Abstract
In this paper, the ability of a commercial starter culture to perform a sausage fermentation is evaluated. Molecular analysis revealed the presence of several strains of the same species contained in the starter culture with different behavior during the fermentation, and the contribution of Lactobacillus curvatus, which was only marginally isolated during the transformation.
Fermentation and drying are among the most ancient ways of preservation of meat products that are otherwise a perishable commodity (10). The use of starter cultures in the field of meat fermentation has been reviewed several times (7, 9, 11). Starter culture selection must take into consideration the ability of the strains to colonize the ecosystem in a fast and effective way. Molecular methods offer a tool for the determination of the ability of a strain to perform a fermentation. The goal of this study was to investigate, using culture-dependent and -independent methods, the bacterial ecology of a fermented sausage produced with a commercial starter culture. The strains used in this study are reported in Table 1. Lactic acid bacteria (LAB) were grown in MRS broth (Oxoid, Milan, Italy) at 30°C for 24 h, and coagulase-negative cocci (CNC) were grown in brain heart infusion broth (Oxoid) at 30°C for 24 h. Fermented sausages were prepared in a local meat factory using traditional techniques. A commercial starter culture containing Staphylococcus carnosus and Lactobacillus plantarum with a total cell concentration of about 109 cells/g (Biostart SL1-200; Wiesby GmbH & Co., Niebull, Germany), previously dissolved in 2 liters of white wine, was used to inoculate a 200-kg batch. The fermented sausages were analyzed in triplicate at 0, 3, 5, 7, 14, and 28 days. Microbiological examinations on the starter culture and on the fermented sausages during maturation, as well as molecular analysis protocols, were performed as previously described (2, 3, 4, 12, 13).
TABLE 1.
Bacterial species used in this study
| Species | Code | Sourcea |
|---|---|---|
| Lactobacillus plantarum | 20174 | DSM |
| Lactobacillus plantarum | 1193 | NCDO |
| Lactobacillus paracasei | 335 | ATCC |
| Lactobacillus curvatus | 20019 | DSM |
| Lactobacillus curvatus | 1432 | LTH |
| Lactobacillus curvatus | L442 | Fermented sausagesb |
| Lactobacillus sakei | 6333 | DSM |
| Lactobacillus casei | 20011 | DSM |
| Staphylococcus xylosus | 6179 | DSM |
| Staphylococcus cohinii | 6669 | DSM |
| Staphylococcus simulans | 20322 | DSM |
| Staphylococcus intermedius | 20373 | DSM |
| Staphylococcus carnosus subsp. carnosus | 20501 | DSM |
| Staphylococcus xylosus | DSA2034 | Fermented sausagesb |
| Staphylococcus carnosus | DSA2016 | Fermented sausagesb |
Abbreviations: ATCC, American Type Culture Collection, Manassas, Virginia; DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany; NCDO, National Collection of Food Bacteria, c/o NCIMB Ltd., Aberdeen, Scotland; LHT, Institut für Lebensmitteltechnologie, Universität Hohenheim, Stuttgart, Germany.
Strains were identified by 16S rRNA gene sequencing.
The microbial trends showed a fast increase of the LAB populations, which provoked a drop in the pH within the first 5 days of fermentation. This fact seemed to inhibit the growth of CNC; in fact, only at day 5 did their counts start to increase significantly. Yeast and enterobacterial counts had already begun to decrease at 5 days, while aerobic bacterial counts presented a negative trend only at the end of the fermentation. Isolated strains were identified by species-specific PCR, denaturing gradient gel electrophoresis (DGGE) analysis, and sequencing of the 16S rRNA gene.
A total of 15 LAB and 15 CNC strains were isolated directly from the starter culture, and 70 LAB and 70 CNC strains were picked up from the fermented sausages during ripening. LAB strains isolated from the starter culture were all identified as L. plantarum. Similarly, the majority of the fermented-sausage isolates were L. plantarum (47 strains). However, Lactobacillus sakei (14 strains), Lactobacillus curvatus (2 strains), and Pediococcus acidilactici (5 strains) were identified as well. CNC isolated from the commercial starter were identified as S. carnosus (9 strains) and Staphylococcus xylosus (6 strains). This last species was not declared on the label of the starter culture and it became the main Staphylococcus species during fermentation (51 strains). S. carnosus (4 strains), Staphylococcus equorum (9 strains), Staphylococcus pasteuri (5 strains), and Staphylococcus saprophyticus (1 strain) were detected at low numbers. L. plantarum and S. xylosus were subjected to random amplified polymorphic DNA (RAPD) analysis with primer M13.
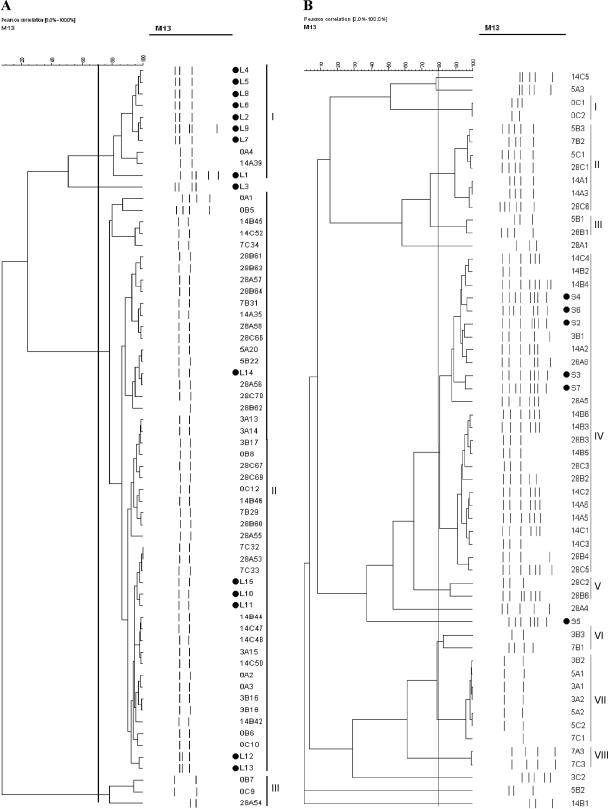
To define the coefficient of similarity, control strains were subjected to RAPD amplification and cluster analysis (data not shown). Coefficients of 70% for LAB and 80% for CNC were used for the analysis of the strains. Three L. plantarum RAPD types were observed (Fig. 1A). However, when the time of isolation was considered, it was determined that only the type included in cluster II was able to conduct the fermentation. A different picture was observed in the case of S. xylosus, for which the strains from the starter culture grouped with strains isolated mainly at 14 and 28 days, thereby highlighting their predominance only in the latter stages of the fermentation. The different behaviors of L. plantarum and S. xylosus could be explained considering the fact that the starter culture was dissolved in white wine, thereby introducing a stress factor (ethanol). This step did not influence the growth dynamics of L. plantarum, able to overcome the ethanol stress (8), while it inhibited the initial development of S. xylosus and suppressed S. carnosus. When the DNA and RNA isolated directly from the starter culture were analyzed by PCR-DGGE and reverse transcriptase PCR-DGGE, only L. plantarum and S. carnosus were present in the migration pattern, while S. xylosus gave a very faint band only at the RNA level (Fig. 2, lane 11).
FIG. 1.
Cluster analysis of the profiles obtained from the Lactobacillus plantarum (A) and Staphylococcus xylosus (B) strains isolated during the fermentation of the sausages; Strains isolated from the starter culture are indicated with letters L and S, respectively, and marked with black dots. Isolates from fermented sausages are identified by code, in which the first number represents the day of isolation, the letter the code of the sample and the second number the progressive number of isolation. Identified clusters are indicated with roman numerals.
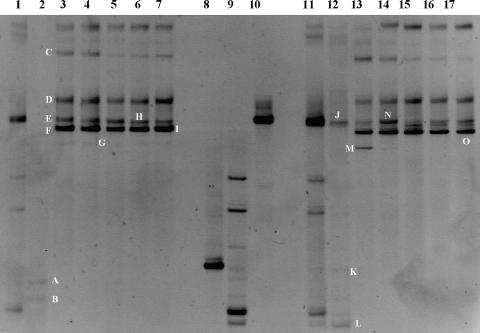
FIG. 2.
Bacterial DGGE profiles of the DNA (lanes 1 to 7) and RNA (lanes 11 to 17) extracted directly from the starter culture and the fermented sausages. Lanes 1 and 11, starter culture envelope; lanes 2 and 12, day zero; lanes 3 and 13, 3 days of fermentation; lanes 4 and 14, 5 days of fermentation; lanes 5 and 15, 7 days of fermentation; lanes 6 and 16, 14 days of fermentation; and lanes 7 and 17, 28 days of fermentation. Bands in lanes 8, 9, and 10 were produced from Staphylococcus xylosus S4, Staphylococcus carnosus S10, and Lactobacillus plantarum L12, respectively, all isolated from the starter culture and thereby representing migration controls. Bands indicated by letters were excised and, after reamplification, subjected to sequencing.
The picture changed significantly when the nucleic acids from the fermented sausages were used in the analysis. The profiles obtained from the DNA and RNA were comparable. No differences were found when the triplicate samples were analyzed. The main differences were detected at day zero, when Staphylococcus sciuri/Staphylococcus pulvereri (band A) and Staphylococcus equorum/Staphylococcus succinus (band B) were detected at the DNA level and L. plantarum (band J) and S. xylosus (band K) were visible when the RNA was analyzed (Fig. 2 and Table 2). From day 3, two bands, E and F, identified as L. plantarum and L. curvatus, respectively, became predominant, and they remained constant throughout the fermentation. However, L. curvatus was rarely isolated on the plates. In our opinion, this disagreement can be explained by considering the random selection of the isolated colonies or assuming the presence of populations of L. curvatus not able to grow on the MRS plates. S. xylosus and other CNC members were detected only at day zero. As previously reported, this may be due to the masking effects by the most abundant nucleic acids present, in this case coming from the LAB populations (3, 13). A possible solution to the problem could be the use of different primers targeting other regions of the 16S rRNA gene. However, it has been reported that only the V1 region allows DGGE differentiation between LAB and CNC without band comigration (3).
TABLE 2.
Sequencing results of the bands cut from the bacterial DGGE gels
| Banda | Size (bp) | Closest relative | % Identity | Sourceb |
|---|---|---|---|---|
| A | 91 | Staphylococcus sciuri/S. pulvereric | 98.9 | AY126231 |
| AY126216 | ||||
| B | 91 | Staphylococcus equorum/S. succinicusc | 98.9 | AF527483 |
| AY126240 | ||||
| C | Heteroduplex | |||
| D | Heteroduplex | |||
| E | 109 | Lactobacillus plantarum | 99.0 | AF404710 |
| F | 113 | Lactobacillus curvatus | 10.0 | AY204894 |
| G | 115 | Lactobacillus sakei | 99.1 | AY204897 |
| H | 109 | Lactobacillus plantarum | 99.0 | AF404710 |
| I | 113 | Lactobacillus curvatus | 99.1 | AY204894 |
| J | 109 | Lactobacillus plantarum | 100 | AF404710 |
| K | 93 | Staphylococcus xylosus | 98.9 | AY126259 |
| L | 91 | Bacillus sp. | 98.8 | AY370628 |
| M | 115 | Lactobacillus sakei | 100 | AY204897 |
| N | 109 | Lactobacillus plantarum | 100 | AF404710 |
| O | 113 | Lactobacillus curvatus | 99.1 | AY204894 |
Only a few studies have exploited molecular methods to assess the capability of selected strains to carry out sausage fermentation (5, 6). In this study it was determined that the starter, declared on the label to contain L. plantarum and S. carnosus, also had S. xylosus, which was the predominant Staphylococcus spies. In addition, the starter culture was formed by a mixture of strains of the same species. Especially for L. plantarum, different RAPD types were determined. Among them only one was able to perform the fermentation process. The application of direct methods allowed understanding of the contribution of L. curvatus, only marginally isolated from the plates, but present throughout the fermentation at both DNA and RNA levels. The ability of the S. xylosus strains contained in the starter to carry out only the second half of the fermentation suggests a need for a change in the production procedures used in processing plants.
Acknowledgments
This study was supported by the Ministry of University, Rome, Italy, action PRIN (ex 40%).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berthier, F., and S. D. Ehrlich. 1998. Rapid species identification within two groups of closely related lactobacilli using PCR primers that target the 16S/23S rRNA spacer region. FEMS Microbiol. Lett. 161:97-106. [DOI] [PubMed] [Google Scholar]
- 3.Cocolin, L., M. Manzano, C. Cantoni, and G. Comi. 2001. Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl. Environ. Microbiol. 67:5113-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cocolin, L., K. Rantsiou, L. Iacumin, R. Urso, C. Cantoni, and G. Comi. 2004. Study of the ecology of fresh sausages and characterization of populations of lactic acid bacteria by molecular methods. Appl. Environ. Microbiol. 70:1883-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Maria, S., A. L. Basso, E. Santoro, L. Grazia, and R. Coppola. 2002. Monitoring of Staphylococcus xylosus DSM 20266 added as starter during fermentation and ripening of soppressata molisana, a typical Italian sausage. J. Appl. Microbiol. 92:158-164. [DOI] [PubMed] [Google Scholar]
- 6.Garriga, M., M. Hugas, P. Gou, M. T. Aymerich, J. Arnau, and J. M. Monfort. 1996. Technological and sensorial evaluation of Lactobacillus strains as starter cultures in fermented sausages. Int. J. Food Microbiol. 32:173-183. [DOI] [PubMed] [Google Scholar]
- 7.Hammes, W. P., and C. Hertel. 1998. New developments in meat starter culture. Meat Sci. 49:S125-S138. [PubMed] [Google Scholar]
- 8.Henick-Kling, T. 1992. Malolactic fermentation, p. 289-326. In G. H. Fleet (ed.), Wine microbiology and biotechnology. Harwood Academic Publishers, Chur, Switzerland.
- 9.Hugas, M., and J. M. Monfort. 1997. Bacterial starter cultures for meat fermentation. Food Chem. 59:547-554. [Google Scholar]
- 10.Incze, K. 1998. Dry fermented sausages. Meat Sci. 49:S169-S177. [PubMed] [Google Scholar]
- 11.Lücke, F. K. 2000. Utilization of microbes to process and preserve meat. Meat Sci. 56:105-115. [DOI] [PubMed] [Google Scholar]
- 12.Rantsiou, K., L. Iacumin, C. Cantoni, G. Comi, and L. Cocolin. 2005. Ecology and characterization by molecular methods of Staphylococcus species isolated from fresh sausages. Int. J. Food Microbiol. 97:277-284. [DOI] [PubMed] [Google Scholar]
- 13.Rantsiou, K., R. Urso, L. Iacumin, C. Cantoni, P. Cattaneo, G. Comi, and L. Cocolin. 2005. Culture-dependent and -independent methods to investigate the microbial ecology of Italian fermented sausages. Appl. Environ. Microbiol. 71:1977-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]