Abstract
Advances in the growth of hitherto unculturable soil bacteria have emphasized the requirement for rapid bacterial identification methods. Due to the slow-growing strategy of microcolony-forming soil bacteria, successful fluorescence in situ hybridization (FISH) requires an rRNA enrichment step for visualization. In this study, catalyzed reporter deposition (CARD)-FISH was employed as an alternative method to rRNA enhancement and was found to be superior to conventional FISH for the detection of microcolonies that are cultivated by using the soil substrate membrane system. CARD-FISH enabled real-time identification of oligophilic microcolony-forming soil bacteria without the requirement for enrichment on complex media and the associated shifts in community composition.
With the advent of novel cultivation strategies, there are increasing requirements for real-time identification methods for bacteria. Many of these cultivation methods result in the selection of slow-growing bacteria that do not allow for traditional signs of growth, such as turbidity or colony formation, to be measured (8, 11, 12, 24). Instead, these bacteria grow in simulated natural environments consisting of minimal levels of substrate, resulting in slow-growing microcolonies that often reach cell densities of only <106 per ml (15, 24). In many cases, these bacteria may be described as oligophiles that select for slow-growth strategies that are detectable only by microscopic visualization (8, 17, 22).
Fluorescence in situ hybridization (FISH) has become one of the most widely used tools in microbial ecology (1, 2). To date, FISH has been used successfully to identify bacteria from eutrophic systems; however, low signal intensities are experienced when identifying oligotrophic aquatic marine or soil bacteria (4, 5, 9, 20). FISH targets rRNA, and the success of hybridization is affected by the cell size, growth rate, cellular ribosome content, accessibility of target sites, and number of rRNA operon copies per cell (3, 4, 13).
Soil substrate membrane system (SSMS) microcultivation on soil extracts enables the growth of micro-CFU (mCFU)-forming soil bacteria, but the identification and isolation of specific bacteria using conventional FISH probes have proven difficult (8). Recently, the modification of FISH protocols to include rRNA enrichment facilitated the successful identification of SSMS-microcultivated bacteria from previously uncultivated lineages, including the candidate division TM7 (8), and from more specialized environments such as methane-oxidizing bacteria (18). Bacterial rRNA enrichment requires a short incubation period on a nutrient-rich medium that increases the rRNA content required for visualization of fluorescence. Alternatively, catalyzed reporter deposition (CARD)-FISH has been reported as an in situ amplification method utilizing horseradish peroxidase, which enhances bacterial cell detection. The method, which has been successfully applied to marine bacteria (14), archaea (10), and bacterial biofilms on seaweed (21) and within benthic organic matter (7), is used here as an alternative to rRNA enrichment.
We employed CARD-FISH, modified for mCFU on polycarbonate membranes, as a rapid-identification method (without the need for rRNA enrichment) for slow-growing oligophilic bacteria. Moreover, we hypothesized that the SSMS community profile could be significantly altered during the microcultivation enrichment process. Here, we report on a shift in morphology of SSMS microcolonies and in the community profile following short-term enrichment on complex media.
Microcultivation using the SSMS.
Seven SSMS replicates were prepared as described previously for three soil samples (8). Soil samples were taken from two sites at Macquarie University (MQA and MQB) and one from Rouse Hill (RH), Sydney, New South Wales, Australia. A subsample of each soil was diluted (1:200) in prefiltered (pore size, 0.2 μm) distilled H20 and vortexed, and 50 μl of each soil inoculum was placed into 10 ml distilled H2O, mixed, and filtered onto 0.2 μm, 25-mm white polycarbonate (PC) membranes (Millipore, North Ryde, Australia). Anopore (0.02 μm) tissue culture inserts (TCIs; Nunc, A/S, Roskilde, Denmark) were filled with 3 g of sieved soil, and microcosms were prepared as described previously (8) and incubated for 8 days at 25°C. To enable visualization of standard FISH signals, three PC membranes from each soil sample were randomly selected for rRNA enrichment. For enrichment, PC membranes were taken off the TCIs on day 8 and placed directly onto 0.1×-strength tryptic soy agar for 8-h enrichment. Three membranes that were not selected for enrichment were kept on the TCIs during this incubation period. Both enriched and standard PC membranes were washed twice by floating on phosphate-buffered saline for 20 min and split for fixation or DNA extraction (MQB). For DNA extraction, half of each PC membrane was placed into a sterile microcentrifuge tube with 500 μl DNA-grade sterile water. Lysates were prepared for PCR (8) and denaturing gradient gel electrophoresis (DGGE) as described previously (19). For FISH preparation, remaining PC membrane halves were fixed in 4% formaldehyde overnight at 4°C, washed twice in phosphate-buffered saline, and then split for comparison of FISH protocols.
Secondary growth of SSMS-selected bacteria and FISH detection.
The seventh PC membrane from the MQB sample was placed into a sterile microcentrifuge tube with 500 μl 0.9% NaCl solution. A portion (10 μl) was then placed into three wells of a 96-well plate which contained 200 μl 0.01× Ravan medium (22). The plate was incubated for 7 days at 25°C. On day 7, 100 μl from each well was placed into a sterile microcentrifuge tube and cells were washed and fixed in 4% formaldehyde (100 μl). Portions (20 μl) were then filtered onto 0.2-μm PC membranes for comparison of FISH protocols.
Correlation of total bacterial staining and conventional FISH.
The FISH protocol optimized for mCFU bacteria on PC membranes was used as described previously (8). The eubacterial probe EUB338-i (1), which targets most eubacteria, was labeled with Alexa534 (Invitrogen) for a direct comparison with CARD-FISH. The eukayotic probe EUK, 5′-ACC AGA CTT GCC CTC C-3′ (6), was used as a negative control for hybridization specificity, and conventional hybridizations were carried out at 46°C for 3 h at the recommended stringencies. For the observation of total bacterial cells, SYBR green II RNA gel stain (Bioscientific, Gymea, Australia) was used as a counterstain and PC membranes were mounted onto slides using DABCO antifade (8). An Olympus Fluoview FV 300 confocal laser scanning microscope was used for the visualization of microcolonies (Olympus, Mount Waverley, Victoria, Australia). Twenty random images were captured per hybridization and were analyzed using Image J software (http://rsb.info.nih.gov/ij).
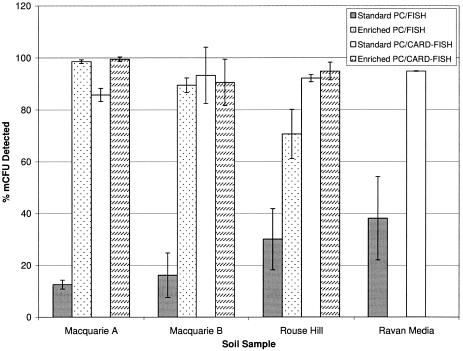
Conventional FISH detected between only 12.6 and 30.1% (standard deviation [SD], 1.7 to 11.8) of bacterial microcolonies when PC membranes were not subjected to rRNA enrichment (Fig. 1). Single-cell analysis of SSMS-selected bacteria grown in diluted media revealed a similar trend with 38% (SD, 16) of cells found positive by conventional FISH. These results are in accordance with previous findings of mCFU soil bacteria isolated from environmental samples (8, 18, 23). In contrast, a significant increase in conventional FISH detection was observed after rRNA enrichment. More than 98% (SD, 0.7) and 70% (SD, 9.5) of total mCFU were visualized for MQB and RH samples, respectively (Fig. 1).
FIG. 1.
Comparison of conventional FISH and CARD-FISH for the detection of bacterial microcolonies grown on standard or enriched PC membranes using the SSMS. Oligophilic bacteria grown in diluted nutrient media following SSMS cultivation were also analyzed using both detection methods.
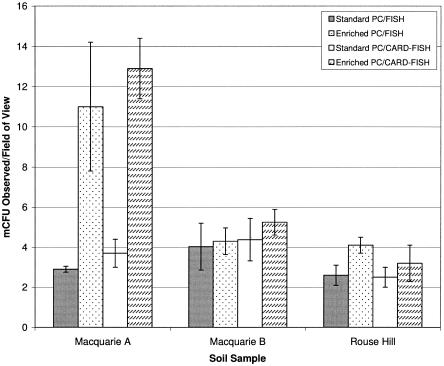
FISH staining of PC membranes revealed a shift in the number of microcolonies present per field of view following enrichment, with up to threefold more mCFU observed for the MQA sample (t test; P, <0.05) (Fig. 2). Microscopic analysis revealed diverse morphological changes between mCFU growing on standard and enriched PC membranes, with an increase in large mCFU morphotypes containing >50 cells per colony observed for MQB and RH samples (Fig. 3). In these cases, there was no significant increase (t test; P, <0.05) in mCFU number per field of view relative to nonenriched membranes (Fig. 2). Moreover, the dramatic increase in microcolonies observed per field of view for the MQA soil sample was linked to the presence of large rod-shaped mCFU which were absent before enrichment (Fig. 3A). Additionally, analysis of single cells grown in oligophilic media revealed that the majority of coccus-shaped bacteria were FISH negative, while all the various rod-shaped bacteria that were observed were FISH positive (images not shown).
FIG. 2.
The total numbers of bacterial microcolonies detected per field of view following SSMS microcultivation on standard and enriched PC membranes, determined by SYBR green staining.
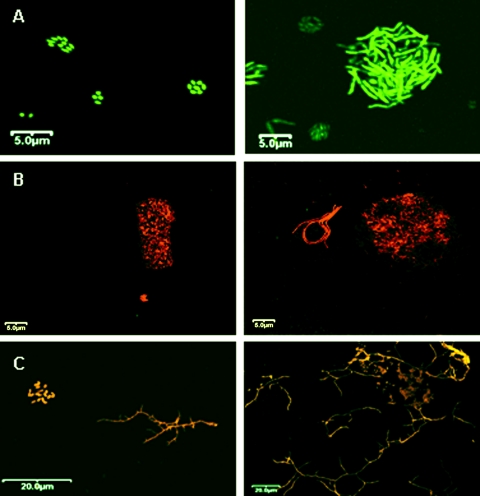
FIG. 3.
Confocal images of standard and enriched membranes from three soil samples following hybridization (red-orange) and total bacterial staining (green) at a magnification of ×600. The left panels refer to standard membranes, and the right panels represent enriched membranes. MQA (A) soil was stained with only SYBR green. MQB (B) soil and RH (C) soil were dual; stained with CARD-FISH (red) and SYBR green. Marked differences in microcolony morphology following rRNA enrichment were observed.
CARD-FISH as an alternative detection method for oligophilic bacteria.
Here, the CARD-FISH method reported for biofilms on seaweed surfaces (21) was modified for SSMS PC membranes. In this case, the horseradish peroxidase probe (Thermo Electron) targeting most bacteria (EUB388-i) was used for hybridizations and Alexa546 tyramides were used for signal amplification (21). As the final step, PC membranes were counterstained with SYBR green. Results show that CARD-FISH facilitated the identification of slow-growing bacterial species without the requirement for rRNA enrichment (Fig. 1). CARD-FISH enabled detection of up to 95.2% (SD, 0.5) of microcolonies on standard membranes relative to 30.1% for conventional FISH (Fig. 1). Bacteria grown in oligophilic media also revealed a marked increase in detection with 94.9% (SD, 0.1) of single cells detected compared with 38% detected by conventional FISH. Additionally, the significant increase in cell detection allowed real-time identification of mCFU without changing the SSMS community through increases in mCFU number and morphology (Fig. 2 and 3).
Community shifts following enrichment on complex media.
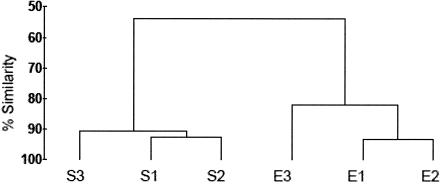
Following the observation that the number of MQB bacterial mCFU observed on PC membranes did not increase significantly after rRNA enrichment (Fig. 2), DGGE was performed to assess the impact of the process on the SSMS bacterial community. 16S rRNA gene fragments were obtained from the DNA lysates by PCR using the primers 341F-GC and 907RC (16), and the products were separated by DGGE using a denaturing gradient of 35 to 55% (19). Amplified bands were determined as present or absent, and the similarity of bacterial communities was determined by cluster analysis using the PRIMER-E program version 5.2.2 (Plymouth). The group average method was used to construct a dendrogram (Fig. 4). It was found that the community structure had shifted in the indigenous community following short-term enrichment on complex media. The number of bands in the DGGE profiles (not shown) fell from 26 for the standard technique to 21 for enriched samples, confirming the enrichment of the predominant species present. Changes to the microbial community following rRNA enhancement confirmed the requirement for a rapid FISH enhancement method that does not alter the SSMS microbial community.
FIG. 4.
Dendrogram generated by cluster analysis of 16S rRNA gene DGGE profiles indicating similarity of SSMS-cultivated bacterial communities from MQB soil grown on standard (S) or enriched (E) polycarbonate membranes.
While cultivation strategies successfully lead to the isolation of slow-growing uncharacterized species, the ability to identify these bacteria using molecular approaches like FISH is difficult because of low signal intensities. The low sensitivity of detection is largely due to the rRNA content of small cells such as bacterioplankton, marine archaea, or soil bacteria (4, 14). This study confirmed that CARD-FISH amplification of the rRNA signal allows for enrichment-independent in situ cell detection of microcolony bacteria on standard membranes and in diluted culture media (Fig. 1). CARD-FISH highlighted the need to source real-time detection methods for oligophilic bacteria, as rapid shifts in microcolony morphologies and communities appear when SSMS soil bacteria undergo short-term enrichment on nutrient-rich media (Fig. 2 to 4). We conclude that CARD-FISH is superior to conventional FISH as an identification method for slow-growing oligophilic and microcolony-forming soil bacteria.
Acknowledgments
Belinda Ferrari was supported by a Macquarie University research fellowship and a Macquarie University New Staff Grant.
We thank Debra Birch from the microscopy unit at Macquarie Univerity for advice and guidance with confocal microscopy imaging.
REFERENCES
- 1.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in-situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behrens, S., C. Ruhland, J. Inacio, H. Huber, A. Fonseca, I. Spencer-Martins, B. Fuchs, and R. Amann. 2003. In situ accessability of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probes. Appl. Environ. Microbiol. 69:1748-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binnerup, S., J. Bloem, B. Hansen, W. Wolters, M. Veninga, and M. Hansen. 2001. Ribosomal RNA content in microcolony forming soil bacteria measured by quantitative 16S rRNA hybridization and image analysis. FEMS Microbiol. Ecol. 37:231-237. [Google Scholar]
- 5.Bouvier, T., and P. A. del Giorgio. 2003. Factors influencing the detection of bacterial cells using fluorescence in situ hybridization (FISH): a quantitative review of published reports. FEMS Microbiol. Ecol. 44:3-15. [DOI] [PubMed] [Google Scholar]
- 6.Deere, D., G. Vesey, M. Milner, K. Williams, N. Ashbolt, and D. Veal. 1998. Rapid method for fluorescent in situ ribosomal RNA labelling of Cryptosporidium parvum. J. Appl. Microbiol. 85:807-818. [DOI] [PubMed] [Google Scholar]
- 7.Fazi, S., S. Amalfitano, J. Pernthaler, and A. Puddu. 2005. Bacterial communities associated with benthic organic matter in headwater stream microhabitats. Environ. Microbiol. 7:1633-1640. [DOI] [PubMed] [Google Scholar]
- 8.Ferrari, B., S. Binnerup, and M. Gillings. 2005. Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl. Environ. Microbiol. 71:8714-8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glockner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishii, K., M. Mußmann, B. J. MacGregor, and R. Amann. 2004. An improved fluorescence in situ hybridization protocol for the identification of bacteria and archaea in marine sediments. FEMS Microbiol. Ecol. 50:203-212. [DOI] [PubMed] [Google Scholar]
- 11.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaeberlein, T., K. Lewis, and S. Epstein. 2002. Isolating ‘uncultivable’ microorganisms in pure culture in a simulated natural environment. Science 296:1127-1129. [DOI] [PubMed] [Google Scholar]
- 13.Moeseneder, M. M., J. M. Arrieta, and G. J. Herendl. 2005. A comparison of DNA- and RNA-based clone libraries from the same marine bacterioplankton community. FEMS Microbiol. Ecol. 51:341-352. [DOI] [PubMed] [Google Scholar]
- 14.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rappe, M. S., S. A. Connon, K. L. Vergin, and S. L. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 16.Schafer, H., L. Bernard, C. Courties, P. Lebaron, P. Servais, R. Pukall, E. Stackebrandt, M. Troussellier, T. Guindulain, J. Vives-Rego, and G. Muyzer. 2001. Microbial community dynamics in mediterranean nutrient-enriched seawater mesocosms: changes in the genetic diversity of bacterial populations. FEMS Microbiol. Ecol. 34:243-253. [DOI] [PubMed] [Google Scholar]
- 17.Simu, K., and A. Hagstrom. 2004. Oligotrophic bacteriplankton with a novel single-cell life strategy. Appl. Environ. Microbiol. 70:2445-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svenning, M. M., I. Wartiainen, A. G. Hestnes, and S. J. Binnerup. 2003. Isolation of a methane oxidising bacteria from soil by use of a soil substrate membrane system. FEMS Microbiol. Ecol. 1509:1-8. [DOI] [PubMed] [Google Scholar]
- 19.Taylor, P. M., P. J. Schupp, P. Dahllof, S. Kjelleberg, and P. Steinberg. 2004. Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ. Microbiol. 6:121-130. [DOI] [PubMed] [Google Scholar]
- 20.Teira, E., T. Reinthaler, A. Pernthaler, J. Pernthaler, and G. J. Herndl. 2004. Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by bacteria and archaea in the deep ocean. Appl. Environ. Microbiol. 70:4411-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tujula, N. A., C. Holmstrom, M. Mußmann, R. Amann, S. Kjelleberg, and G. R. Crocetti. 2005. A CARD-FISH protocol for the identification and enumeration of epiphytic bacteria on marine algae. J. Microbiol. Methods [Online.] [DOI] [PubMed]
- 22.Watve, M., V. Shejval, C. Sonawane, M. Rahalkar, A. Matapurkar, Y. Shouche, M. Patole, N. Phadnis, A. Champhenkar, K. Damle, S. Karandikar, V. Kshirsagar, and M. Jog. 2000. The ‘K’ selected oligophilic bacteria: a key to uncultured diversity? Curr. Sci. 78:1535-1542. [Google Scholar]
- 23.Winding, A., S. Binnerup, and J. Sørensen. 1994. Viability of indigenous soil bacteria assayed by respiratory activity and growth. Appl. Environ. Microbiol. 60:2869-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zengler, K., G. Toledo, M. Rappe, J. Elkins, E. J. Mathur, J. M. Short, and M. Keller. 2002. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA 99:15681-15686. [DOI] [PMC free article] [PubMed] [Google Scholar]