxleyi blooms. Limnol. Oceanogr. 50:1020-1024. [Google Scholar]
- 21.Leustek, T., M. N. Martin, J. Bick, and J. P. Davies. 2000. Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu. Rev. Plant Physiol. 51:141-165. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen, B., R. M. Bowers, T. M. Wahlund, and B. A. Read. 2005. Suppressive subtractive hybridization of and differences in gene expression content of calcifying and noncalcifying cultures of Emiliania huxleyi strain 1516. Appl. Environ. Microbiol. 71:2564-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogawa, N., J. DeRisi, and P. O. Brown. 2000. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell 11:4309-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paasche, E. 2002. A review of the coccolithophorid Emiliania huxleyi (Prymnesiophyceae), with particular reference to growth, coccolith formation, and calcification-photosynthesis interactions. Phycologia 40:503-529. [Google Scholar]
- 25.Paasche, E. 1998. Roles of nitrogen and phosphorus in coccolith formation in Emiliania huxleyi (Prymnesiophyceae). Eur. J. Phycol. 33:33-42. [Google Scholar]
- 26.Paasche, E., and S. Brubak. 1994. Enhanced calcification in the coccolithophorid Emiliania huxleyi (Haptophyceae) under phosphorus limitation. Phycologia 33:324-330. [Google Scholar]
- 27.Palenik, B., B. Brahamsha, F. Larimer, M. Land, L. Hauser, P. Chain, J. Lamberdin, W. Regala, E. Allen, J. McCarren, I. Paulsen, A. Dufresne, F. Partensky, E. Webb, and J. Waterbury. 2003. The genome of a motile marine Synechococcus. Nature 424:1037-1042. [DOI] [PubMed] [Google Scholar]
- 28.Palenik, B., and S. E. Henson. 1997. The use of amides and other organic nitrogen sources by the phytoplankton Emiliania huxleyi. Limnol. Oceanogr. 42:1544-1551. [Google Scholar]
- 29.Palenik, B., and J. Koke. 1995. Characterization of a nitrogen-regulated protein identified by cell-surface biotinylation of a marine phytoplankton. Appl. Environ. Microbiol. 61:3311-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pérez-Castiñeira, J. R., R. Gómez-García, R. L. López-Marqués, and M. L. A. Serrano. 2001. Enzymatic systems of inorganic pyrophosphate bioenergetics in photosynthetic and heterotrophic protists: remnants or metabolic cornerstones? Int. Microbiol. 4:135-142. [DOI] [PubMed] [Google Scholar]
- 31.Rao, N. N., S. Liu, and A. Kornberg. 1998. Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J. Bacteriol. 180:2186-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riegman, R., W. Stolte, A. A. M. Noordeloos, and D. Slezak. 2000. Nutrient uptake and alkaline phosphatase activity of Emiliania huxleyi (Prymnesiophyceae) during growth under N and P limitation in continuous cultures. J. Phycol. 36:87-96. [DOI] [PubMed] [Google Scholar]
- 33.Saha, S., A. B. Sparks, C. Rago, V. Akmaev, C. J. Wang, B. Vogelstein, K. W. Kinzler, and V. E. Velculescu. 2002. Using the transcriptome to annotate the genome. Nat. Biotechnol. 19:508-511. [DOI] [PubMed] [Google Scholar]
- 34.Scala, S., N. Carels, A. Falciatore, M. L. Chiusano, and C. Bowler. 2002. Genome properties of the diatom Phaeodactylum tricornutum. Plant Physiol. 129:993-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein, L. D., C. Mungall, S. Shu, M. Caudy, M. Mangone, A. Day, E. Nickerson, J. E. Stajich, T. W. Harris, A. Arva, and S. Lewis. 2002. The generic genome browser: a building block for a model organism system database. Genome Res. 12:1599-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stekel, D. J., Y. Git, and F. Falciani. 2000. The comparison of gene expression from multiple cDNA libraries. Genome Res. 10:2055-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taroncher-Oldenburg, G., and D. M. Anderson. 2000. Identification and characterization of three differentially expressed genes, encoding S-adenosylhomocysteine hydrolase, methionine aminopeptidase, and a histone-like protein, in the toxic dinoflagellate Alexandrium fundyense. Appl. Environ. Microbiol. 66:2105-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Velculescu, V. E., L. Zhang, B. Vogelstein, and K. W. Kinzler. 1995. Serial analysis of gene expression. Science 270:484-487. [DOI] [PubMed] [Google Scholar]
- 39.Wahlund, T. M., A. R. Hadaegh, R. Clark, B. Nguyen, M. Fanelli, and B. Read. 2004. Analysis of expressed sequence tags from calcifying cells of marine coccolithophorid (Emiliania huxleyi). Mar. Biotechnol. 6:278-290. [DOI] [PubMed] [Google Scholar]
- 40.Wahlund, T. M., X. Zhang, and B. Read. 2004. Expressed sequence tag profiles from calcifying and non-calcifying cultures of Emiliania huxleyi. Micropaleontology 50:145-155. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.