Abstract
Dibenzothiophene is a sulfur heterocycle found in crude oils and coal. The biodegradation of dibenzothiophene through the Kodama pathway by Pseudomonas sp. strain BT1d leads to the formation of three disulfides: 2-oxo-2-(2-thiophenyl)ethanoic acid disulfide, 2-oxo-2-(2-thiophenyl)ethanoic acid-2-benzoic acid disulfide, and 2,2′-dithiodibenzoic acid. When provided as the carbon and sulfur source in liquid medium, 2,2′-dithiodibenzoic acid was degraded by soil enrichment cultures. Two bacterial isolates, designated strains RM1 and RM6, degraded 2,2′-dithiodibenzoic acid when combined in the medium. Isolate RM6 was found to have an absolute requirement for vitamin B12, and it degraded 2,2′-dithiodibenzoic acid in pure culture when the medium was supplemented with this vitamin. Isolate RM6 also degraded 2,2′-dithiodibenzoic acid in medium containing sterilized supernatants from cultures of isolate RM1 grown on glucose or benzoate. Isolate RM6 was identified as a member of the genus Variovorax using the Biolog system and 16S rRNA gene analysis. Although the mechanism of disulfide metabolism could not be determined, benzoic acid was detected as a transient metabolite of 2,2′-dithiodibenzoic acid biodegradation by Variovorax sp. strain RM6. In pure culture, this isolate mineralized 2,2′-dithiodibenzoic acid, releasing 59% of the carbon as carbon dioxide and 88% of the sulfur as sulfate.
Dibenzothiophene (DBT) is a common sulfur heterocycle in petroleum, and its biodegradation has been studied as the model organosulfur compound in crude oils. There are three pathways for DBT biodegradation which have been described (reviewed in references 6 and 32). In a unique pathway, van Afferden et al. (47) reported the angular dioxygenation of DBT by Brevibacterium sp. DO producing DBT sulfoxide, DBT sulfone, and benzoate as major intermediates during DBT mineralization. The most extensively studied pathway, the biodesulfurization pathway for DBT (22, 25, 26, 27, 31, 37), is inhibited by the presence of sulfate and is not likely to contribute to DBT biodegradation in sulfate-containing environments such as the oceans (32).
In the Kodama pathway (28, 29), one of the homocyclic rings of DBT is broken, and the sulfur remains in an organic form as 3-hydroxy-2-formylbenzothiophene (HFBT). There have been no reports of the complete mineralization of DBT by bacteria using the Kodama pathway. However, HFBT can be mineralized (7), and benzothiophene-2,3-dione was detected in acidified HFBT-degrading cultures (7), and in DBT-degrading cultures (5, 34). Bressler and Fedorak (8) proposed an abiotic mechanism for the formation of benzothiophene-2,3-dione from HFBT. At neutral pH, the thiophene ring of benzothiophene-2,3-dione would open to form 2-mercaptophenylglyoxylate (15), as shown in Fig. 1.
FIG. 1.
Formation of disulfides from abiotic reactions beginning with benzothiophene-2,3-dione, according to Bressler and Fedorak (8).
Bressler and Fedorak (8) identified three disulfides, including two novel products from DBT biodegradation and a third that was previously reported (18), in cultures degrading DBT through the Kodama pathway. 2-Mercaptophenylglyoxylate abiotically dimerizes as shown in Fig. 1. After abiotic losses of two carbon and oxygen atoms, 2,2′-dithiodibenzoic acid is formed (Fig. 1).
The fate of these disulfides is unknown, and our objective was to isolate a bacterial strain capable of biodegrading 2,2′-dithiodibenzoic acid.
MATERIALS AND METHODS
Chemicals.
2,2′-Dithiosalicylic acid (2,2′-dithiodibenzoic acid, 95%; Lancaster Synthesis Windham, NH) contained 1% (wt/wt) benzoic acid as a contaminant (48). Benzoic acid-free 2,2′-dithiodibenzoic acid was prepared by abiotic dimerization of 2-mercaptobenzoic acid (Aldrich, Milwaukee, WI).
2-Oxo-2-(2-thiophenyl)ethanoic acid disulfide was prepared by adding 20 mg of benzothiophene-2,3-dione (23), dissolved in 1 ml of dichloromethane, to warm, sterile sulfate-free mineral medium (SFMM; see below).
4,4′-Dithiodibenzoic acid was formed from the oxidation of 4-mercaptobenzoic acid (Aldrich) in SFMM. Reacting 2-mercaptobenzoic acid with 4-mercaptobenzoic acid in SFMM yielded three disulfides. Comparison of the high-pressure liquid chromatography (HPLC) retention times and UV spectra with those of authentic standards showed that two of the disulfides were 2,2′-dithiodibenzoic acid and 4,4′-dithiodibenzoic acid. The third disulfide was presumed to be the mixed disulfide, 2,4′-dithiodibenzoic acid.
Combining 2-mercaptobenzoic acid and mercaptoacetic acid (Aldrich) in SFMM yielded a mixture of 2,2′-dithiodibenzoic acid, dimercaptoacetic acid disulfide, and a mixed disulfide (HOOC-C6H4S-SCH2COOH).
Sulfitolysis (12) of 2,2′-dithiodibenzoic acid was done by mixing 0.082 mmol of this disulfide with 1.2 mmol of Na2SO3 in 100 ml of SFMM. The formation of the sulfitolysis product, 2-(S-sulfo)benzoic acid (HOOC-C6H4S-SO3H), was confirmed by LC-MS in the Mass Spectrometry Laboratory in the Department of Chemistry, University of Alberta. The product gave a M− ion of m/z 233.0, as expected.
Media and general incubation conditions.
The SFMM (pH 7) contained (per liter): K2HPO4, 0.5 g; NaCl, 1.0 g; MgCl2.6H2O, 0.1 g; NaNO3, 2.4 g; and 1 ml of a trace metals solution (17). In later studies, 1 ml of a filter-sterilized vitamin solution (13) was also added to each liter of SFMM. The SFMM, 200 ml in 500-ml Erlenmeyer flasks containing 100 mg of CaCO3 (to buffer the medium near neutrality), was sterilized by autoclaving at 121°C and 15 lb/in2. All cultures were incubated with shaking at 200 rpm in the dark at 28°C. Samples were stored at −20°C prior to HPLC analysis. To prepare inocula, pure cultures were grown on PCA plates (Becton Dickinson, Sparks, MD); each plate was then washed with 4 ml of sterile phosphate buffer (3 mM, pH 7.3), and 1 ml of the cell suspension was added to 200 ml of medium.
Biodegradation of 2,2′-dithiodibenzoic acid and related compounds.
2,2′-Dithiodibenzoic acid was added to SFMM from a filter-sterilized solution that contained 200 mg of 2,2′-dithiodibenzoic acid dissolved in 20 ml of 0.1 M NaOH.
To prepare enrichment cultures, 10-g samples of six different garden soils were added individually to flasks of SFMM. The most active enrichment culture that degraded 2,2′-dithiodibenzoic acid contained garden soil collected from around marigold (Tagetes sp.) roots in Edmonton, Alberta. Two bacterial isolates from this culture, designated RM1 and RM6, were studied in detail.
Isolate RM6 was inoculated into vitamin-supplemented SFMM with different carbon or carbon and sulfur sources and observed for growth. The substrates: glucose, sodium benzoate, sodium acetate, naphthalene, DBT (Fluka, Buch, Switzerland), and benzyl disulfide, phenyl disulfide, benzyl sulfide, phenyl sulfide, octyl sulfide, n-dodecyl sulfide, benzothiophene, or benzenethiol (Aldrich) were tested individually at a concentration of 1 mmol/liter. For substrates tested as carbon sources in vitamin-supplemented SFMM, including 100 mg of salicylic acid/liter or 100 mg of catechol/liter, 2 mM sulfate was added. 2-Oxo-2-(2-thiophenyl)ethanoic acid disulfide was provided as a carbon and sulfur source and as a sulfur source with 3 mmol acetate/liter as a carbon source.
The mixture of 2,4′-dithiodibenzoic acid, 4,4′-dithiodibenzoic acid, and the 2-mercaptobenzoic acid-mercaptoacetic acid mixed disulfide was added to SFMM as substrates for isolate RM6. Isolate RM6 was also inoculated into SFMM with 100 mg of 2-sulfobenzoic acid/liter (Aldrich) or with both 100 mg of 2-sulfobenzoic acid/liter and 100 mg of 2,2′-dithiodibenzoic acid/liter.
To confirm that benzoic acid was a metabolite, isolate RM6 was inoculated in triplicate into vitamin-supplemented SFMM that contained 2,2′-dithiodibenzoic acid formed from 2-mercaptobenzoic acid. When benzoic acid was detected, the culture was acidified with concentrated HCl to pH <2, extracted with dichloromethane, and the extract was evaporated to dryness. The extract was derivatized by using MTBSTFA (Aldrich) (43) and analyzed by gas chromatography-mass spectrometry (GC-MS).
Isolate RM6 was inoculated into vitamin-supplemented SFMM to determine the extent of mineralization of 2,2′-dithiodibenzoic acid. The medium contained 200 mg of 2,2′-dithiodibenzoic acid/liter, and 15 ml was added to each of 21 sterile 60-ml serum bottles. The medium was also prepared without 2,2′-dithiodibenzoic acid, and 15 ml was added to each of 21 sterile 60-ml serum bottles to account for carbon dioxide and sulfate carried over from the inoculum. Isolate RM6 was grown in vitamin-supplemented SFMM with 2,2′-dithiodibenzoic acid as the carbon and sulfur source, and after 6 days, 2 ml of the culture was added to each serum bottle. The bottles were capped with sterile butyl rubber stoppers. The headspace gas (43 ml) contained sufficient O2 to allow complete mineralization of the disulfide. At each sampling time, three cultures were acidified with concentrated HCl to pH <2 for carbon dioxide analyses. Culture supernatant samples were analyzed for disulfide and sulfate.
Analytical methods.
Disulfides, benzoic acid, salicylic acid, and catechol were analyzed by using an Agilent (Wilmington, DE) 1100 series HPLC with a UV-visible diode array detector connected to an Agilent Chemstation operating software for LC 3D systems. An analytical LiChrosopher 100 RP-18 column (5-μm particle size, 125 mm by 4 mm; Agilent) was used and the mobile phase (flowing at 1.5 ml/min) contained acetonitrile-water (35:65) with phosphoric acid (5 ml/liter) and 1 M KH2PO4 buffer (5 ml/liter). 2-Sulfobenzoic acid was analyzed by using an acetonitrile-water (10:90) mobile phase (flowing at 1 ml/min) containing acetic acid (5 ml/liter) and 1 M KH2PO4 (1 ml/liter). In all cases, the effluent was monitored at 240 nm.
Mineralization experiments were monitored for carbon dioxide (9) and sulfate (30) production. The GC-MS method was described previously (7). A spot test with cadmium acetate was used to detect H2S in some culture supernatants.
Identification of isolates.
Isolates RM1 and RM6 were identified by using the Biolog system (Biolog, Inc., Hayward, CA). Isolate RM6 was inoculated into Biolog plates with or without the vitamin mixture.
Genomic DNA was extracted from isolate RM6 by a bead-beating lysis method with 10% sodium dodecyl sulfate and chloroform (19). The 16S rRNA gene fragment of isolate RM6 was amplified by PCR using the oligonucleotide primers PB36 (5′-AGRGTTTGATCMTGGCTCAG-3′) and PB38 (5′-GKTACCTTGTTACGACTT-3′), corresponding to positions 8 to 27 and 1492 to 1509, respectively (10). The PCR was carried out in a final volume of 50 μl using 5 μl of genomic DNA, 0.5 μM concentrations of each forward and reverse primer; 50 mM Tris-HCl (pH 9); 1.5 mM magnesium chloride; 0.4 mM dGTP, dCTP, dATP, and dTTP; 5% dimethyl sulfoxide; and 0.5 U Taq polymerase (Roche Diagnostics, Laval, Quebec, Canada). Reactions were performed by using a Mastercycler Eppendorf thermocycler (Eppendorf, Hamburg, Germany) for 30 cycles (39 s at 93°C, 60 s at 54°C, and 120 s at 73°C) after an initial denaturation of 4 min at 95°C, followed by a final extension at 73°C for 10 min.
The PCR product was cleaned up according to the manufacturer directions from the Roche High Pure PCR Clean-Up kit (Roche Diagnostics). BigDye Terminator (Applied Biosystems) was incorporated into sequences using the primers PB36, PB38, 16S.1, 16S.2, 16S.3, 16S.4, and 16S.5 (Table 1). Sequencing reactions were resolved on an ABI 3100 Genetic Analyzer (Applied Biosystems).
TABLE 1.
Full complement of primers used to obtain near full-length 16S rRNA gene sequences
| Primer | Sequencea | Positionb |
|---|---|---|
| PB36 | 5′-AGR GTT TGA TCM TGG CTC AG-3′ | 8-27 |
| 16S.1 | 5′-ACT CCT ACG GGA GGC AGC AG-3′ | 360-379 |
| 16S.2 | 5′-GTA TTA CCG CGG CTG CTG GCA-3′ | 559-539 |
| 16S.3 | 5′-GGA TTA GAT ACC CKG GTA GTC C-3′ | 808-829 |
| 16S.4 | 5′-GGT TAA GTC CCG CAA CGA GC-3′ | 1125-1144 |
| 16S.5 | 5′-GCT CGT TGC GGG ACT TAA CC-3′ | 1144-1125 |
| PB38 | 5′-GKT ACC TTG TTA CGA CTT-3′ | 1509-1492 |
R represents A or G, M denotes A or C, and K represents G or T
Brosius et al. (10).
Sequence data assembly, analysis, and storage were done by using the Canadian Bioinformatics Resource (http://www.cbr.nrc.ca.login.ezproxy.library.ualberta.ca). Raw sequence data were assembled into contigs with PreGap4 (version 1.1) and Gap4 (version 4.6) in the Staden software package (release 2000.0; J. Bonfield, K. Beal, M. Betts, M. Jordan, and R. Staden, 2000). Contig sequences were compared to known other sequences in GenBank by using the FASTA and BLASTn search tools (2).
The 16S rRNA gene sequence for isolate RM6 has been deposited in the GenBank database under accession number DQ118732.
RESULTS
Biodegradation of 2,2′-dithiodibenzoic acid.
A 2,2′-dithiodibenzoic acid-degrading soil enrichment culture yielded six morphologically different colonies on PCA plates. Together, two of the bacterial isolates, RM1 and RM6, were capable of degrading 2,2′-dithiodibenzoic acid in SFMM. This mixture consumed all of the 2,2′-dithiodibenzoic acid (initial concentration 100 mg/liter) in 10 days.
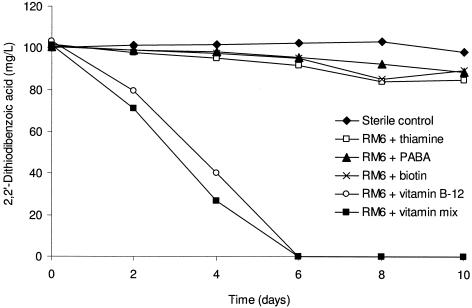
Initially, growth of isolate RM6 could not be obtained independently from isolate RM1 in SFMM. Supplementation with a mixed vitamin solution allowed isolate RM6 to degrade 2,2′-dithiodibenzoic acid, revealing that this bacterium required growth factors not present in the SFMM. When filter-sterilized supernatant from cultures of isolate RM1 grown on glucose or benzoate was added to cultures of isolate RM6, this isolate was capable of biodegrading 2,2′-dithiodibenzoic acid. Each of the four vitamins contained in the vitamin mixture was tested individually as supplements in the SFMM. Isolate RM6 degraded 2,2′-dithiodibenzoic acid in 6 days when the vitamin mixture or vitamin B12 was provided (Fig. 2).
FIG. 2.
Biodegradation of 2,2′-dithiodibenzoic acid by isolate RM6 in the presence of a vitamin mixture and four individual vitamins. PABA is p-aminobenzoic acid.
Identification and characterization of the isolates.
The Biolog system identified isolate RM1 as a member of the genus Rhodococcus and isolate RM6 as a member of the genus Variovorax when the test wells were supplemented with vitamin B12. 16S rRNA gene sequence analysis showed that isolate RM6 is 99% similar to Variovorax isolates WDL1 and 55 (GenBank accession numbers AF538929 and AY238498, respectively).
Variovorax sp. strain RM6 was capable of using glucose, benzoate, acetate, and salicylate as carbon sources if sulfate and vitamin B12 were supplied in SFMM. None of a number of compounds—naphthalene, catechol, benzothiophene, DBT, benzenethiol, benzyl sulfide, phenyl sulfide, octyl sulfide, n-dodecyl sulfide, benzyl disulfide, or phenyl disulfide—served as a growth substrate for isolate RM6 in vitamin-supplemented SFMM.
Isolate RM6 could not use 2-oxo-2-(2-thiophenyl)ethanoic acid disulfide as a sulfur or carbon and sulfur source in vitamin-supplemented SFMM. Isolate RM6 did not degrade 2,4′-dithiodibenzoic acid or 4,4′-dithiodibenzoic acid. However, it did degrade the mixed disulfide formed from 2-mercaptobenzoic acid and mercaptoacetic acid. It also grew with 2-sulfobenzoic acid (100 mg/liter) provided as a carbon and sulfur source, degrading 2-sulfobenzoic acid in 20 days. Isolate RM6 degraded a mixture of 2-sulfobenzoic acid and 2,2′=dithiodibenzoic acid (100 mg/liter each) in 4 days.
Benzoic acid as an intermediate in the biodegradation of 2,2′-dithiodibenzoic acid.
When vitamin supplement-SFMM was prepared with 2,2′-dithiodibenzoic acid formed from the abiotic dimerization of 2-mercaptobenzoic acid, the medium was free of benzoic acid. However, after 2 days of incubation with Variovorax sp. strain RM6, a metabolite was detected by HPLC analysis, and it had the same retention time and UV scan as benzoic acid. This metabolite was not detected in the sterile control.
Triplicate cultures were established with Variovorax sp. strain RM6 degrading 2,2′-dithiodibenzoic acid. The transient peak tentatively identified as benzoic acid was observed after 2 days (Fig. 3). Cultures of strain RM6 degrading 2,2′-dithiodibenzoic acid were acidified, extracted, and derivatized with MTBSTFA yielding a peak with the same retention time and mass spectrum as derivatized benzoic acid. The most abundant ions were m/z 179 (100%), 135 (11%), 105 (20%), 77 (21%), and 51 (8%). Thus, confirming that benzoic acid was produced during the biodegradation of 2,2′-dithiodibenzoic acid by strain RM6.
FIG. 3.
Benzoic acid formed in cultures of Variovorax sp. strain RM6 degrading 2,2′-dithiodibenzoic acid formed from 2-mercaptobenzoic acid dimerization. The datum points represent means of triplicate cultures, and the error bars are one standard deviation from the mean. Some error bars are smaller than the symbols.
Mineralization experiments.
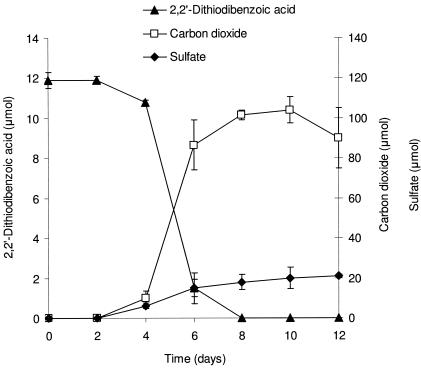
2,2′-Dithiodibenzoic acid, 12 μmol, was provided in vitamin-supplemented SFMM and degraded by Variovorax sp. strain RM6 in 8 days (Fig. 4). After 12 days of incubation, approximately 90 μmol of carbon dioxide and 20 μmol of sulfate were released.
FIG. 4.
Mineralization of 2,2′-dithiodibenzoic acid by Variovorax sp. stain RM6 in SFMM. The datum points represent means of triplicate cultures and the error bars are one standard deviation from the mean. Some error bars are smaller than the symbols.
A similar experiment with the sulfitolysis product 2-(S-sulfo)benzoic acid as the carbon and sulfur source showed no mineralization after 28 days of incubation.
DISCUSSION
2,2′-Dithiodibenzoic acid has been previously reported in the literature. It has been clinically compared to aspirin as a medication for arthritis patients (14) and is a decomposition product of thiomersal (46), an antibacterial and antifungal agent used in ophthalmic solutions and opened multidose vial vaccines (1). Bressler and Fedorak (8) reported that 2,2′-dithiodibenzoic acid could be used as a sole carbon and sulfur source by garden soil enrichment cultures but provided no additional information. Many of the soils used in the present study were taken from the root zones of marigolds because these plants, and other members of the family Asteraceae (Compositae), produce a variety of sulfur-containing metabolites (11).
Variovorax sp. strain RM6 is capable of degrading 2,2′-dithiodibenzoic acid in pure culture if vitamin B12 is provided in the SFMM. Isolate RM1 likely produces vitamin B12, an absolute requirement for the growth of isolate RM6 in defined medium, because a mixture of these two bacteria degrades 2,2′-dithiodibenzoic acid. Vitamin B12 contains cobalt and is required for microbial biosynthesis of methionine (45). Although many B12-enzymes perform rearrangements of functional groups (21), its role in the biodegradation of 2,2′-dithiodibenzoic acid is unknown. Hansen et al. (24) reported that Pseudomonas sp. strain RW611 has an absolute requirement for vitamin B12 when it uses 2-sulfobenzoic acids as its sole sulfur source, but the specific role of the vitamin was not reported.
Variovorax sp. strain RM6 did not use many other compounds as carbon or carbon and sulfur sources. The disulfide-degrading isolate did not attack DBT, implying that a mixed culture would be necessary for DBT mineralization. Interestingly, strain RM6 could not degrade 2-oxo-2-(2-thiophenyl)ethanoic acid disulfide (Fig. 1) or use it as a sulfur source if acetate was provided in the vitamin-supplemented SFMM (48). The biodegradation of 2,2′-dithiodibenzoic acid by strain RM6 is very specific, because it does not degrade 2,4′-dithiodibenzoic acid or 4,4′-dithiodibenzoic acid. However, strain RM6 does degrade the 2-mercaptobenzoic acid-mercaptoacetic acid mixed disulfide.
Analyses of acid extracts of laboratory cultures biodegrading benzothiophene, methylbenzothiophene, DBT, and methylDBTs produce a variety of 2,3-diones (5, 15, 33, 34, 39, 40). When benzothiophene-2,3-dione and 7-methylbenzothiophene-2,3-dione were combined in liquid medium, seven different disulfides were formed (8) by the mechanism shown in Fig. 1. Thus, during the biodegradation of a crude oil that contains many different substituted thiophenes, one would expect a vast array of disulfides to be formed. Based on the narrow range of disulfides used by strain RM6, it may not play a major role in the degradation of the predicted array of disulfides in biodegraded petroleum.
Various investigations have focused on the biodegradation of disulfide-containing macromolecules in insoluble biological materials such as wool (41, 44) and feathers (3, 4, 35). However, the metabolism of disulfides is generally poorly understood and only a few studies with compounds such as diphenyl disulfide (16), dibenzyl disulfide (16), diallyl disulfide (20) and dimethyl disulfide (42) have identified sulfur-containing metabolites.
No sulfur-containing intermediates were detected in our cultures that degraded 2,2′-dithiodibenzoic acid. Benzoic acid was detected during the biodegradation of this disulfide by Variovorax sp. strain RM6 (Fig. 3). By analogy to reports in the literature, three different hypothetical pathways for 2,2′-dithiodibenzoic acid biodegradation, that would produce benzoic acid as an intermediate, might involve (a) initial reduction of the disulfide bond, (b) initial oxidation of the disulfide sulfur atoms, or (c) sulfitolysis of the disulfide.
Smith and Kelly (42) proposed a pathway for the aerobic biodegradation of dimethyl disulfide by Thiobacillus thioparus strain E6 that began with reductive cleavage of the disulfide to give two molecules of methanethiol. These authors postulated that an oxygen-requiring reaction released H2S from methanethiol and that the H2S was rapidly oxidized to sulfate. Although there was stoichiometric conversion of the disulfide sulfur to sulfate, Smith and Kelly (42) could not detect H2S in their cultures. Similarly, sulfate was released when Variovorax sp. strain RM6 degraded 2,2′-dithiodibenzoic acid (Fig. 4), but we were unable to detect H2S in these cultures.
Tan and Parkin (46) studied the abiotic decomposition of thiomersal and found that 2-mercaptobenzoic acid was produced, which dimerized to form 2,2′-dithiodibenzoic acid. They detected 2-sulfinobenzoic acid from the abiotic reaction of 2,2′-dithiodibenzoic acid with the ethylmercuric ion, released from thiomersal. Oxidation of sulfinobenzoic acid to 2-sulfobenzoic acid was catalyzed by 50 ppb Cu2+ ion, and a trace amount of benzoic acid was produced through these abiotic reactions (46). Although Variovorax sp. strain RM6 degrades 2-sulfobenzoic acid, this acid was not detected as an intermediate of 2,2′-dithiodibenzoic acid biodegradation. In addition, benzoic acid was not detected in cultures degrading 2-sulfobenzoic acid. Thus, we have no evidence that 2-sulfobenzoic acid is an intermediate in the degradation of 2,2′-dithiodibenzoic acid by strain RM6.
Sulfitolysis is a mechanism by which disulfide bonds in wool are broken by some fungi (35). This reaction occurs in the presence of sulfite (generated by the fungi) and under alkaline conditions cleaving the disulfide in cystine to S-sulfocysteine and cysteine (35, 38). In general, the reaction can be written as R1-S-S-R2 + SO3 = → R1-S-SO3− + −S-R2. We incubated a solution of 2,2′-dithiodibenzoic acid with sulfite to prepare 2-(S-sulfo)benzoic acid which was not degraded by Variovorax sp. strain RM6. Thus, it appears that this bacterium does not utilize extracellular sulfitolysis to degrade 2,2′-dithiodibenzoic acid.
Benzoic acid, which was reported as an intermediate in DBT mineralization (47), was the only intermediate that we detected from the biodegradation of 2,2′-dithiodibenzoic acid. Strain RM6 mineralizes 2,2′-dithiodibenzoic acid, releasing 59% ± 6% of the carbon as carbon dioxide and 88% ± 5% of the sulfur as sulfate (Fig. 4). These finding are in agreement with the general assumption that aerobic heterotrophic activity releases ca. 50% of the substrate carbon as carbon dioxide (36). Within experimental error, Smith and Kelly (42) observed stoichiometric conversion of the sulfur in dimethyl disulfide to sulfate, slightly more than that observed from 2,2′-dithiodibenzoic acid (88% ± 5%). Although sulfite has been detected as a transient intermediate of DBT mineralization (47), we did not analyze for sulfite because of the high yield of sulfate in our mineralization experiment.
This investigation demonstrated that 2,2′-dithiodibenzoic acid is biodegradable by soil microorganisms. The results of our studies suggest that if DBT degradation in the environment occurs by some microorganisms using the Kodama pathway, yielding disulfides, other microorganisms such as Variovorax sp. strain RM6 may complete the overall mineralization of DBT.
Acknowledgments
This study was funded by the Natural Sciences and Engineering Research Council of Canada and the Department of Biological Sciences (University of Alberta).
We thank Don Morgan for LC-MS analyses and Julia Foght for providing helpful discussions and Biolog supplies.
REFERENCES
- 1.Abuqaddom, A. I., R. M. Darwish, and H. Muti. 2003. The effects of some formulation factors used in ophthalmic preparations on thiomersal activity against Pseudomonas aeruginosa and Staphylococcus aureus. J. Appl. Microbiol. 95:250-255. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Böckle, B., and R. Müller. 1997. Reduction of disulfide bonds by Streptomyces pactum during growth on chicken feathers. Appl. Environ. Microbiol. 63:790-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Böckle, B., B. Galunsky, and R. Müller. 1995. Characterization of a keratinolytic serine protease from Streptomyces pactum DSM 40530. Appl. Environ. Microbiol. 61:3705-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohonos, N., T. W. Chou, and R. J. Spanggord. 1977. Some observations on biodegradation of pollutants in aquatic systems. Jpn. J. Antibiot. 30(Suppl.):275-285. [PubMed] [Google Scholar]
- 6.Bressler, D. C., and P. M. Fedorak. 2000. Bacterial metabolism of fluorene, dibenzofuran, dibenzothiophene, and carbazole. Can. J. Microbiol. 46:397-409. [PubMed] [Google Scholar]
- 7.Bressler, D. C., and P. M. Fedorak. 2001. Purification, stability and mineralization of 3-hydroxy-2-formylbenzothiophene, a metabolite of dibenzothiophene. Appl. Environ. Microbiol. 67:821-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bressler, D. C., and P. M. Fedorak. 2001. Identification of disulfides from the biodegradation of dibenzothiophene. Appl. Environ. Microbiol. 67:5084-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bressler, D. C., B. K. Leskiw, and P. M. Fedorak. 1999. Biodegradation of benzothiophene sulfones by a filamentous bacterium. Can. J. Microbiol. 45:360-368. [PubMed] [Google Scholar]
- 10.Brosius, J., T. L. Dull, D. D. Sleeter, and H. Noller. 1981. Gene organization and primary structure of a rRNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 11.Christensen, L. P., and J. Lam. 1990. Acetylenes and related compounds in Cynareae. Photochemistry 29:2753-2785. [Google Scholar]
- 12.Cole, R. D. 1967. Sulfitolysis. Methods Enzymol. 11:206-208. [Google Scholar]
- 13.Collins, M. D., and F. Widdel. 1986. Respiratory quinines of sulphate-reducing and sulphur-reducing bacteria: a systematic investigation. Syst. Appl. Microbiol. 8:8-18. [Google Scholar]
- 14.Dequeker, J., E. Stevens, and L. Wuyts. 1980. A controlled trial of magnesium dithiosalicylate compared with aspirin in rheumatoid arthritis. Curr. Med. Res. Opin. 6:589-592. [DOI] [PubMed] [Google Scholar]
- 15.Eaton, R. W., and J. D. Nitterauer. 1994. Biotransformation of benzothiophene by isopropylbenzene-degrading bacteria. J. Bacteriol. 176:3992-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faison, B. D., T. M. Clark, S. N. Lewis, C. Y. Ma, D. M. Sharkey, and C. A. Woodward. 1991. Degradation of organic sulfur compounds by a coal-solubilizing fungus. Appl. Biochem. Biotechnol. 28/29:237-251. [DOI] [PubMed] [Google Scholar]
- 17.Fedorak, P. M., and D. Grbić-Galić. 1991. Aerobic microbial cometabolism of benzothiophene and 3-methylbenzothiophene. Appl. Environ. Microbiol. 57:932-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finkel'stein, Z. I., B. P. Baskunov, L. N. Vavilova, and L. A. Golovleva. 1997. Microbial transformation of dibenzothiophene and 4,6-dimethyldibenzothiophene. Microbiologiya 66:481-487. [Google Scholar]
- 19.Foght, J. M., J. Aislabie, S. Turner, C. E. Brown, J. Ryburn, D. J. Saul, and W. Lawson. 2004. Culturable bacteria in subglacial sediments and ice from two southern hemisphere glaciers. Microb. Ecol. 47:329-340. [DOI] [PubMed] [Google Scholar]
- 20.Germain, E., J. Ager, C. Ginies, M.-H. Sess, and C. Teyssier. 2002. In vivo metabolism of diallyl disulphide in the rat: identification of two new metabolites. Xenobiotica 32:1127-1138. [DOI] [PubMed] [Google Scholar]
- 21.Gottschalk, G. 1986. Bacterial metabolism, 2nd ed., p. 245-247. Springer-Verlag, Inc., New York, N.Y.
- 22.Gray, K. A., O. S. Pogrebinsky, T. Mrachko, L. Xi, D. J. Monticello, and C. H. Squires. 1996. Molecular mechanisms of biocatalytic desulfurization of fossil fuels. Nat. Biotechnol. 14:1705-1709. [DOI] [PubMed] [Google Scholar]
- 23.Hannoun, M., N. Blazevic, D. Kolbah, M. Mihalic, and F. Kajfez. 1982. α-Phenylpropionic acid derivatives. Synthesis and dethiation of 5-benzoylbenzo-[b]thiophene-3-carboxylic acid. J. Heterocyclic Chem. 19:1131-1136. [Google Scholar]
- 24.Hansen, C., P. Fortnagel, and R. M. Wittich. 1992. Initial reactions in the mineralization of 2-sulfobenzoate by Pseudomonas sp. RW611. FEMS Microbiol. Lett. 92:35-40. [Google Scholar]
- 25.Isbister, J. D., R. Wyza, J. Lippold, A. DeSouza, and G. Anspach. 1988. Bioprocessing of coal, p. 281-293. In G. S. Omenn (ed.), Environmental biotechnology, reducing risks from environmental chemicals through biotechnology. Plenum Press, Inc., New York, N.Y.
- 26.Kilbane, J. J., and K. Jackowski. 1992. Biodesulfurization of water-soluble coal-derived material by Rhodococcus rhodochrous IGTS8. Biotechnol. Bioeng. 40:1107-1114. [DOI] [PubMed] [Google Scholar]
- 27.Kayser, K. J., B. A. Bielaga-Jones, K. Jackowski, O. Odusan, and J. J. Kilbane. 1993. Utilization of organosulfur compounds by axenic and mixed cultures of Rhodococcus rhodochrous IGTS8. J. Gen. Microbiol. 139:3123-3129. [Google Scholar]
- 28.Kodama, K., S. Nakatani, K. Umehara, K. Shimizu, Y. Minoda, and K. Yamada. 1970. Microbial conversion of petro-sulfur compounds. Isolation and identification of products from dibenzothiophene. Agric. Biol. Chem. 34:1320-1324. [Google Scholar]
- 29.Kodama, K., K. Umehara, K. Shimizu, S. Nakatani, Y. Minoda, and K. Yamada. 1973. Identification of microbial products from dibenzothiophene and its proposed oxidation pathway. Agric. Biol. Chem. 37:45-50. [Google Scholar]
- 30.Kolmert, Å., Wikström, P., and K. B. Hallberg. 2000. A fast and simple turbidimetric method for the determination of sulfate in sulfate-reducing bacterial cultures. J. Microbiol. Methods 41:179-184. [DOI] [PubMed] [Google Scholar]
- 31.Krawiec, S. 1990. Bacterial desulfurization of thiophenes: screening techniques and some speculations regarding biochemical and genetic bases. Dev. Ind. Microbiol. 31:103-114. [Google Scholar]
- 32.Kropp, K. G., and P. M. Fedorak. 1998. A review of the occurrence, toxicity, and biodegradation of condensed thiophenes found in petroleum. Can. J. Microbiol. 44:605-622. [DOI] [PubMed] [Google Scholar]
- 33.Kropp, K. G., J. A. Gonçalves, J. T. Andersson, and P. M. Fedorak. 1994. Bacterial transformations of benzothiophene and methylbenzothiophenes. Environ. Sci. Technol. 28:1348-1356. [DOI] [PubMed] [Google Scholar]
- 34.Kropp, K. G., J. T. Andersson, and P. M. Fedorak. 1997. Bacterial transformations of three dimethyldibenzothiophenes by pure and mixed bacterial cultures. Environ. Sci. Technol. 31:1547-1554. [Google Scholar]
- 35.Kunert, J. 1989. Biochemical mechanism of keratin degradation by the actinomycete Streptomyces fradiae and the fungus Microsporum gypseum: a comparison. J. Basic Microbiol. 29:597-604. [Google Scholar]
- 36.Maier, R. M., I. L. Pepper, and C. P. Gerba. 2000. Environmental microbiology, p. 243-244. Academic Press, Inc., New York, N.Y.
- 37.Monticello, D. J., D. Bakker, and W. R. Finnerty. 1985. Plasmid-mediated degradation of dibenzothiophene by Pseudomonas species. Appl. Environ. Microbiol. 49:756-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruffin, O., S. Andrieu, G. Biserte, and J. Biguet. 1976. Sulphitolysis in keratinolysis: biochemical proof. Sabouraudia 14:181-184. [DOI] [PubMed] [Google Scholar]
- 39.Saftić, S., P. M. Fedorak, and J. T. Andersson. 1992. Diones, sulfoxides, and sulfones from the aerobic cometabolism of methylbenzothiophenes by Pseudomonas strain BT1. Environ. Sci. Technol. 26:1759-1764. [Google Scholar]
- 40.Saftić, S., P. M. Fedorak, and J. T. Andersson. 1993. Transformations of methyldibenzothiophenes by three Pseudomonas isolates. Environ. Sci. Technol. 27:2577-2584. [Google Scholar]
- 41.Shrivastava, J. N., V. K. Ghawana, A., and Kumar. 1996. Biodegradation of wool by species Trichophyton simmi and Aspergillus niger. Mycoses 39:483-487. [DOI] [PubMed] [Google Scholar]
- 42.Smith, N. A., and D. P. Kelly. 1988. Mechanism of oxidation of dimethyl disulphide by Thiobacillus thioparus strain E6 source of energy. J. Gen. Microbiol. 134:3031-3039. [Google Scholar]
- 43.St. John., W. P., J. Rughani, S. A. Green, and G. D. McGinnis. 1998. Analysis and characterization of naphthenic acids by gas chromatography-electron impact mass spectrometry of tert-butyldimethylsilyl derivatives. J. Chromatogr. A 807:241-251. [Google Scholar]
- 44.Stahl, W. H., B. McQue, G. R. Mandels, and R. G. H. Siu. 1949. Studies on the microbiological degradation of wool. I. Sulfur metabolism. Arch. Biochem. 20:422-432. [PubMed] [Google Scholar]
- 45.Stupperich, E. 1993. Recent advances in elucidation of biological corrinoid functions. FEMS Microbiol. Rev. 12:349-366. [DOI] [PubMed] [Google Scholar]
- 46.Tan, M., and J. E. Parkin. 2000. Route of decomposition of thiomersal (thimerosal). Int. J. Pharm. 208:23-34. [DOI] [PubMed] [Google Scholar]
- 47.van Afferden, M., S. Schacht, J. Klein, and H. G. Trüper. 1990. Degradation of dibenzothiophene by Brevibacterium sp. DO. Arch. Microbiol. 153:324-328. [Google Scholar]
- 48.Young, R. F. 2005. Aerobic biodegradation of disulfides produced from dibenzothiophene metabolites. MSc. thesis. Department of Biological Sciences, University of Alberta, Edmonton, Alberta, Canada.