Abstract
The use of Bacillus thuringiensis as a biopesticide is a viable alternative for insect control since the insecticidal Cry proteins produced by these bacteria are highly specific; harmless to humans, vertebrates, and plants; and completely biodegradable. In addition to Cry proteins, B. thuringiensis produces a number of extracellular compounds, including S-layer proteins (SLP), that contribute to virulence. The S layer is an ordered structure representing a proteinaceous paracrystalline array which completely covers the surfaces of many pathogenic bacteria. In this work, we report the identification of an S-layer protein by the screening of B. thuringiensis strains for activity against the coleopteran pest Epilachna varivestis (Mexican bean beetle; Coleoptera: Coccinellidae). We screened two B. thuringiensis strain collections containing unidentified Cry proteins and also strains isolated from dead insects. Some of the B. thuringiensis strains assayed against E. varivestis showed moderate toxicity. However, a B. thuringiensis strain (GP1) that was isolated from a dead insect showed a remarkably high insecticidal activity. The parasporal crystal produced by the GP1 strain was purified and shown to have insecticidal activity against E. varivestis but not against the lepidopteran Manduca sexta or Spodoptera frugiperda or against the dipteran Aedes aegypti. The gene encoding this protein was cloned and sequenced. It corresponded to an S-layer protein highly similar to previously described SLP in Bacillus anthracis (EA1) and Bacillus licheniformis (OlpA). The phylogenetic relationships among SLP from different bacteria showed that these proteins from Bacillus cereus, Bacillus sphaericus, B. anthracis, B. licheniformis, and B. thuringiensis are arranged in the same main group, suggesting similar origins. This is the first report that demonstrates that an S-layer protein is directly involved in toxicity to a coleopteran pest.
Chemical insecticides cause many environmental problems. Their exhaustive use has resulted in pollution of soils and underground water. Moreover, a number of synthetic insecticides have lost their efficacy against certain pests due to the development of resistance in these insects. The use of Bacillus thuringiensis as a biopesticide is a viable alternative for insect control in agriculture and other areas (i.e., disease vectors) and will intensify crop production in an economically sustainable and environmentally friendly way. The Cry toxins produced by B. thuringiensis are highly specific; harmless to humans, vertebrates, and plants; and completely biodegradable, so they cause no residual toxic products to accumulate in the environment (26). To date, over 200 cry gene sequences have been determined and classified into 44 families and different subclasses (7). Additionally, B. thuringiensis produces a number of extracellular compounds, such as phospholipases, proteases, and chitinases, and other toxins, such as β-exotoxin and vegetative insecticidal proteins, that may contribute to virulence (26).
The S-layer is an ordered structure of proteinaceous paracrystalline arrays which completely covers the surfaces of many archaea and bacteria (3, 25) and constitutes up to 15% of the total cell protein. The function of S-layer proteins (SLP) has not been accurately defined. It was proposed that these proteins are involved in cell integrity and shape maintenance (3). Also, it has been hypothesized that they may be involved in macromolecule exchange with the environment since they are the outermost cell envelope component (3). In some gram-negative pathogens, they have been implicated in virulence and resistance to complement-mediated killing (3, 23). In Bacillus cereus, the SLP promotes interactions with human leukocytes and with the host, contributing to pathogenicity (14). It has been proposed that in Bacillus anthracis, the S-layer and the capsule might cooperate in the interaction with the host (21).
Several S-layer genes have been cloned, and their amino acid sequences have revealed that they have low similarity except in the cell wall-targeting S-layer homology domain, which is involved in the anchoring of these proteins to the peptidoglycan-linked polysaccharides (25). Thus, different types of SLP may have different functions. SLP from Bacillus species have a molecular mass of between 66 and 255 kDa. In B. anthracis, two different SLP (Sap and EA1) have been described previously (18, 21). The presence of these proteins is not required for normal encapsulation of the bacilli (19). These proteins appear sequentially in a growth phase-dependent manner, with the synthesis of Sap preceding that of EA1 (21). In B. thuringiensis subsp. galleria, an SLP termed SlpA is similar to the Sap of B. anthracis. The construction of a null mutant with a mutation in the corresponding gene revealed that the mutant strain does not express another S-layer gene (20). A parasporal body protein named CTC, described to occur in B. thuringiensis subsp. finitimus (GenBank accession number AY460125), shows high identity with the S-layer protein EA1 from B. anthracis (27). CTC has a molecular size of 100 kDa and forms a parasporal body during the sporulation phase of growth. So far, the role of SLP of B. thuringiensis in virulence against insect pests has never been described.
In this work, we report the screening of B. thuringiensis strains for activity against the coleopteran pest Epilachna varivestis (Mexican bean beetle; Coleoptera: Coccinellidae). This is a serious pest in various legume crops in North America and in other crops in Asia and Africa, such as cucurbits, solanaceae, beans, maize, sorghum, rice, wheat, cotton, sesame, lettuce, soybean, and cowpea (12). We found that B. thuringiensis strains expressing known Cry toxins active against coleopteran species (Cry3A, Cry3Ba, Cry3Bb, Cry3Ca, and Cry7Aa) were not active against this pest. Two B. thuringiensis strain collections containing unidentified Cry proteins and B. thuringiensis strains isolated from dead insects were used to search for insecticide activity against this pest. One B. thuringiensis strain (GP1) was selected, and the parasporal crystal was characterized. Our results show that the GP1 strains produced an SLP that confers toxicity to the E. varivestis coleopteran pest.
MATERIALS AND METHODS
Bacterial strains.
B. thuringiensis strains that express the different Cry3 or Cry7 toxins were kindly supplied by J. Van Rie, Bayer BioScience N.V., Belgium. B. thuringiensis strains from the Mexican collection (4) and from the Plant Genetic System (now Bayer BioScience N.V.) collection were also used in this study. Novel B. thuringiensis strains were isolated from dead insect samples by the acetate selection method (28). The E. varivestis corpses were washed twice with sodium hypochlorite 1% and with sterile water before homogenization and B. thuringiensis strain isolation.
Bioassays.
The bioassays against E. varivestis were performed with spore-crystal suspensions or with pure crystal protein using the leaf dip technique. Squares of 4 cm by 4 cm from the Phaseolus vulgaris plant leaves were dipped in the toxin diluents and allowed to dry. Different concentrations of each toxin were tested with five larvae at the first instar stage per treated leaf. Four repetitions per concentration were done. Mortality and leaf damage were determined after 6 days. Bioassays against first-instar larvae of Manduca sexta or Spodoptera frugiperda were done in artificial diet assays as described previously (4). Bioassays against Aedes aegypti larvae were done in 100 ml H2O as described previously (13).
Purification of the crystal inclusion present in the GP1 strain.
The GP1 strain was grown in petri dishes containing solid nutrient broth sporulation medium (15). The spore-crystal mixture was collected in 5 ml of sterile water and centrifuged for 10 min at 10,000 rpm. The supernatant was recovered, and the pellet containing only spores was discarded. This step was repeated five times in order to eliminate all spores from the suspension. Finally, the crystal inclusions were recovered by centrifugation at 19,000 rpm for 30 min. The crystal proteins were solubilized in 50 mM Na2CO3, pH 10.5, with 0.2% β-mercaptoethanol and purified by anion-exchange chromatography in a Q-Sepharose column, using fast protein liquid chromatography (Pharmacia) (11).
N-terminal sequencing.
N-terminal sequencing of the crystal protein produced by the GP1 strain was performed at the Microchemistry Facility of Harvard University (Cambridge, Massachusetts) after separation on 7% sodium dodecyl sulfate (SDS) gels and transfer onto polyvinylidene difluoride membranes (Millipore Co., Bedford, MA) in a semidry transfer chamber as directed by the manufacturer. N-terminal sequencing was also carried out from a trypsin fragment of the GP1 crystal protein that was purified by size exclusion high-performance liquid chromatography (11).
Antibody production.
A New Zealand White rabbit was immunized with the crystal protein produced by the GP1 strain by subcutaneous injections. One milligram of GP1 SLP in phosphate-buffered saline was emulsified with Freund's complete adjuvant and injected at five sites on the back of the rabbit. The rabbit was boosted three times at 15-day intervals with 1 mg of the GP1 SLP mixed with incomplete Freund's adjuvant. A sample of the blood was collected 40 days after the primary immunization.
Determination of the DNA sequence of the SLP gene produced by the GP1 strain.
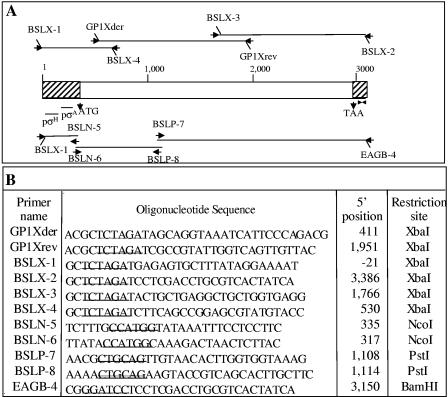
Based on the N-terminal sequencing of the crystal protein and of an internal tryptic fragment of the crystal protein, two PCR primers, GP1Xder and GP1Xrev, that amplify a 1,536-bp PCR product were designed (Fig. 1). PCRs were performed by standard techniques (24) with Pfu DNA polymerase (Stratagene, La Jolla, CA). Total DNA was extracted from the GP1 strain (22) and used as a template in all PCRs. The PCR product was sequenced in the in-house automated sequencing facilities of the Instituto de Biotecnología, UNAM (Cuernavaca, México). BLAST analysis of the sequence identified two related sequences of S-layer genes (GenBank accession numbers U38842 and X99724). Alignment of these sequences demonstrated that they had similar 5′ and 3′ ends. Four PCR primers were designed from the 5′- and 3′-terminal ends of these S-layer genes and from the internal sequence of the GP1-amplified fragment. Primers BSLX-1 and BSLX-4 amplified a 553-bp 5′-end fragment, and primers BSLX-3 and BSLX-2 amplified a 1,372-bp 3′-end fragment (Fig. 1). Each PCR fragment was digested with XbaI (New England BioLabs, Beverly, MA) and cloned into pBluescript SK (Stratagene, La Jolla, CA) previously digested with the XbaI restriction enzyme. The ligation products were purified by extraction with phenol-chloroform, precipitated with ethanol, and electroporated into electrocompetent Escherichia coli TG1 cells (16). Transformant colonies were grown on LB agar plates and supplemented with ampicillin (100 μg/ml), and selected colonies were mixed with glycerol (20%) and stored at −70°C. These plasmids were named GP1 SL1, GP1 SL2, and GP1 SL3, and their DNA inserts were sequenced at the in-house automated sequencing facilities of the Instituto de Biotecnología, UNAM.
FIG. 1.
Diagrammatic representation of S-layer gene fragments and PCR primers. (A) Representation of the S-layer gene of the GP1 strain and locations of oligonucleotides used as PCR primers (arrows); (B) sequences of oligonucleotides used in PCR amplification. Underlined sequences correspond to restriction sites.
Cloning and expression of GP1 SLP in B. thuringiensis.
The complete putative S-layer gene was reconstituted by cloning three PCR fragments into the pHT315 plasmid (16). The first PCR product containing the promoter region was obtained with BSLX-1 and BSLN-5. Primer BSLX-1 has an extra 10 bp at the 5′ end containing an XbaI restriction site, and primer BSLN-5 has an internal NcoI restriction site (Fig. 1). The second PCR fragment was obtained with primer BSLN-6, which contains an internal NcoI restriction site, and primer BSLP-8, which includes the unique PstI restriction site of the GP1 SL gene. Finally, the third PCR fragment was amplified with primer BSLP-7, which also includes the unique PstI restriction site, and primer EAGB-4, which is similar to BSLX-2 but has 8 extra bp containing the BamHI restriction site at the 5′ end (Fig. 1). Each PCR product was purified and digested with the corresponding restriction enzymes and subcloned separately into pBluescript KS. The DNA fragments obtained from these plasmids were purified, ligated, and inserted into the pHT315 plasmid previously digested with XbaI and BamHI. The product of the ligation reaction was directly transformed in the acrystalliferous B. thuringiensis strain 407 (16), which was kindly provided by Didier Lereclus (Pasteur Institute, France) and was grown at 30°C in LB supplemented with 7.5 μg/ml erythromycin. The resulting plasmid was named pHT-GP1. The B. thuringiensis strain containing pHT-GP1 was grown in petri dishes containing solid sporulation medium supplemented with erythromycin. The spore-crystal mixture was collected in 2 ml sterile water and used in Western blot experiments. Detection was done with anti-GP1 SL polyclonal antibody (dilution, 1/10,000; 1 h) and visualized with a goat anti-rabbit antibody coupled with horseradish peroxidase (Sigma, St. Louis, MO) (dilution, 1/7,500; 1 h), followed by SuperSignal chemiluminescent substrate (Pierce, Rockford, IL) as described by the manufacturers.
Chemical extraction of S-layer protein.
The GP1 strain, the B. thuringiensis strain containing pHT-GP1, and the acrystalliferous B. thuringiensis strain 407 were grown in brain heart infusion broth medium (Difco) until an optical density at 600 nm of 0.9 was reached in order to have exclusively vegetative cells. Cells were pelleted by centrifugation (10 min at 10,000 rpm). Pellets were washed and resuspended in 1/50 of the initial volume of 1, 1.5, or 2 M guanidinium hydrochloride (pH 2.5), as described by Luckevich and Beveridge (17), to extract specifically the cell surface-anchored proteins. The samples were centrifuged for 10 min at 10,000 rpm, and the pellets containing the bacterial cells and the supernatants containing the extracted proteins were then precipitated by adding trichloroacetic acid to a final concentration of 10% (20 min at −20°C). The precipitated proteins were then centrifuged for 10 min at 10,000 rpm, washed two times with H2O, and resuspended in 0.03 N NaOH. An equal volume of 2× Laemmli sample loading buffer was added. Samples were boiled for 5 min, and equal amounts of protein were loaded onto two SDS-10% polyacrylamide gel electrophoresis (PAGE) gels. One gel was stained with Coomassie brilliant blue, and the duplicated gel was electrotransferred onto polyvinylidene difluoride membranes for Western blot detection using anti-GP1 SL polyclonal antibody as described above.
Amino acid sequence alignment and phylogenetic analysis.
An amino acid sequence alignment of 33 different SLP (Table 1) was done with the ClustalW program. The genetic distances among the sequences were calculated with the PROTDIST program in J. Felsenstein′s PHYLIP 3.5 phylogeny inference package with the Dayhoff PAM matrix (9). The program FITCH was then used to estimate phylogenies from the distance matrix data under the additive tree model. The phylogenetic analyses were carried out 100 times by using the bootstrapping tool, which generates multiple data sets, in order to get a strict consensus tree. The consensus phylogenetic trees were computed by the CONSENSE program (9).
TABLE 1.
S-layer proteins used in the phylogenetic analysis
| Species | Gene | No. of amino acids | GenBank accession no. |
|---|---|---|---|
| Bacillus anthracis | sap | 814 | Z36946 |
| Bacillus anthracis | eag | 862 | X99724 |
| Bacillus cereus | S-layer like | 530 | AE017000 |
| Bacillus cereus | S-layer like | 486 | AE0170006N |
| Bacillus licheniformis | olpA | 874 | U38842 |
| Bacillus thuringiensis | GP1 SL | 863 | AY956311 |
| B. thuringiensis galleriae | slpA | 821 | AJ249446 |
| B. thuringiensis finitimus | ctc2 | 862 | AY460125 |
| B. thuringiensis mexicanensis | cryTKD | 823 | D86346 |
| Bacillus thuringiensis | ctc | 816 | AJ012290 |
| Bacillus sphaericus | slaP | 1,176 | M28361 |
| Bacillus sphaericus | sbpA | 1,268 | AF211170 |
| Geobacillus stearothermophilus | sbsA | 1,228 | X71092 |
| Geobacillus stearothermophilus | sbsB | 920 | X98095 |
| Geobacillus stearothermophilus | sbsC | 1,099 | AF055578 |
| Brevibacillus brevis | mwp | 1,053 | M19115 |
| Brevibacillus brevis | hwp | 1,116 | D90050 |
| Campylobacter fetus | sapA2 | 1,109 | S76860 |
| Campylobacter fetus | sapA | 933 | J05577 |
| Campylobacter fetus | sapA1 | 920 | L15800 |
| Campylobacter fetus | sapB2 | 1,112 | AF048699 |
| Campylobacter fetus | sapB | 936 | U25133 |
| Clostridium thermocellum | sdbA | 631 | U49980 |
| Halobacterium halobium | csg | 852 | J02767 |
| Haloferax volcanii | 827 | M62816 | |
| Lactobacillus acidophilus | slpA | 444 | X89375 |
| Lactobacillus helveticus | slpH1 | 439 | X91199 |
| Lactobacillus helveticus | slpH2 | 439 | X92752 |
| Lactobacillus crispatus | cbsA | 440 | AF001313 |
| Rickettsia prowazekii | spaP | 1,612 | M37647 |
| Rickettsia rickettsii | p120 | 1,300 | X16353 |
| Rickettsia typhi | slpT | 1,645 | L04661 |
| Serratia marcescens | slaA | 1,004 | AB007125 |
RESULTS
Isolation of B. thuringiensis strains active against Epilachna varivestis.
The toxicity of control B. thuringiensis strains containing coleopteran-specific Cry toxins (Cry3s and Cry7Aa) was analyzed. These strains showed no toxicity against E. varivestis. We then tested 241 native B. thuringiensis strains that are part of two different strain collections, one (144 strains) from Mexico (4) and the other (97 strains) from different parts of the world, supplied by Bayer BioScience. From this screening, only moderately toxic strains were found (Table 2).
TABLE 2.
Selected Bacillus thuringiensis strains from collections that showed activity against Epilachnia varivestis larvae
| Strain | cry gene | % Mortality
|
|
|---|---|---|---|
| 100 ng/cm2 of treated leaf | 1,000 ng/cm2 of treated leaf | ||
| IB281 | cry3B | 5 | 35 |
| IB312 | 10 | 60 | |
| IB476 | cry3A | 30 | 80 |
| IB209 | cry8C | 12 | 50 |
| IB293 | cry8C | 8 | 40 |
| IB477 | 5 | 35 | |
| 2134N | cry1Aa, cry1Ac | 10 | 50 |
| 306B | cry1Aa | 10 | 55 |
Finally, the toxicity of four B. thuringiensis strains that were isolated from corpses of E. varivestis larvae was assayed. These strains produced similar point crystals composed of a 100-kDa protein and had identical total protein patterns (data not shown). The four strains showed 100% mortality when tested at 100 and 1,000 ng/cm2 of treated leaf.
One of these four strains was selected and named GP1, and the crystal inclusion was purified by successive centrifugation followed by solubilization and anion-exchange chromatography (Fig. 2). The bioassays against E. varivestis larvae performed with spore-crystal mixtures of this strain showed a 50% lethal concentration (LC50) of 16 ng/cm2 of treated bean plant leaf (95% confidence interval, 7 to 25 ng/cm2), and the bioassays performed with pure crystal protein presented an LC50 of 8.6 ng/cm2 of treated leaf (95% confidence interval, 4 to 14 ng/cm2). In contrast, neither the pure crystal nor the spore-crystal mixture of this strain (up to 10,000 ng/cm2 of artificial diet) showed toxic activity against the first-instar larvae of the lepidopteran insects Manduca sexta and Spodoptera frugiperda, and neither of the 10,000-ng/ml mixtures was toxic against the fourth-instar larvae of the dipteran Aedes aegypti.
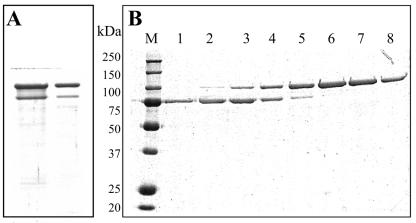
FIG. 2.
SDS-PAGE analysis of the fractions from Q-Sepharose chromatography of the crystal inclusion protein of the GP1 strain. (A) Protein profile of the spore-crystal mixture. (B) Elution profiles of anion-exchange chromatography with a linear gradient of 50 to 400 mM NaCl, in 50 mM Na2CO3 (pH 10.5), 0.2% β-mercaptoethanol. Lanes 1 to 8, fraction samples subjected to SDS-PAGE and stained with Coomassie brilliant blue; lane M, molecular masses (kDa) of standard marker proteins.
The GP1 strain was characterized by PCRs using general and specific primers for the cry1, cry3, cry5, cry7, cry8, cry9, cry11, cry13, cry14, and cyt1A genes (4, 5, 6). All PCRs were negative (data not shown). The 16S rRNA gene of the GP1 strain was then amplified using the primers designed by Aguino de Muro and Priest (2). BLAST analysis of the 16S DNA sequence confirmed that the GP1 strain belongs to the B. thuringiensis group.
Characterization of the GP1 protein found in the crystal inclusion.
The 100-kDa protein produced by the GP1 strain was purified by anion-exchange chromatography and transferred to Immobilon PSQ, and the amino-terminal sequence of the protein was obtained. Sequence analysis suggested that the GP1 protein could be an S-layer protein, as the amino-terminal sequence (AGKSFPDVPAGH) corresponded to the first 12 amino acids after the signal peptide of an SLP precursor from B. licheniformis OlpA (GenBank accession number U38842) and B. anthracis EA1 (X99724). Trypsin digestion of the pure GP1 protein was performed, and a fragment of 50 kDa was purified by size exclusion high-performance liquid chromatography. An internal sequence of 19 amino acids (KLPVTFVTTDQYGDPYGAN) that was also found in OlpA and EA1 SLP was obtained.
PCR primers (GP1Xder and GP1Xrev) (Fig. 1) were designed from the N-terminal sequence of the GP1 protein and from the internal tryptic fragment of this protein. These primers were used for the amplification of an internal fragment that was sequenced. BLAST analysis of the resulting sequence showed high scores with sequences of two S-layer genes, olpA and eag. Using this information and the DNA sequence alignment of these genes, novel PCR primers were designed to amplify two other overlapping PCR products. One included 500 bp upstream of the ATG codon in order to have the putative promoter (Fig. 1), and the other included 200 bp after the putative stop codon (Fig. 1). The sequence of the complete GP1 SL gene was obtained (GenBank accession number AY956311). The open reading frame found in this sequence contained 2,589 nucleotides; it was preceded by a Shine-Dalgarno sequence (AGGAGG). A palindromic structure was observed 22 nucleotides downstream from the TTA stop codon. Comparison of the N termini of the sequenced protein and of the amino acid sequence deduced from the nucleotide sequence confirmed that this protein is also synthesized as a prepolypeptide with a 29-amino-acid-long signal peptide. Three S-layer homology domain motifs are present in this sequence; the first was observed from residues 34 to 76 (FPDVPAGHWGLDSINYLVDKGAIEGKPDGTYAPAEEIDRASAA), the second from residues 95 to 136 (FKDAKNHWASKYIAAVEKAGVVRGDGKENFSPDKKIDRASFA), and the third from positions 162 to 198 (DHWGEEKANILIHLGLSEGTGGNKWEPNKSVSRAEAA).
Finally, the complete GP1 SL gene was cloned directly into the B. thuringiensis acrystalliferous strain 407 by amplifying three PCR fragments that overlap in NcoI and PstI restriction sites (Fig. 1). It was not possible to obtain E. coli transformant cells with this construction. Other authors have reported that in many cases, the cloning of the S-layer gene in E. coli cells with its regulatory region could lead to problems (20, 27). The resulting B. thuringiensis strain expressed the GP1 SL protein, as judged by immunodetection of the protein using a polyclonal antibody raised against the pure GP1 protein (Fig. 3). A sporulated culture of this strain showed 100% mortality of E. varivestis larvae when assayed at 100 and 1,000 ng/cm2 of treated leaf. This was in contrast to what was found with the control strain transformed with the pHT315 shuttle vector that does not express the GP1 SL protein; it did not show toxicity to E. varivestis larvae. The crystal inclusion was purified from the transformant strain 407pHT-GP1, and bioassays against E. varivestis larvae showed a lethal concentration (LC50) of 10 ng/cm2 of treated leaf (95% confidence interval, 5 to 19 ng/cm2).
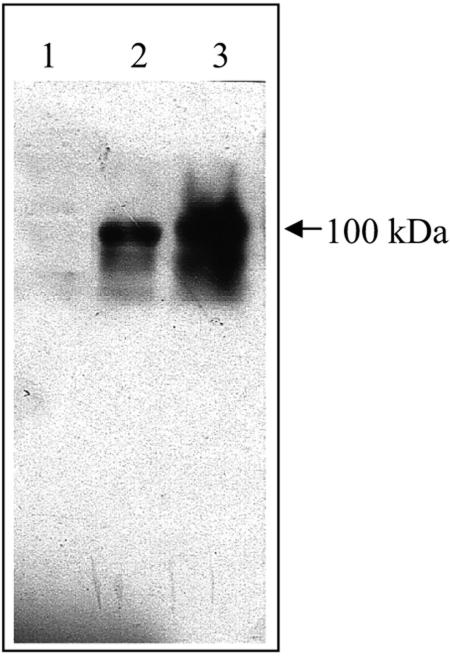
FIG. 3.
Western blot analysis of GP1 S-layer protein. Lane 1, control strain, B. thuringiensis 407 transformed with pHT315 plasmid; lane 2, B. thuringiensis 407 strain transformed with pHT-GP1 plasmid; lane 3, GP1 strain. Samples were subjected to SDS-PAGE, and the GP1 SL protein was detected by Western blotting with an anti-GP1 SL polyclonal antibody.
Expression of the GP1 SL protein in the GP1 strain.
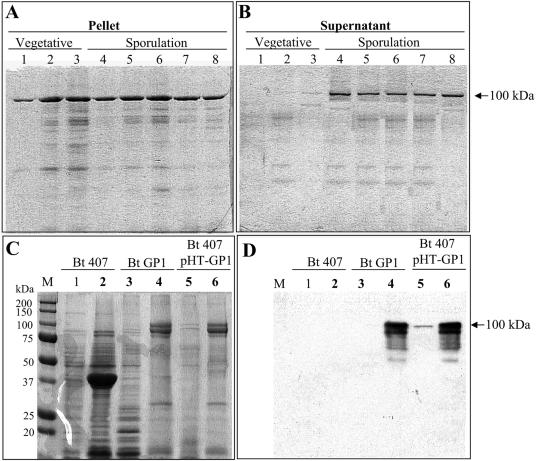
The expression of the GP1 SL protein during growth in SP sporulation medium was then analyzed. Figure 4A and B show that this protein was expressed during the vegetative (lanes 1 to 3) and the sporulation (lanes 4 to 8) phases of growth. During the vegetative phase of growth, the protein was associated with the bacteria since all the protein detected by Western blot analysis was found in the bacterial pellet obtained after the centrifugation of the culture (Fig. 4A, lanes 1 to 3). However, during the sporulation phase, the GP1 SL protein was found associated with the bacteria (Fig. 4A, lanes 4 to 8) and also found in the supernatants of centrifuged cultures (Fig. 4B). Luckevich and Beveridge (17) described a specific extraction procedure for the SLP of B. thuringiensis subsp. galleria. We used this method with the purpose of testing whether the GP1 SL protein has the ability to bind to the cell surface from the original strain and in the transformant strain as other SLP do. Figure 4C and D show the treatment of vegetative cells with 2 M chaotropic agent at a low pH, which resulted in the specific extraction of the GP1 SL protein as judged by the SDS-PAGE (Fig. 4C) and Western blot analysis (Fig. 4D) of the extracted protein. The GP1 SL protein was present only in the GP1 strain and in the B. thuringiensis 407 transformant strain; this protein was absent in the 407 acrystalliferous strain that, in contrast, showed a protein of 37 kDa (Fig. 4C). Similarly, as reported by Luckevich and Beveridge (17), the GP1 SL protein was extracted from the vegetative cells only with the treatment of 2 M chaotropic agent; treatments with lower concentrations of the chaotropic agent (1 to 1.5 M) did not extract the protein from the bacterial cells (data not shown).
FIG. 4.
Expression of the GP1 SL protein in the GP1 strain. (A and B) Analysis of the expression of the GP1 SL protein during growth in SP sporulation medium determined by Western blotting using a specific anti-GP1 SL antibody. (A) Detection of the GP1 SL protein attached to the bacteria in the pellet of centrifuged cultures (10 min at 10,000 rpm). (B) Detection of soluble protein secreted into the supernatant of the same centrifuged cultures. Lane 1, 6 h of growth; lane 2, 9 h of growth; lane 3, 12 h of growth; lane 4, 15 h of growth; lane 5, 18 h of growth; lane 6, 21 h of growth; lane 7, 24 h of growth; lane 8, 36 h of growth. (C and D) Extraction of S-layer proteins from vegetative cells by using 2 M guanidine hydrochloride (pH 2.5). The proteins that remained attached to the cell wall or were released into the supernatant after chemical extraction were analyzed by SDS-PAGE, the gels for which were stained with Coomassie brilliant blue (C), and by Western blot detection using anti-GP1 SL polyclonal antibody (D). The proteins that remained attached to the cell wall are presented in even-numbered lanes, and the proteins extracted from the cells are presented in odd-numbered lanes. Lanes 1 and 2, control strain, B. thuringiensis 407; lanes 3 and 4, GP1 strain; lanes 5 and 6, B. thuringiensis 407 transformed with pHT-GP1 plasmid containing the GP1 SL gene. Bt, B. thuringiensis.
Western blot detection of the GP1 SL protein was also performed in other B. thuringiensis strains, such as B. thuringiensis subsp. kurstaki HD1 and HD73, B. thuringiensis subsp. israelensis HD567, B. thuringiensis subsp. aizawai HD137, B. thuringiensis subsp. tolworthi HD125, and B. thuringiensis subsp. morrisoni tenebrionis. These studies showed that this protein is not produced in these strains (data not shown). PCR analysis of these strains, using primers GP1Xder and GP1Xrev, confirmed that these B. thuringiensis strains do not harbor the GP1 SL coding sequence (data not shown).
Phylogenetic relationships of GP1 SL proteins with other S-layer proteins.
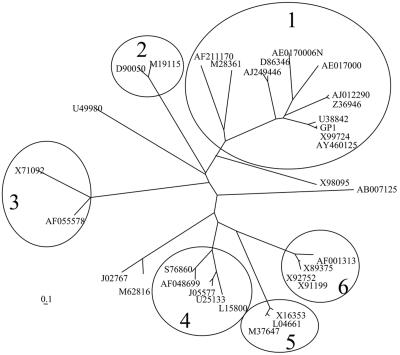
An amino acid sequence alignment of 33 different SLP was used to analyze the phylogenetic relationship of these proteins. We have estimated the phylogenies from the distance matrix data under the additive tree model, according to which the distances are expected to equal the sums of the branch lengths between the species, by using the Fitch and Margoliash method (10) and the least squares criterion. Figure 5 shows the phylogeny of the SLP sequences. The encircled branches represent branches with a solid topology, since they were found in more than 95% of the obtained trees. This analysis demonstrated that SLP are highly variable in forming independent clusters. The SLP from Bacillus cereus, B. sphaericus, B. anthracis, B. licheniformis, and B. thuringiensis are arranged in one main group (Fig. 5, group 1). The SLP of other bacterial species are clustered together in separate branches, indicating the relationship among the members of each branch in the evolutionary process.
FIG. 5.
Unrooted phylogenic tree of S-layer protein sequences. Encircled branches correspond to branches with solid topology and to different groups of bacteria. Groups: 1, the Bacillus sp. group; 2, Brevibacillus brevis; 3, Geobacillus stearothermophilus; 4, Campylobacter fetus; 5, Rickettsia sp.; 6, Lactobacillus sp. The numbers of the S-layer proteins correspond to their accession numbers in GenBank.
The GP1 SL protein is highly related to the EA1 protein from B. anthracis and to the CTC2 protein reported for B. thuringiensis subsp. finitimus, with 92% identity at the nucleotide sequence level with both proteins. The GP1 SL protein showed 63% identity with the OlpA protein from B. licheniformis (GenBank accession number u38842). In relation to the rest of the SLP found in the Bacillus group, the sequence identity is quite low, ranging from 27% to 15% identity.
DISCUSSION
We isolated and characterized B. thuringiensis strain GP1, which produces a parasporal crystal composed of a 100-kDa S-layer protein that is highly active against the coleopteran pest Epilachna varivestis. The pure crystal protein preparation of this strain is responsible for the insecticidal activity against E. varivestis, and this activity appears to be specific since this strain is not toxic to the larvae of the two lepidopteran species and the one dipteran species examined here. It was shown for the first time that an SLP is directly implicated in virulence against the target organism. The complete GP1 SL gene from a B. thuringiensis strain was successfully cloned, and it was demonstrated that this gene encodes a protein active against this pest.
The GP1 SL protein of the GP1 strain shows 92% identity at the primary sequence level with the EA1 protein of B. anthracis. In B. anthracis, the expression of SLP is dependent on different sigma factors (21). In addition, in the case of the EA1 protein, the regulatory system is quite complex, since the Sap protein (the other SLP) is required for the temporal control of EA1, as it acts as a transcriptional repressor of the eag gene. The result of this regulation is that the sap gene is expressed during the exponential phase, whereas eag expression starts in the stationary phase. The synthesis of EA1 displaces Sap from the rigid cell wall layer; thus, Sap could be found both cell associated and secreted into the culture medium. EA1, in contrast, is associated only with the sporulated cells (21). In the case of the GP1 SL protein of the GP1 strain, this protein is associated with the bacteria during both growth phases but is also found in the medium of sporulated cultures. Further studies are needed to understand the regulation of the GP1 SL protein in B. thuringiensis.
The SLP comprise a family of proteins found on the surfaces of very different bacterial species that live under different environmental conditions and have different targets. These proteins are modular in structure, consisting of two different functional domains. In this work, the evolutionary relationship of the SLP family was analyzed. Since SLP represents an abundant cellular protein group, it has been proposed that these metabolically expensive products must provide organisms with some advantage of selection in different habitats (3). The phylogenetic studies reported here showed that SLP distribute in different branches that group the different bacteria analyzed. These data support the divergence of these proteins, which were adapted and selected for acting under particular living conditions. In the case of B. thuringiensis, the insecticidal activity is an advantage in gaining access to the nutrients of the hemocoel in a sensitive insect. The phylogenetic study showed that the SLP described here is highly related to the EA1 protein from B. anthracis (19) and also to the CTC protein found in B. thuringiensis subsp. finitimus (27), which has a molecular size of 100 kDa and forms a parasporal body during the sporulation phase of growth. However, this is the first time that a putative SLP has been directly implicated in the virulence of B. thuringiensis strains against their targets. The only evidence that links SLP of B. thuringiensis with virulence is the report that the expression of S-layer genes is controlled by plcR. PlcR is a transcriptional activator of several extracellular virulence factor genes, such as degradative enzymes and enterotoxins (1).
Screening programs have identified thousands of different B. thuringiensis strains, all of which have a limited host range but together span a wide range of insect orders (Lepidoptera, Diptera, Coleoptera, Hymenoptera, Homoptera, Orthoptera, and Mallophaga) and even other organisms, such as nematodes, mites, and protozoans. The toxins produced by B. thuringiensis belong to different protein groups (8) that could be divided into at least four or five fundamentally distinct homology groups (three Cry domain toxins, Vip, Cyt, Mtx-like, and Bin-like). Some of the toxins produced by B. thuringiensis share similarities to toxins produced by other bacteria, as in the case of the binary toxins produced by B. sphaericus, which have some homology with the Cry35 and Cry36 proteins produced by B. thuringiensis strains. This work will add a new protein to the list of insecticidal proteins produced by B. thuringiensis. This work also showed that classical screening strategies using B. thuringiensis strains isolated from soil samples could be laborious and inefficient at isolating strains highly active against a selected pest. The strategy for isolating B. thuringiensis strains from dead insect bodies seems to be highly effective at identifying active strains.
Finally, it would be relevant to test whether other S-layer proteins produced by B. thuringiensis have some insecticidal activity against other insect species. This knowledge could be important for future control of insects. However, the mechanism of action of this SLP is unknown, and future work is needed to describe it.
Acknowledgments
We thank Lizbeth Cabrera, Rene Hernandez-Vargas, and Oswaldo López for technical assistance.
This work was supported in part by INCO contract IC18CT980303 and CONACyT G36505-N.
REFERENCES
- 1.Agaisse, H., M. Gominet, O. A. Okstad, A.-B. Kolsto, and D. Lereclus. 1999. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 32:1043-1053. [DOI] [PubMed] [Google Scholar]
- 2.Aguino de Muro, M., and F. G. Priest. 1993. Phylogenetic analysis of Bacillus sphaericus and development of an oligonucleotide probe specific for mosquito-pathogenic strains. FEMS Microbiol. Lett. 112:205-210. [DOI] [PubMed] [Google Scholar]
- 3.Beveridge, T. J., P. H. Pouwels, M. Sára, A. Kotiranta, K. Lounatmaa, K. Kari, E. Kerosuo, M. Haapasalo, E. M. Egelseer, I. Schocher, U. B. Sleytr, L. Morelli, M.-L. Callegari, J. F. Nomellini, W. H. Bingle, J. Smit, E. Leibovitz, M. Lemaire, I. Miras, S. Salamitou, P. Beguin, H. Ohayon, P. Gounon, M. Matuschek, K. Sahm, H. Bahl, R. Grogono-Thomas, J. Dworkin, M. J. Blaser, R. M. Woodland, D. G. Newell, M. Kessel, and S. F. Koval. 1997. Functions of S-layers. FEMS Microbiol. Rev. 20:99-149. [DOI] [PubMed] [Google Scholar]
- 4.Bravo, A., S. Sarabia, L. Lopez, H. Ontiveros, C. Abarca, A. Ortiz, M. Ortiz, L. Lina, F. J. Villalobos, G. Peña, M.-E. Nuñez-Valdez, M. Soberón, and R. Quintero. 1998. Characterization of cry genes in a Mexican Bacillus thuringiensis strain collection. Appl. Environ. Microbiol. 64:4965-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ceron, J., L. Covarrubias, R. Quintero, A. Ortiz, M. Ortiz, E. Aranda, L. Lina, and A. Bravo. 1994. PCR analysis of the cryI insecticidal crystal family genes from Bacillus thuringiensis. Appl. Environ. Microbiol. 60:353-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerón, J., A. Ortíz, R. Quintero, L. Güereca, and A. Bravo. 1995. Specific PCR primers directed to identify cryI and cryIII genes within a Bacillus thuringiensis strain collection. Appl. Environ. Microbiol. 61:3826-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crickmore, N., D. R. Zeigler, E. Schnepf, J. Van Rie, D. Lereclus, J. Baum, A. Bravo, and D. H. Dean. 2005. Bacillus thuringiensis toxin nomenclature. [Online.] http://www.biols.susx.ac.uk/Home/Neil_Crickmore/Bt/index.html.
- 8.de Maagd, R., A. Bravo, C. Berry, N. Crickmore, and H. E. Schnepf. 2003. Structure, diversity and evolution of protein toxins from spore-forming entomopathogenic bacteria. Annu. Rev. Genet. 37:409-433. [DOI] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1993. PHYLIP—Phylogeny Interference Package, version 3.5c. Department of Genetics, University of Washington, Seattle.
- 10.Fitch, W. M., and E. Margoliash. 1967. Construction of phylogenetic trees. Science 155:279-284. [DOI] [PubMed] [Google Scholar]
- 11.Güereca, L., and A. Bravo. 1999. The oligomeric state of Bacillus thuringiensis Cry toxins in solution. Biochim. Biophys. Acta 1429:342-350. [DOI] [PubMed] [Google Scholar]
- 12.Hill, D. S. 1983. Agricultural insect pests of the tropics and their control, p. 438. Cambridge University Press, Cambridge, United Kingdom.
- 13.Ibarra, J. E., M. C. del Rincón, S. Ordúz, D. Noriega, G. Benintende, R. Monnerat, L. Regis, C. M. F. de Oliveira, H. Lanz, M. H. Rodriguez, J. Sánchez, and A. Bravo. 2003. Diversity of Bacillus thuringiensis strains from Latin America with insecticidal activity against different mosquito species. Appl. Environ. Microbiol. 69:5269-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotiranta, A., M. Haapasalo, K. Kari, E. Kerosuo, I. Olsen, T. Sorsa, J. H. Muerman, and K. Lounatmaa. 1998. Surface structure, hydrophobicity, phagocytosis and adherence to matrix protein of Bacillus cereus cells with and without the crystalline surface protein layer. Infect. Immunol. 66:4895-4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lereclus, D., H. Agaisse, M. Gominet, and J. Chaufaux. 1995. Overproduction of encapsulated insecticidal crystal proteins in a Bacillus thuringiensis spoOA mutant. Bio/Technology 13:67-71. [DOI] [PubMed] [Google Scholar]
- 16.Lereclus, D., O. Arantes, J. Chaufaux, and M.-M. Lecadet. 1989. Transformation and expression of a cloned δ-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol. Lett. 60:211-218. [DOI] [PubMed] [Google Scholar]
- 17.Luckevich, M. D., and T. J. Beveridge. 1989. Characterization of a dynamic S layer on Bacillus thuringiensis. J. Bacteriol. 171:6656-6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mesnage, S., E. Tosi-Couture, M. Mock, P. Gounon, and A. Fouet. 1997. Molecular characterization of the Bacillus anthracis main S-layer component: evidence that it is the major cell-associated antigen. Mol. Microbiol. 23:1147-1155. [DOI] [PubMed] [Google Scholar]
- 19.Mesnage, S., E. Tosi-Couture, P. Gounon, M. Mock, and A. Fouet. 1998. The capsule and S-layer: two independent and yet compatible macromolecular structures in Bacillus anthracis. J. Bacteriol. 180:52-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mesnage, S., M. Haustant, and A. Fouet. 2001. A general strategy for identification of S-layer genes in the Bacillus cereus group: molecular characterization of such a gene in Bacillus thuringiensis subsp. galleria NRRL 4045. Microbiology 147:1343-1351. [DOI] [PubMed] [Google Scholar]
- 21.Mignot, T., S. Mesnage E. Couture-Tosi, M. Mock, and A. Fouet. 2002. Developmental switch of S-layer protein synthesis in Bacillus anthracis. Mol. Microbiol. 43:1615-1627. [DOI] [PubMed] [Google Scholar]
- 22.Msadek, T., F. Kunst, D. Henner, A. Klier, G. Rapoport, and R. Dedonder. 1990. Signal transduction pathway controlling synthesis of a class of degradative enzymes in Bacillus subtilis: expression of the regulatory genes and analysis of mutations in degS and degU. J. Bacteriol. 172:824-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pei, Z., and M. J. Blaser. 1990. Pathogenesis of Campylobacter fetus infections. Role of surface array proteins in virulence in a mouse model. J. Clin. Investig. 85:1036-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Sára, M., and U. B. Sleytr. 2000. S-layer proteins. J. Bacteriol. 182:859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun, M., C. G. Zhu, and Z. Yu. 2001. Cloning of parasporal body protein gene resembling to S-layer protein genes from Bacillus thuringiensis CTC strain. Acta Microbiol. Sin. 41:141-147. [PubMed] [Google Scholar]
- 28.Travers, R. S., P. A. W. Martin, and C. F. Reichelderfer. 1987. Selective process for efficient isolation of soil Bacillus spp. Appl. Environ. Microbiol. 53:1263-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]