Abstract
The gene for glycine betaine transmethylase (gbt) was identified in Pseudomonas aeruginosa strain Fildes III by biochemical, physiological, and molecular approaches. Based on sequence analysis, the knockout gene corresponded to an open reading frame (ORF) named PA3082 in the genome of P. aeruginosa PAO1. The translated product of this ORF displayed similarity to transferases of different microorganisms. Mutation in gbt blocked the utilization of choline and glycine betaine as carbon and nitrogen sources.
Choline and its degradation product glycine betaine (N,N,N-trimethylglycine) support the growth of Pseudomonas aeruginosa in media of iso-osmolarity, serving as both carbon and nitrogen sources (7). Additionally, in a hyperosmolar medium, P. aeruginosa utilizes choline not only as the sole carbon and nitrogen source but also as an osmoprotectant, accumulating glycine betaine as the main osmolite compound (6). Thus, glycine betaine plays a central role in P. aeruginosa, since it is catabolized to allow cellular growth and is accumulated at a high intracellular concentration to confer an osmoprotection action. The latter condition suggests that the accumulation of glycine betaine may occur by a decreased activity of glycine betaine transmethylase (EC 2.1.1.5) (GBT), the enzyme responsible for the demethylation of glycine betaine to N,N-dimethylglycine (DMG), as was reported for Rhizobium meliloti (9). The genes involved in the osmoprotection of P. aeruginosa (betA, betB, betI,and betT1, encoding choline dehydrogenase, betaine aldehyde dehydrogenase, a putative regulatory protein, and a choline transporter, respectively) had been localized from nucleotides 6047363 to 6053193 of the PAO1 complete genome (10), but those involved in glycine betaine degradation have not yet been localized. In this study, we report the identification of a genomic region that encodes a product required in the catabolic pathway of choline, specifically in the demethylation of glycine betaine to DMG.
Isolation and characterization of the TN5::751 mutant of P. aeruginosa.
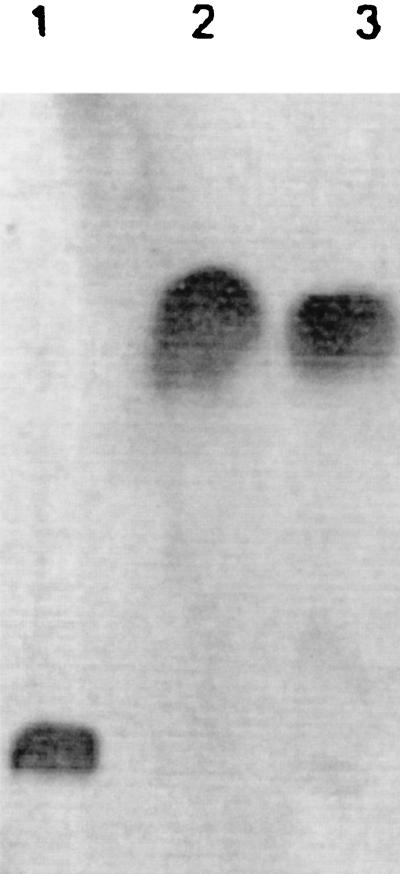
To identify the locus required for the utilization of glycine betaine as a carbon and nitrogen source in P. aeruginosa, strain PRS (a streptomycin-resistant [Strr] derivative of the wild-type strain Fildes III) was subjected to random mutagenesis with the Tn5::751 transposon, encoding kanamycin resistance (Kmr) and trimethoprim resistance (Tpr) (8). Approximately 3,200 colonies capable of growing in nutritive medium with streptomycin, kanamycin, and trimethoprim (1,000, 250, and 500 μg · ml−1, respectively) were screened for growth on iso-osmolar high-phosphate basal salt medium (HPi-BSM) (7) containing succinate plus NH4Cl or on choline as a carbon and nitrogen source. One strain that did not grow on choline was selected and designated ALS-96. Studies of growth on plates, subsequently confirmed to occur in liquid media (Table 1), showed that this strain was not capable of growing on either choline or glycine betaine under iso-osmolar or hyperosmolar conditions. Nevertheless, its growth was similar to that of the wild-type strain on succinate-NH4Cl or on choline derivatives such as DMG or sarcosine. The doubling times obtained under iso-osmolar and hyperosmolar conditions were similar to those obtained previously with the wild-type strain (Table 1). Given that the inability of ALS-96 to grow on choline may be due to a failure to take up choline or an incapacity to metabolize choline to glycine betaine, we performed studies with HPi-BSM-succinate-NH4Cl hyperosmolar medium with the addition of 1 mM exogenous [methyl-14C]choline as an osmoprotectant. Under such osmotic-stress conditions, both the mutant and the wild-type strains grew similarly (data not shown), thus confirming the integrity of the uptake system. In addition, the metabolic transformation of choline was confirmed by thin-layer chromatography analysis, since glycine betaine was the main labeled soluble osmoprotector extracted from strain ALS-96 (Fig. 1, lane 3), as was earlier demonstrated for the wild-type strain (2).
TABLE 1.
Growth and doubling times of the P. aeruginosa PRS and ALS-96 strains under iso-osmolar and hyperosmolar culture conditions
| Addition to HPi-BSM culture mediumb |
P. aeruginosa PRSa
|
P. aeruginosa ALS-96a
|
||||||
|---|---|---|---|---|---|---|---|---|
| Iso-osmolar
|
Hyperosmolar
|
Iso-osmolar
|
Hyperosmolar
|
|||||
| A660c | DTd | A660 | DT | A660 | DT | A660 | DT | |
| Succinate-NH4Cl | 1.30 | 0.35 | 0.60 | >8 | 1.21 | 0.45 | 0.39 | >10 |
| DMG | 1.10 | 0.62 | 0.65 | >4 | 0.85 | 0.70 | 0.27 | >7 |
| Sarcosine | 0.90 | 0.80 | 0.45 | >7 | 0.76 | 0.95 | 0.20 | >8 |
| Choline or glycine betaine | 1.10 | 1.00 | 1.00 | 1.7 | NGe | NGe | ||
Values are means from three independent experiments with less than 3% variation.
Cells were grown at 37°C in the presence of a 20 mM concentration of each compound as the carbon and nitrogen source in HPi-BSM under iso-osmolar or hyperosmolar (without or with the addition of 0.7 M NaCl, respectively) culture conditions.
Turbidity at 660 nm, determined when the cultures reached the late exponential phase.
DT, doubling time expressed in hours.
NG, no growth observed in 60 h.
FIG. 1.
Fate of [methyl-14C]choline. The ALS-96 mutant strain was grown to the mid-exponential phase (optical density at 600 nm, 0.5) in hyperosmolar HPi-BSM medium with 1 mM choline-[methyl-14C]choline as the osmoprotectant. Cells were harvested and treated as described previously (2). Soluble metabolites were analyzed by thin-layer chromatography (methanol-NH4OH, 75:25 [0.8 M]). The radiolabeled compounds were visualized by autoradiography. Lane 1, [methyl-14C]choline as a standard; lane 2, [methyl-14C]glycine betaine as a standard; lane 3, ALS-96 extract.
The growth tests and the accumulation of glycine betaine by the ALS-96 strain suggested that the mutant was impaired in the enzyme involved in the production of DMG from glycine betaine. This was confirmed by measuring GBT activity, determined by quantifying the production of [methyl-14C]methionine from [methyl-14C]glycine betaine and l-homocysteine. The standard assay contained 40 mM K2HPO4 (pH 7.5), 7 mM l-homocysteine, 7 mM [methyl-14C]glycine betaine (5,000 dpm/μmol), and 2 to 4 mg of protein in a final volume of 0.5 ml. The resulting [methyl-14C]methionine was isolated by ion-exchange chromatography and quantified by liquid scintillation counting (4). The GBT activity was measured in cell extracts of the wild-type strain grown on HPi-BSM medium with choline (415 ± 1.3 nmol of methionine produced h−1 · mg of protein−1) or DMG (75 ± 2.8 nmol of methionine produced h−1 · mg of protein−1) as a carbon and nitrogen source. No activity was detected in a cell extract of the mutant strain grown in DMG.
Localization and cloning of the DNA fragment affected by the TN5::751 insertion and complementation of the mutant strain by wild-type.
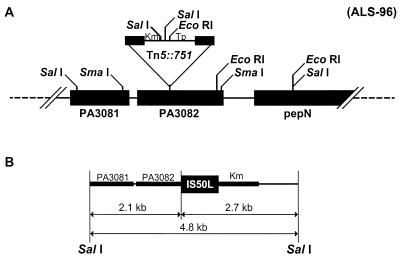
Southern hybridization experiments revealed a single insertion of Tn5::751 in the genome of the mutant strain ALS-96. A SalI restriction site was found 2.1 kb from the left border of the Tn5::751 insertion (Fig. 2). Digestion of Tn5::751 with SalI separates the two resistance gene markers (Km and Tp) (Fig. 2A). The total DNA of the mutant strain ALS-96 was digested with SalI, and the resulting DNA was purified and ligated into the ampicillin-resistant (Apr) pBluescript II SK plasmid, digested with the same enzyme, and used to transform Escherichia coli DH5α. Ten identical Apr and Kmr clones were obtained, and one was selected for further studies. The transformant E. coli strain carried a recombinant plasmid named pJM1 (8.0 kb) that had a 4.8-kb insert carrying the Kmr marker (2.7 kb) and the P. aeruginosa DNA adjacent to it (2.1 kb).
FIG. 2.
(A) Restriction map of the region of DNA around the inactivated gbt gene by insertion of Tn5::751 in the genome of P. aeruginosa Fildes III. The locations of PA3081, PA3082, and pepN (PA3083) are also shown in the complete genome of P. aeruginosa PAO1 published previously (10). (B) The 4.8-kb SalI-SalI DNA fragment containing the 2.1 kb of genomic DNA adjacent to the 2.7 kb of Tn5::751, which contains IS50L and the kanamycin resistance marker is shown.
The DNA sequence of the insert was determined and found to be almost the same as that recently determined by the Pseudomonas Genome Sequencing Project (10) in the P. aeruginosa genome project website (http://www.pseudomonas.com). The SalI fragment cloned in pJM1 revealed 98% identity to the PAO1 sequence named PA3081. The analysis of the sequence revealed that the transposon had interrupted the locus named PA3082 (Fig. 2A). Sequence analysis of this locus revealed the two overlapping potential ORFs, one of them (ORF1) being 2,526 bp in length and the other (ORF3) being 1,763 bp in length. Both of them were amplified by PCR with appropriate primers and cloned into vector pGEM-T Easy (Promega). The ApaI-SpeI fragments containing the PCR products were subcloned into the ApaI- and SpeI-digested broad-host-range gentamicin-resistant (Gmr) plasmid vector pBBR1MCS-5 (5), giving rise to the p5-orf1 and p5-orf3 plasmids.
Plasmids p5-orf1 and p5-orf3 were mobilized to P. aeruginosa ALS-96 by conjugation (3). Colonies capable of growing on Luria-Bertani agar with TMP, KAN, and GEN (500, 200, and 20 μg · ml−1, respectively) were selected and tested for their capacity to grow on HPi-BSM medium with glycine betaine as the sole carbon and nitrogen source. All transconjugants degraded glycine betaine. This result clearly shows that the corresponding wild-type DNA included in the p5-orf3 plasmid was sufficient to complement the mutation.
The corresponding knockout gene was named gbt for glycine betaine transmethylase. The gbt nucleotide sequence has a G+C content of 64.8%, which is typical for the genes of P. aeruginosa (11). The corresponding translated protein, GBT, would contain 548 amino acid residues. Comparison of this ORF product with sequences in the GenBank database (BLASTP) (1) showed sequence similarity with different transferases, like a putative O-methyltransferase (PKSA) of Streptomyces collinus (29% identity and 36% similarity; E = 3e−11) (GenBank accession no. AF293354), with a uridylyl transferase of P. aeruginosa PAO1 (27% identity and 34% similarity; E = 1e−07) (accession no. AB024601), and with an adenylyltransferase of Chlamydomonas reinhardtii (32% identity and 39% similarity; E = 1e−06) (accession no. X91736). The ClustalW alignment of sequences from GBT and O-methyltransferase of S. collinus does not show a specific conserved domain, but the homology between them is extended over the entire protein.
In conclusion, the results of this study strongly suggest that P. aeruginosa ALS-96 is impaired in GBT activity but not in either the production of glycine betaine from choline or the uptake of choline from the culture medium. The complementation test described above confirms that the P. aeruginosa ALS-96 phenotype was clearly due to the presence of a Tn5::751 insertion in the gbt sequence located within the ORF that corresponded to PA3082 in the unmutated genome of P. aeruginosa PAO1. Although the gbt gene is unlinked to those genes involved in the first steps of choline utilization (10), all of them may be coordinately regulated in response to the osmotic conditions of the culture.
Acknowledgments
We are grateful to L. Actis for providing the E. coli (pTGL166:: Tn5::751) strain, to M. Garrido for helpful discussions, to Mariela Woelker for technical assistance, and to language consultant Iliana A. Martínez.
Financial support was provided by the Universidad Nacional de Río Cuarto, Agencia Córdoba Ciencia de la Provincia de Córdoba, and CONICET, all of the Republica Argentina. J.L.B., C.E.D., and A.T.L. are Career Members of the CONICET.
REFERENCES
- 1.Altschul, S. F., F. Stephen, T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casale, C. H., A. T. Lisa, G. I. Lucchesi, and C. E. Domenech. 1994. The production of labeled betaine by incubation of osmolyte-free Pseudomonas aeruginosa with radioactive choline. Curr. Microbiol. 29:295-299. [Google Scholar]
- 3.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 4.Finkelstein, J., and S. Mudd. 1967. Trans-sulfuration in mammals. J. Biol. Chem. 242:873-880. [PubMed] [Google Scholar]
- 5.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800-802. [PubMed] [Google Scholar]
- 6.Lisa, A. T., C. H. Casale, and C. E. Domenech. 1994. Cholinesterase, acid phosphatase, and phospholipase C of Pseudomonas aeruginosa under hyperosmotic conditions in a high-phosphate medium. Curr. Microbiol. 28:71-76. [DOI] [PubMed] [Google Scholar]
- 7.Lisa, A. T., M. N. Garrido, and C. E. Domenech. 1983. Induction of acid phosphatase and cholinesterase activities in Pseudomonas aeruginosa and their in vitro control by choline, acetylcholine and betaine. Mol. Cell. Biochem. 50:149-155. [DOI] [PubMed] [Google Scholar]
- 8.Rella, M., A. Mercenier, and D. Haas. 1985. Transposon insertion mutagenesis of P. aeruginosa with a Tn5 derivative: application to physical mapping of the arc gene cluster. Gene 33:293-303. [DOI] [PubMed] [Google Scholar]
- 9.Smith, L. T., J. A. Pocard, T. Bernard, and D. Le Rudulier. 1988. Osmotic control of glycine betaine biosynthesis and degradation in Rhizobium meliloti. J. Bacteriol. 170:3142-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 11.West, S. E., and B. H. Iglewski. 1988. Codon usage in Pseudomonas aeruginosa. Nucleic Acids Res. 16:9323-9335. [DOI] [PMC free article] [PubMed] [Google Scholar]