Abstract
Thirteen different serotypes of the food-borne pathogen Listeria monocytogenes have been described. Serotype 4b strains are most often associated with illness, and serotype 1/2a strains are most often isolated from foods and processing plants. Different abilities to respond to stresses have been described for serotype 4b and 1/2a strains. One of the common enrichment protocols used to test foods for the presence L. monocytogenes is described in the U.S. Food and Drug Administration (FDA) Bacterial Analytical Manual (BAM). We compared three strains of L. monocytogenes serotype 4b and five strains of serotype 1/2a in direct competition with each other in two-strain mixed cultures by using the FDA BAM enrichment protocol, which includes both enrichment broth and selective agar, with and without added food to mimic the conditions that occur during attempts to isolate Listeria species from contaminated foods. Using a colony immunoblot procedure and analyzing over 112,000 colonies, we observed differences in strain fitness, but these differences were not attributable to serotype or genetic lineage.
The food-borne pathogen Listeria monocytogenes is commonly associated with deli meats, soft cheeses, unpasteurized dairy products, produce, and processed foods. It is a hardy organism that can withstand extreme salt concentrations and drying and can grow at refrigeration temperatures. Susceptible individuals who ingest contaminated foods can develop listeriosis, a systemic disease that can manifest as meningitis, liver failure, septicemia, and spontaneous abortion. If they receive a dose that is high enough, healthy individuals can develop febrile gastroenteritis (20). About 2,500 cases of listeriosis occur each year in the United States, and there are roughly 500 deaths (13). Although 13 different serotypes of L. monocytogenes have been described, the overwhelming majority of cases of illness stem from three serotypes, serotypes 1/2a, 1/2b, and 4b. Most of the outbreaks have been attributed to serotype 4b; however, in surveys of foods or food-processing plants for L. monocytogenes, serotype 1/2a strains are found most often (10).
Variable traits of L. monocytogenes serotype 4b and 1/2a strains have been described previously. Buncic et al. (5) showed that at 4°C, serotype 1/2a strains survived exposure to bacteriocins better than serotype 4b strains. Additionally, serotype 4b strains survived heat treatments immediately following cold storage better than serotype 1/2a strains. Genome sequence comparisons between a serotype 1/2a strain and multiple serotype 4b strains revealed serotype-specific genes and showed that the serotype 1/2a strain genome is slightly larger than the serotype 4b strain genomes (14). Analysis of the genotypic differences between the serotypes has led to subtyping of the species into three genetic lineages, based on ribotyping, virulence gene analysis, and mixed-genome microarray analysis (1, 2, 9, 21, 22). Bruhn et al. (4) recently described a bias in favor of lineage 2 strains (which include serotype 1/2a strains) over lineage 1 strains (which include serotype 4b strains) in the commonly used enrichment broth media, University of Vermont Listeria enrichment media I and II (UVM I and II).
Another of the standard protocols used to enrich L. monocytogenes from food samples is the U.S. Food and Drug Administration (FDA) Bacterial Analytical Manual (BAM) method (8). The BAM L. monocytogenes enrichment protocol includes a nutrient-rich primary enrichment medium, buffered Listeria enrichment broth (BLEB), into which food samples are inoculated. After a 4-h incubation to allow for the recovery of stressed cells, two antibiotics (nalidixic acid and cycloheximide) and a toxic dye (acriflavine) are added. The cycloheximide is used to reduce fungal growth, the nalidixic acid eliminates many gram-negative bacteria, and the acriflavine eliminates many gram-positive bacteria, especially fecal enterococci. The culture is then incubated for two more days before it is plated on one of four approved selective and differential agar media. The entire process can take a few days to a week. Many food diagnostic laboratories use either this method or a variation of it to isolate potential L. monocytogenes colonies for further analysis by phenotypic and/or molecular methods. Prior to establishment of the antibiotic enrichment protocols in the late 1980s, L. monocytogenes enrichment was a simple, but lengthy, process that took advantage of the ability of the species to grow at 4°C. Enrichments were done in rich medium in a refrigerator and could take months. No single enrichment protocol is best, as studies that have compared different enrichment protocols as well as direct plating of contaminated foods side by side have shown that some strains are recovered by one method and not by another (11, 19). Studies have also shown that use of two independent enrichment protocols yields more isolates than use of one protocol alone (18, 19).
The antibiotic enrichments have made isolation easier. However, since genetic and phenotypic differences between serotypes have been established, it is reasonable to think that these differences could extend to the enrichment process, a stressful environment that demands coping with antibiotics and competition with normal food microbiota. Since serotype 1/2a strains are isolated from foods more often than serotype 4b strains and since the two serotypes have physiological and genetic differences, we speculated that differences in fitness related to serotype might also occur in common selective enrichment protocols. Our hypothesis was that serotype 1/2a strains are not more common in foods but rather are more fit in the enrichment protocol and thus outcompete any serotype 4b strains that may also be present. We tested this hypothesis with a set of eight L. monocytogenes strains using the FDA BAM enrichment protocol. We selected both serotype 1/2a strains and serotype 4b strains that had been isolated from patients, animals, food, and silage. Some of the strains were cultured originally before the standard antibiotic enrichment protocols were developed and were included to determine how well they competed with more recent strains isolated by antibiotic enrichment. We inoculated enrichment broth media with mixed cultures of serotype 1/2a and 4b strains with and without added foods to determine if either serotype survived the enrichment protocol better than the other. The colonies of the two serotypes were differentiated with a colony immunoblot assay that we developed previously (16). This study differs from a study described recently (4) because a different enrichment medium was used, foods were added to the enrichment process, the entire enrichment protocol (liquid enrichment and plating on selective agar) was used, and a much larger number of colonies were assessed.
MATERIALS AND METHODS
Strains and media.
The L. monocytogenes strains used in this study are listed in Table 1; whether antimicrobial agents were used in the original isolation of the strains is also indicated. Strains were grown in tryptic soy broth without dextrose supplemented with 0.6% yeast extract (TSYE medium) (Difco, Becton Dickinson,Franklin Lakes, NJ) at 30°C overnight for use in the enrichment protocol. For enrichment, the media used were those recommended in the FDA BAM for L.monocytogenes (8), and they included buffered Listeria enrichment broth (Difco) and modified Oxford (MOX) medium (Difco). Phosphate-buffered saline (PBS) contained 150 mM NaCl and 10 mM sodium phosphate (pH 7.2). Strains were serotyped by enzyme-linked immunosorbent assay-based serotyping (16), and the genetic lineage was determined by multiplex PCR as described previously (21).
TABLE 1.
Strains used in this study
| Strain | Other designation | Description | Original isolation involved antibiotic supplements | Lineage | Reference or source |
|---|---|---|---|---|---|
| RM2387 | None | Serotype 4b, isolated from mint survey, 2000 | Yes | 1 | 6 |
| RM2199 | F2379 | Serotype 4b, patient outbreak isolate, Jalisco cheese, 1985 | No | 1 | D. Portnoy, University of California, Berkeley |
| RM3176 | TS27/L.4738 | Serotype 4b, patient outbreak isolate, coleslaw, 1981 | No | 1 | M. Wiedmann, Cornell University |
| RM3106 | MB58 | Serotype 1/2a, chicken processing plant, 2002 | Yes | 2 | R. Meinersmann, USDA Agricultural Research Service |
| RM3175 | TS14/F6900 | Serotype 1/2a, patient isolate, sporadic, hot dog | No | 2 | M. Wiedmann, Cornell University |
| RM3364 | EGD-e | Serotype 1/2a, rabbit dung isolate, 1926 | No | 2 | Institut Pasteur |
| RM2985 | 32490A | Serotype 1/2a, bulk milk, 2003 | Yes | 2 | M. Borucki, USDA, Agricultural Research Service |
| RM3316 | CWD243 | Serotype 1/2a, silage, 1990 | Yes | 2 | C. Donnelly, University of Vermonta |
See reference 17.
Enrichment procedure, differentiation of colony serotypes, and enumeration of strains.
The protocol used for enrichment was exactly the protocol described in the FDA BAM for L. monocytogenes (8), except that the MOX agar plates were incubated for 3 days instead of 2 days to allow better immunoblotting from the strains that produced the smallest colonies. Briefly, 225-ml portions of BLEB were prepared in 500-ml Erlenmeyer flasks. When food was added, 25 g of food was weighed, emulsified with a portion of the BLEB in a sterile Waring blender, and poured into the flask. Iceberg lettuce was purchased on the morning of use from a local grocery store. The outer leaves were removed, and the remainder was cut and weighed for use. A medium cheddar cheese, sealed in plastic, was purchased at a local store. None of the foods were washed before use. L. monocytogenes cultures were grown overnight in TSYE medium with shaking at 30°C. The A600 of each culture was determined, and the cultures were diluted in PBS so that the densities matched the densities of the least dense cultures, in order to obtain equivalent cell densities. The cultures were serially diluted in PBS, and 100 μl of a 10−4 dilution was added to the appropriate enrichment flasks, resulting in, on average, 40 to 100 CFU/ml of each L. monocytogenes strain in the flasks. Serial dilutions were plated on TSYE agar plates to ensure that equal densities were used for inocula. Mixed-culture enrichments were prepared by adding mixtures of strains at a 1:1 ratio directly to flasks containing BLEB or BLEB with food. The BLEB enrichment cultures were incubated at 30°C for 4 h, and then the cultures were supplemented with acriflavine HCl (10 mg/liter), nalidixic acid (40 mg/liter), and cycloheximide (50 mg/liter) as described in the BAM protocol (8). The cultures were incubated for a total of 48 h before dilution plating onto MOX agar. The MOX agar plates were incubated at 35°C for 3 days. If single-strain enrichment was done, at this point the colonies were counted, and the numbers of CFU/ml of each strain in the various enrichments were calculated. If mixed-culture enrichment was done, the colonies on the MOX agar plates were transferred to a nitrocellulose membrane, and colony immunoblotting was performed as described previously (16). The blots were probed sequentially with 1:1,000 dilutions of type 1 or type 4 Listeria O antiserum (Difco), and this was followed by detection of the primary antibody by probing with alkaline phosphatase-conjugated goat anti-rabbit antibody and visualization of the secondary antibody binding with either SIGMAFAST 5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium, which stains colonies blue, or SIGMAFAST Fast Red TR/naphthol AS-MX, which stains colonies red (Sigma-Aldrich, St. Louis, MO). The order of use of the antisera (type 1 or type 4) and the color reagent (red or blue) used to visualize the binding of the secondary antibody was random, with the result that colonies of one serotype were differentiated from colonies of the other serotype by color. After the immunoblot procedure, the colonies of each color (serotype) were counted, and the percentage that each serotype contributed to the total L. monocytogenes colony count was determined.
Doubling times and adaptation to supplement stress.
Doubling times of cultures in different media were measured by monitoring the A600 of cultures growing in BLEB and BLEB containing BAM supplements. Cultures were grown overnight in the same medium in which they were to be monitored, diluted 1:50 into fresh medium, and incubated at 30°C with shaking at 100 rpm, and the A600 was monitored. The doubling time was calculated by determining the time that it took for the A600 value to double during the logarithmic phase. The time that it took to respond to chemical stress was determined by diluting a BLEB-grown culture into fresh BLEB, allowing it to grow for 90 min, and then adding the BAM supplements. The A600 values of the cultures were monitored and compared to see if there were differences in growth or adaptation to the antimicrobial agents. The experiments were done at least twice, and the average values and standard deviations were calculated.
RESULTS
Doubling times of strains and adaptation to supplement stress.
Doubling times were calculated for cultures grown in BLEB and BLEB containing the BAM supplements nalidixic acid, cycloheximide, and acriflavine (Table 2). The culture doubling times were 8 to 26 min longer in BLEB containing the supplements than in plain BLEB. For most of the strains the doubling times in BLEB were approximately 70 min. Strain RM3175 was the slowest grower in BLEB, with a doubling time of 80 min. In BLEB containing supplements, the doubling times ranged from 77 min (RM2199) to more than 90 min (RM3176, RM3175, RM3364, and RM2985). The rate of doubling was not dependent on the serotype of the strains, since two serotype 1/2a strains and two serotype 4b strains had doubling times of ≥90 min in BLEB containing supplements.
TABLE 2.
Doubling times of cultures of L. monocytogenes in BLEB and in BLEB containing antibiotic and chemical supplements
| Strain | Serotype | Doubling times (min) (mean ± SD)
|
|
|---|---|---|---|
| BLEB | BLEB + supplements | ||
| RM2387 | 4b | 70.0 ± 3.3 | 85.4 ± 1.5 |
| RM2199 | 4b | 69.0 ± 2.8 | 77.0 ± 0.2 |
| RM3176 | 4b | 68.1 ± 6.7 | 93.9 ± 2.0 |
| RM3106 | 1/2a | 69.6 ± 3.3 | 85.1 ± 0.7 |
| RM3175 | 1/2a | 80.6 ± 1.3 | 90.1 ± 0.4 |
| RM3364 | 1/2a | 71.3 ± 5.6 | 90.9 ± 2.7 |
| RM2985 | 1/2a | 65.4 ± 3.0 | 90.6 ± 3.3 |
| RM3316 | 1/2a | 66.6 ± 3.0 | 87.0 ± 3.0 |
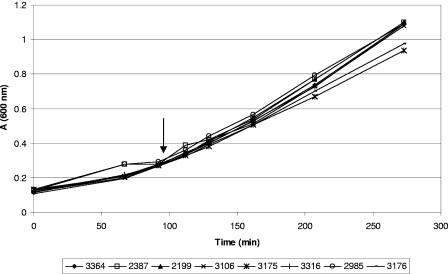
As part of the BAM protocol, chemical supplements are added after 4 h of incubation in order to allow repair of stressed cells. Differences in strain fitness could have resulted from variable responses to addition of these chemicals. To see if there were differences in the responses to supplement addition, growth of the cultures was monitored in plain BLEB for 90 min, and then supplements were added. Figure 1 shows the results of one of these experiments. After addition of the supplements there was little difference in the growth of the strains, as shown by the curves for the eight strains. No additional lag phase was observed in response to addition of the chemical supplements.
FIG. 1.
Growth of cultures in BLEB before and after addition of BAM supplements. The time of addition of the supplements is indicated by an arrow. See text for details.
Fitness of strains in single and mixed-culture enrichments.
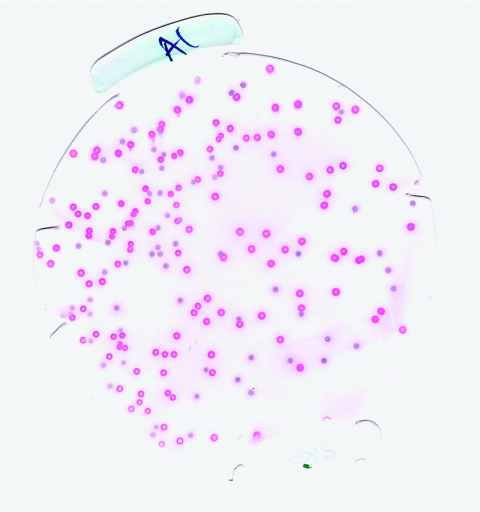
The starting cultures for inoculation of the enrichment broth media were diluted to the same A600, and the presence of equivalent numbers of CFU/ml was confirmed by serial dilution and plate counting. In all experiments, the enrichment broth media were inoculated with a serotype 4b strain and a serotype 1/2a strain at a ratio of 1:1. Figure 2 shows the results of a typical immunoblot experiment. MOX agar plates were incubated for 3 days at 35°C instead of the 2 days recommended by the BAM, because some of the L. monocytogenes colonies were too small to blot well after 48 h as recommended in the BAM protocol. In fact, some colonies, especially those of strain RM3175, were only pinpoint size even after 48 h of growth. Generally but not exclusively, the colonies of the serotype 4b strains on MOX agar plates were larger than those of the serotype 1/2a strains. An example of the difference in colony size is shown in the immunoblot in Fig. 2.
FIG. 2.
Typical immunoblot with mixed serotype colonies of L. monocytogenes. The pink colonies are RM2387 colonies (serotype 4b), and the blue colonies are RM3364 colonies (serotype 1/2a).
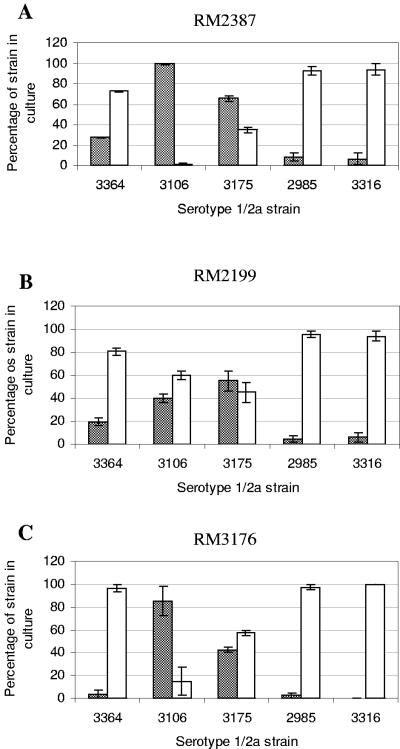
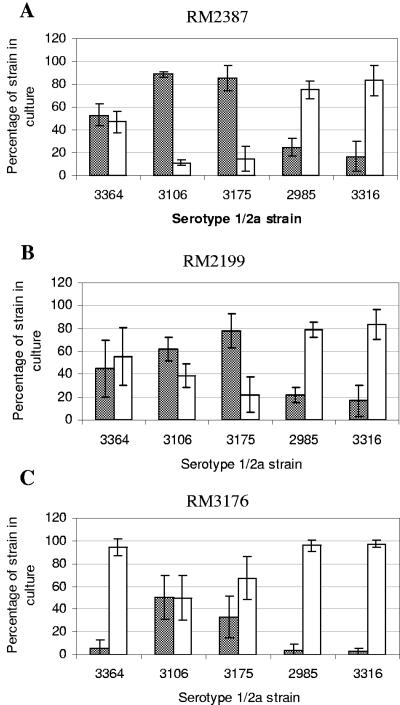
The fates of the serotype 4b and 1/2a strains in direct competition in the BAM enrichment protocol are shown in Fig. 3 to 5. Each experiment was done at least three times, and the graphs show averages and standard deviations. A total of 112,000 colonies were assessed, and between 744 and 8,805 colonies were counted for each strain pair for each enrichment in all the replicate experiments. When a strain pair is indicated below by two strain designations with a slash between them (e.g., RM2387/RM3106), the first strain is the serotype 4b strain and the second strain is the serotype 1/2a strain. In general, the standard deviations were smaller in the enrichments without added food (Fig. 3). Although culture bias in the enrichment protocol was observed in some experiments, it was not strictly associated with either the 4b or 1/2a serotype. Strain RM2387 (serotype 4b), originally isolated from mint by enrichment with antimicrobial agents, was very competitive in enrichments with the serotype 1/2a strains RM3106 (isolated from a chicken processing plant with the use of antimicrobial agents) and RM3175 (isolated from a human who had eaten contaminated hot dogs). Serotype 4b strain RM2387 consistently accounted for nearly 100% of the colonies on MOX agar plates derived from the enrichments that included either of these two serotype 1/2a strains in plain media (Fig. 3) and in media with both cheese and lettuce (Fig. 4 and 5). The results shown in Table 2 indicate that the doubling times of strain RM2387 (serotype 4b) in plain BLEB and BLEB containing supplements were equivalent to those of strain RM3106 (serotype 1/2a) and less than those of strain RM3175 (serotype 1/2a). Serotype 1/2a strains RM3175 and RM3106 were more competitive with the two other serotype 4b strains used in the study, strains RM2199 and RM3176.
FIG. 3.
Percentages of serotype 4b and 1/2a strains resulting from mixed enrichments in BLEB without added food. Shaded bars, serotype 4b strains; open bars, serotype 1/2a strains. The serotype 1/2a strains are indicated on the x axes. (A) RM2387 versus serotype 1/2a strains; (B) RM2199 versus serotype 1/2a strains; (C) RM3176 versus serotype 1/2a strains.
FIG. 5.
Percentages of serotype 4b and 1/2a strains resulting from mixed enrichments in BLEB with lettuce. Shaded bars, serotype 4b strains; open bars, serotype 1/2a strains. The serotype 1/2a strains are indicated on the x axes. (A) RM2387 versus serotype 1/2a strains; (B) RM2199 versus serotype 1/2a strains; (C) RM3176 versus serotype 1/2a strains.
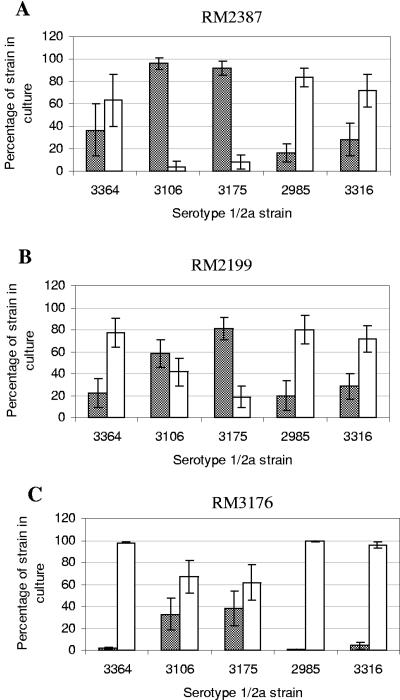
FIG. 4.
Percentages of serotype 4b and 1/2a strains resulting from mixed enrichments in BLEB with cheese. Shaded bars, serotype 4b strains; open bars, serotype 1/2a strains. The serotype 1/2a strains are indicated on the x axes. (A) RM2387 versus serotype 1/2a strains; (B) RM2199 versus 1/2a strains; (C) RM3176 versus 1/2a strains.
Serotype 1/2a strains RM3175 and RM3106 were recovered at or near statistically equivalent levels when they were paired with serotype 4b strain RM2199 in media alone (Fig. 3). The recovery of RM3106 was statistically equivalent to that of RM2199 in both cheese and lettuce enrichments, whereas RM3175 colonies comprised only about 20% of the colonies recovered for the RM2199/RM3175 pair in the enrichments with food. The third serotype 4b strain tested, RM3176, was more fit than the serotype 1/2a strain RM3106 in plain media; only 15% of the colonies recovered for the pair were RM3106 colonies. However, in enrichments with food, equal amounts of strains RM3176 and RM3106 were recovered. For the RM3176/RM3175 pair, the serotype 1/2a strain RM3175 had a slight edge over the serotype 4b strain in plain media, but when food was added, the levels of recovery of the two strains were statistically equivalent.
Whereas RM3106 and RM3175 were the two serotype 1/2a strains that were the least competitive in the mixed enrichments overall with the serotype 4b strains, the two serotype 1/2a strains that consistently proved to be more fit in the enrichments in comparisons with all three serotype 4b strains were milk isolate RM2985 and silage isolate RM3316. The latter two strains were both isolated originally with antimicrobial supplements. RM2985 and RM3316 consistently dominated the L. monocytogenes populations recovered from the different enrichments in which they were used (Fig. 3 to 5). In pairs with the serotype 4b strains incubated in plain medium, RM2985 and RM3316 each accounted for over 90% of the colonies recovered. When foods were added, the recovery of serotype 4b strains RM2387 and RM2199 was slightly better in pairs with serotype 1/2a strains RM2985 and RM3316 than in plain medium (approximately 20% to 30%). The third serotype 4b strain, RM3176, however, was still poorly recovered from the food enrichments when it was paired with either RM2985 or RM3316. In these pairs, only 0% to 4% of the colonies recovered were RM3176 colonies. While RM2985 and RM3316 were among the strains that were recovered best from all enrichments, their doubling times were equivalent to those of many of the other strains in this study, including the three serotype 4b strains (Table 2).
RM3364 was the serotype 1/2a strain that exhibited intermediate fitness in enrichment mixtures with the serotype 4b strains compared with the “high-fitness” serotype 1/2a strains RM2985 and RM3316 and the “low-fitness” serotype 1/2a strains RM3106 and RM3175. Strain RM3364 (serotype 1/2a), which was isolated originally in 1926 without the use of antimicrobial agents, was more fit than any of the serotype 4b strains in enrichments containing media without added food, accounting for 60% to 96% of the colonies on the MOX agar plates (Fig. 3). RM3364 was still recovered at levels of 94% to 98% when it was paired with RM3176 in enrichments with cheese and lettuce. However, the other two serotype 4b strains, RM2387 and RM2199, competed better with RM3364 in the food enrichments. For the RM2387/RM3364 pair in cheese and lettuce, statistically equivalent numbers of each strain were recovered. Statistically equivalent numbers of RM2199 (serotype 4b) and RM3364 (serotype 1/2a) were recovered when this strain pair was grown in enrichments with cheese; however, with lettuce only 20% of the colonies recovered were serotype 4b strain RM2199 colonies (Fig. 4 and 5).
Table 3 shows the log CFU/ml of each strain recovered from all the types of enrichments when the strain was either in coculture or was subjected to the enrichment process in a single culture. Each strain grew to equivalent levels when it was inoculated in monoculture into plain medium or cheese or lettuce enrichments; the levels were roughly 9.5 to 9.6 log CFU/ml in plain medium, 8.5 to 8.8 log CFU/ml in enrichments with cheese, and 9.5 to 10 CFU/ml in enrichments with lettuce. Therefore, strains that were poor competitors in mixed cultures, such as serotype 1/2a strains RM3106 and RM3175 and serotype 4b strain RM3176, were recovered at the same levels as strains that were good competitors when they were enriched as monocultures. In coculture enrichments, when the values for log CFU/ml recovered of each strain were averaged, the number was lower and the standard deviation was greater than when the strains were enriched in single cultures.
TABLE 3.
Numbers of CFU/ml of strains present in different enrichments when they were cultured alone and in coculturea
| Strain | Log CFU/ml in medium alone
|
Log CFU/ml with cheese
|
Log CFU/ml with lettuce
|
|||
|---|---|---|---|---|---|---|
| Single culture | Coculture | Single culture | Coculture | Single culture | Coculture | |
| RM2387 | 9.6 ± 0.01 | 8.4 ± 1.1 | 8.8 ± 0.2 | 8.2 ± 1.0 | 9.8 ± 0.3 | 9.1 ± 0.6 |
| RM2199 | 9.6 ± 0.02 | 8.2 ± 1.1 | 8.7 ± 0.4 | 8.1 ± 1.0 | 10.0 ± 0.4 | 8.9 ± 0.4 |
| RM3176 | 9.5 ± 0.1 | 6.2 ± 3.2 | 8.8 ± 0.3 | 6.1 ± 3.2 | 9.6 ± 0.4 | 6.9 ± 3.0 |
| RM3106 | 9.6 ± 0.02 | 8.4 ± 1.3 | 8.8 ± 0.3 | 7.9 ± 0.9 | 10.0 ± 0.4 | 7.8 ± 2.8 |
| RM3175 | 9.6 ± 0.05 | 9.0 ± 0.2 | 8.5 ± 0.02 | 7.7 ± 1.1 | 9.5 ± 0.03 | 8.5 ± 0.7 |
| RM3364 | 9.6 ± 0.04 | 9.2 ± 0.4 | 8.5 ± 0.1 | 8.2 ± 0.9 | 9.6 ± 0.3 | 9.4 ± 0.5 |
| RM2985 | 9.6 ± 0.02 | 8.5 ± 1.1 | 8.5 ± 0.1 | 8.3 ± 1.0 | 9.8 ± 0.5 | 9.6 ± 0.3 |
| RM3316 | 9.6 ± 0.01 | 8.6 ± 1.1 | 8.8 ± 0.3 | 8.5 ± 0.8 | 9.7 ± 0.2 | 9.2 ± 0.3 |
The values are averages ± standard deviations from at least two experiments with replicates in each experiment.
DISCUSSION
In addition to the obvious difference between serotypes, different cell surface antigens and physiological differences related to bacteriocin resistance and the ability to recover from temperature stress have been reported for serotype 1/2a and 4b strains (5). To begin to determine if serotype 1/2a strains are more commonly isolated from environmental samples because they are more fit in enrichment protocols, we tested serotype 4b-serotype 1/2a strain mixtures using the FDA BAM enrichment protocol with and without addition of foods to mimic regular enrichments for L. monocytogenes that might occur in testing laboratories. Therefore, the L. monocytogenes competition and/or fitness data for the strains were compared with each other and also with data obtained in the presence of the indigenous flora and chemicals on the foods. Strains that represented both serotypes and were isolated from similar sources (e.g., patients, dairy products, and plants) were selected. Patient serotype 4b isolates were also selected based on the foods from which the outbreaks originated (cheese and coleslaw). We hypothesized that serotype 1/2a strains would be more fit in the commonly used BAM enrichment procedure, but our analysis of over 112,000 colonies isolated from multiple mixed-strain enrichment experiments did not support this hypothesis.
Although only a limited number of strains were tested here, it is clear that some strains were more competitive or more fit than others, but the difference was not correlated with serotype. Two serotype 1/2a strains, RM2985 and RM3316, were high-fitness strains in any pair and in any food (or nonfood) source, whereas other serotype 1/2a strains (e.g., RM3175 and RM3106) exhibited lower fitness. The least fit strain, RM3175, had the shortest doubling time of the strains in this study, and this may be one reason for its low fitness overall in the FDA BAM protocol. However, RM3106, another low-fitness serotype 1/2a strain, had doubling times equivalent to those of the serotype 4b strains tested, and the very-high-fitness serotype 1/2a strain RM2985 had one of the longer doubling times in BLEB containing supplements. Therefore, differences in fitness cannot be attributed to differences in growth rate alone. Potential differences in fitness could be attributed to the ability of a strain to survive the entire 48-h enrichment process; however, as shown in Table 3, all the strains produced similar numbers of cells in monoculture enrichments. Differences in the total number of cells of a given strain were seen only when the cells were mixed in a coculture, Therefore, the individual abilities to survive the 48-h enrichment culture do not explain the differences seen in coculture. These data suggest that there were fitness differences in coculture as a result of competition between L. monocytogenes strains, as well as any complexities added by the presence of foods. Another area of potential difference could be the plating efficiency of the strains on MOX agar, but in our experience L. monocytogenes strains have the same plating efficiency on MOX agar as on the nonselective medium TSYE agar (data not shown).
For the comparison of the coculture enrichments with plain media to the coculture enrichments with added foods, several aspects are worth noting. First, the variability in the data was smaller with plain media. This is understandable considering that the addition of food adds factors that could affect growth and/or fitness, including resident food microbiota, nutrients, inhibitors, and chemicals (e.g., pesticides and preservatives). Second, in some instances the fitness of strain pairs changed with the addition of foods. The serotype 4b strain RM2199 was more fit than some of the serotype 1/2a strains with food in the enrichments. The serotype 1/2a strain RM3364 was recovered at higher levels in plain medium than in the food enrichments when it was paired with the serotype 4b strain RM2387. RM3364 was also more fit in pairings with serotype 4b strain RM2199 in lettuce than in cheese. It should be noted that RM2199, although a human isolate, resulted from an outbreak associated with contaminated cheese. Additional variables in the enrichment, such as the presence of food and/or another L. monocytogenes strain, seemed to challenge the fitness of a strain.
The three strains that were most fit were serotype 1/2a strains RM2985 and RM3316 and serotype 4b strain RM2387, all of which were environmental isolates (isolated from bulk milk, silage, and mint, respectively) that were isolated originally using antimicrobial enrichments (Table 1). Several investigators have described persistent or recurrent strains of L. monocytogenes that are routinely found in processing plants or agricultural environments (7, 10, 12, 15). Presumably, these persistent strains are well suited to their environments and also potentially to the enrichment procedures with which they are routinely found. The very fit strain RM2985 has been identified as a persistent strain in a previous study (3).
Another aspect to consider when competitive fitness is assessed is how the strains were originally isolated. Environmental and food isolates obtained since the late 1980s are most likely to have been isolated using antimicrobial agent-based enrichments similar to that used here. We included patient isolates in part because they were more likely to have been isolated by methods not requiring selective enrichment culture, especially if they were isolated from normally sterile body fluids. Similarly, RM3364 was included since it is an old isolate, not found with the present antimicrobial enrichments, to see if it could compete in the current protocols. While the original antimicrobial agent-based enrichments were developed and tested with older strains, the colony immunoblot method provided a way to test these strains directly in mixed cultures with recent isolates with a large number of colonies to analyze. The serotype 1/2a strain RM3364 proved to be a midlevel competitor with the serotype 4b strains overall, doing very well in plain media and with foods. On the other hand, none of the human isolates (serotype 4b strains RM2199 and RM3176 and serotype 1/2a strain RM3175), which were also isolated from non-antimicrobial agent enrichments, were among the fittest isolates in this study. Therefore, with the limited number of strains tested here, it is difficult to conclude that the FDA BAM enrichment protocol is inherently biased against the strains not isolated with the use of antimicrobial agents. The possibility that clinical isolates might be less fit in enrichment culture than nonclinical isolates is an avenue for future studies.
A previous study reported a bias toward serotype 1/2a strains over serotype 4b strains in UVM I and UVM II, which are enrichment media that are different from the medium used here (4). BLEB has a slightly higher nutrient content (higher concentrations of tryptone and yeast extract, with addition of soytone, dextrose, and pyruvate) than UVM I and II. Additionally, BLEB contains twice as much nalidixic acid as both UVM media and less acriflavine (BLEB, 10 ml/liter acriflavine; UVM I, 12 mg/liter; UVM II, 25 mg/liter). Bruhn et al. (4) speculated that acriflavine might be a cause of the bias in the UVM media; however, when we tested our strains in BLEB containing either 10 or 12 mg/liter acriflavine, there was no difference in growth of any of the strains with the two different acriflavine concentrations (data not shown). Therefore, the lack of bias seen in the present study may be related to nutrient factors present in BLEB that are absent or present at lower concentrations in UVM and/or to the strains used in the studies. The lack of serotype or lineage bias seen here with the BLEB enrichments suggests that BLEB might facilitate isolation of more L. monocytogenes strains present in food than the UVM I and UVM II dual-enrichment protocol. We modified the BAM protocol in one area and incubated the MOX agar plates for 3 days instead of 2 days, so that the colonies of the slowly growing strain RM3175 would be visible. Since RM3175 was originally a human clinical isolate, we recommend that the BAM method should be amended to allow slowly growing, potentially disease-causing isolates to grow on selective agar plates. Nevertheless, with the limited number of strains tested here, we observed that certain strains of L. monocytogenes were more likely to be isolated than others with the FDA BAM enrichment protocol but that the difference was not attributable to serotype or lineage. This may have been due to differences in fitness that are related to the ability of a strain to survive in a given niche and that do not necessarily make it ideally suited to survive enrichment culture when it is competing with a more fit strain.
Acknowledgments
We thank M. Borucki, C. Donnelly, R. Meinersmann, D. Portnoy, and M. Wiedmann for strains. We thank J. Duhé for technical assistance and J. Palumbo for helpful discussions and critical reading of the manuscript. We also thank the reviewers for very helpful suggestions.
This research was funded by the U.S. Department of Agriculture (Agricultural Research Service CRIS project number 5325-42000-040-00D).
REFERENCES
- 1.Bibb, W. F., B. Schwartz, B. G. Gellin, B. D. Plikaytis, and R. E. Weaver. 1989. Analysis of Listeria monocytogenes by multilocus enzyme electrophoresisand application of the method to epidemiologic investigations. Intl. J. Food Microbiol. 8:233-239. [DOI] [PubMed] [Google Scholar]
- 2.Borucki, M. K., M. J. Krug, W. T. Muraoka, and D. R. Call. 2003. Discrimination among Listeria monocytogenes isolates using a mixed genome DNA microarray. Vet. Microbiol. 92:351-362. [DOI] [PubMed] [Google Scholar]
- 3.Borucki, M. K., J. D. Peppin, D. White, F. J. Loge, and D. R. Call. 2003. Variation in biofilm formation among strains of Listeria monocytogenes. Appl. Environ. Microbiol. 69:7336-7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruhn, J. B., B. F. Vogel, and L. Gram. 2005. Bias in the Listeria monocytogenes enrichment procedure: lineage 2 strains outcompete lineage 1 strains in University of Vermont selective enrichments. Appl. Environ. Microbiol. 71:961-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buncic, S., S. M. Avery, J. Rocourt, and M. Dimitrijevic. 2001. Can food-related environmental factors induce different behaviour in two key serovars, 4b and 1/2a, of Listeria monocytogenes? Int. J. Food Microbiol. 65:201-212. [DOI] [PubMed] [Google Scholar]
- 6.Gorski, L., J. D. Palumbo, and R. E. Mandrell. 2003. Attachment of Listeria monocytogenes to radish tissue is dependent upon temperature and flagellar motility. Appl. Environ. Microbiol. 69:258-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harvey, J., and A. Gilmour. 2001. Characterization of recurrent and sporadic Listeria monocytogenes isolates from raw milk and nondairy foods by pulsed-field gel electrophoresis, monocin typing, plasmid profiling, and cadmium and antibiotic resistance determination. Appl. Environ. Microbiol. 67:840-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hitchins, A. D. 2003. Detection and enumeration of Listeria monocytogenes in foods. In Bacterial analytical manual online. [Online.] U.S. Food and Drug Administration, Washington, D.C. http://www.cfsan.fda.gov/∼ebam/bam-10.html.
- 9.Jeffers, G. T., J. L. Bruce, P. L. McDonough, J. Scarlett, K. J. Boor, and M. Wiedmann. 2001. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147:1095-1104. [DOI] [PubMed] [Google Scholar]
- 10.Kathariou, S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811-1829. [DOI] [PubMed] [Google Scholar]
- 11.Loncarevic, S., W. Tham, and M.-L. Danielsson-Tham. 1996. The clones of Listeria monocytogenes detected in food depend on the method used. Lett. Appl. Microbiol. 22:381-384. [DOI] [PubMed] [Google Scholar]
- 12.Lunden, J. M., T. J. Autio, A.-M. Sjoberg, and H. J. Korkeala. 2003. Persistent and nonpersistent Listeria monocytogenes contamination in meat and poultry processing plants. J. Food Prot. 66:2062-2069. [DOI] [PubMed] [Google Scholar]
- 13.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulson, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norton, D. M., M. A. McCamey, K. L. Gall, J. M. Scarlett, K. J. Boor, and M. Wiedmann. 2001. Molecular studies on the ecology of Listeria monocytogenes in the smoked fish processing industry. Appl. Environ. Microbiol. 67:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palumbo, J. D., M. K. Borucki, R. E. Mandrell, and L. Gorski. 2003. Serotyping of Listeria monocytogenes by enzyme-linked immunosorbent assay and identification of mixed-serotype cultures by colony immunoblotting. J. Clin. Microbiol. 41:564-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perry, C., and C. W. Donnelly. 1990. Incidence of Listeria monocytogenes in silage and its subsequent control by specific and nonspecific antagonism. J. Food Prot. 53:642-647. [DOI] [PubMed] [Google Scholar]
- 18.Pritchard, T. J., and C. W. Donnelly. 1999. Combined secondary enrichment of primary enrichment broths increases Listeria detection. J. Food Prot. 62:532-535. [DOI] [PubMed] [Google Scholar]
- 19.Ryser, E. T., S. M. Arimi, M. M.-C. Bunkuki, and C. W. Donnelly. 1996. Recovery of different Listeria ribotypes from naturally contaminated, raw refrigerated meat and poultry products with two primary enrichment media. Appl. Environ. Microbiol. 62:1781-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlech, W. F., III. 2000. Epidemiology and clinical manifestations of Listeria monocytogenes infection, p. 473-479. In V. A. Fishetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 21.Ward, T. J., L. Gorski, M. K. Borucki, R. E. Mandrell, J. Hutchins, and K. Pupedis. 2004. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 186:4994-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiedmann, M., J. L. Bruce, C. Keating, A. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in their pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]