Abstract
Shipworms (wood-boring bivalves of the family Teredinidae) harbor in their gills intracellular bacterial symbionts thought to produce enzymes that enable the host to consume cellulose as its primary carbon source. Recently, it was demonstrated that multiple genetically distinct symbiont populations coexist within one shipworm species, Lyrodus pedicellatus. Here we explore the extent to which symbiont communities vary among individuals of this species by quantitatively examining the diversity, abundance, and pattern of occurrence of symbiont ribotypes (unique 16S rRNA sequence types) among specimens drawn from a single laboratory-reared population. A total of 18 ribotypes were identified in two clone libraries generated from gill tissue of (i) a single specimen and (ii) four pooled specimens. Phylogenetic analysis assigned all of the ribotypes to a unique clade within the γ subgroup of proteobacteria which contained at least five well-supported internal clades (phylotypes). By competitive quantitative PCR and constant denaturant capillary electrophoresis, we estimated the number and abundance of symbiont phylotypes in gill samples of 13 individual shipworm specimens. Phylotype composition varied greatly; however, in all specimens the numerically dominant symbiont belonged to one of two nearly mutually exclusive phylotypes, each of which was detected with similar frequencies among specimens. A third phylotype, containing the culturable symbiont Teredinibacter turnerae, was identified in nearly all specimens, and two additional phylotypes were observed more sporadically. Such extensive variation in ribotype and phylotype composition among host specimens adds to a growing body of evidence that microbial endosymbiont populations may be both complex and dynamic and suggests that such genetic variation should be evaluated with regard to physiological and ecological differentiation.
Lyrodus pedicellatus is a member of the family Teredinidae, a group of worm-like, wood-boring marine bivalves that are commonly known as shipworms and that include many ecologically and economically important consumers of wood in marine environments (5, 30). This species burrows in wood for both food and shelter and is the only marine invertebrate shown to grow and reproduce on a diet composed solely of wood (18). The ability of shipworms to consume wood is thought to depend on gill-borne, intracellular endosymbiotic bacteria which provide enzymes, including cellulase and nitrogenase, critical for digestion of wood and supplementation of the host’s nitrogen-deficient diet (5, 31). Anatomically, the symbiont-bearing gills and cells of shipworms closely resemble those observed in sulfur- and/or methane-oxidizing bivalves and gastropods from deep-sea hydrothermal vents, cold-water seeps, and other marine habitats (5). In contrast to these other symbioses, which are thought to contain one (8, 9, 12, 13, 15) symbiont ribotype or in a few cases two (7, 16, 32) symbiont ribotypes (types denoted by unique 16S rRNA sequences), individual specimens of L. pedicellatus have been shown to harbor as many as four phylogenetically distinct bacterial ribotypes, two of which are observed to co-occur within some host cells (6). The ribotypes identified to date are closely related and include one, Teredinibacter turnerae, that has been grown and characterized in pure culture (10, 20, 31).
The previous detection of multiple symbiont ribotypes in L. pedicellatus raises questions regarding the extent to which symbiont communities differ within this single host species. Specifically, does symbiont community structure vary among individuals or are these communities under tight selection or regulation? To address this question, we have quantitatively examined the diversity, abundance, and frequency of occurrence of ribotypes present in specimens drawn from a population of L. pedicellatus reared under controlled conditions in a long-term laboratory culture.
MATERIALS AND METHODS
Specimen preparation and DNA extraction.
Specimens of L. pedicellatus were reared in the laboratory in wooden dowels as described previously (6). Gills were removed, washed in sterile filtered seawater, blotted dry on clean filter paper, weighed, and frozen in liquid nitrogen. DNA extracts were prepared from the frozen tissues as previously described (6). The specimens used here are distinct from those used in previously published investigations.
T. turnerae was isolated from gill tissue of laboratory-reared L. pedicellatus by the method of Waterbury et al. (31) as modified by Distel et al. (6), and DNA was extracted as described in reference 6.
PCR clone libraries.
Bacterial 16S rRNA genes were amplified by PCR (26) with Bacteria domain-specific primers 27f and 1492r (22) and the high-fidelity polymerase PfuTurbo (Stratagene, La Jolla, CA). Each PCR mixture (50-μl total volume) contained 50 ng of bulk nucleic acid extracted from gill tissue, 1× PFU buffer, 250 μM each deoxynucleoside triphosphate, 1.25 U of Pfu polymerase, and each primer at 200 nM. Cycling parameters were 2 min at 94°C, followed by 25 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C, followed by 8 min at 72°C. To eliminate potential heteroduplex artifacts, 5 μl of the completed PCR mixture was diluted 1:10 into a fresh PCR mixture and three cycles of reconditioning PCR were performed (28).
Amplified 16S rRNA gene fragments were cloned into plasmid vector pCR-Blunt II-TOPO (Invitrogen, Carlsbad, CA) and transformed into Escherichia coli (TOP10) cells according to the manufacturer’s instructions. Two 16S rRNA clone libraries were created and screened, one constructed from the whole gill of a single specimen (single-specimen library [SSL]) and the second from the pooled gills of four specimens (multiple-specimen library [MSL]) collected from a single lab-reared population of this host species. Plasmids were purified and prepared for sequencing with a RevPrep Orbit automated plasmid purification workstation (GeneMachines, San Carlos, CA).
Sequence analysis.
Cloned 16S rRNA gene fragments were sequenced bidirectionally with primers 27f, 1392r, 926f, 530f (22), and 519r2 (5′-GTGCCAGCMGCCGCGGTAAT-3′) with the ABI PRISM Big Dye Terminator cycle sequencing kit, Amplitaq DNA polymerase, and an ABI 3730X1 DNA analyzer. Raw ABI sequence data were edited manually with Sequencher software v.3.1.2 (Gene Codes Corp., Ann Arbor, MI). For phylogenetic analyses, sequences were aligned manually with SeqLab (GCG v.10.1; Accelrys Inc.), taking into consideration secondary-structure information (3). Gaps were inserted to compensate for length variation in inferred loops and helices. Sites within regions of uncertain alignment were identified and eliminated from further analyses. In addition, nucleotide substitutions at positions conserved in greater than 98% of the known sequences and substitutions that result in mispairings that interrupt conserved helices (3) were treated as putative nucleotide misincorporation errors and were disregarded in all further analyses, leaving a final character set of 1,234 nucleotide positions. To identify putative chimeras, phylogenetic analyses were performed independently on 5′ and 3′ portions of the sequence alignments. Putative chimeras were considered confirmed if (i) different parent ribotypes were identified for the 3′ and 5′ ends, (ii) these identifications were supported by multiple sequence differences in both ends, (iii) all differences could be explained by a single recombination event, and (iv) both parent ribotypes occurred multiple times in the clone libraries.
Phylogenetic analyses were performed with algorithms contained in PAUP* (version 4.0b10) (27). Evolutionary-distance trees were inferred by using the minimum evolution optimality criterion with distance correction by HKY85. In all analyses, branch swapping was by tree bisection-reconnection with characters weighted equally (weight = 1) and gaps treated as missing data. Bootstrap analyses were performed by the full heuristic search option with 1,000 replicates.
Oligonucleotide design, synthesis, and labeling.
Based on the sequence diversity encountered in clone libraries in this study and a previous study (6), a region of the 16S rRNA gene 115 bp in length (nucleotide positions 948 to 1063, E. coli numbering [3]) was identified that (i) contained sufficient variability to differentiate symbiont phylotypes (clades containing several ribotypes) and (ii) was bounded by priming sites conserved among all members of the symbiont clade. For constant denaturant capillary electrophoresis (CDCE), primers 1063r (E. coli numbering) and 948f were synthesized with a 54-bp GC-rich region and a fluorescein isothiocyanate label, respectively, at their 5′ ends (Table 1). Primers were synthesized and polyacrylamide gel electrophoresis purified by Integrated DNA Technologies (Coralville, IA).
TABLE 1.
CDCE primer sequences
| Primer or sequence | Nucleotide sequencea |
|---|---|
| 948f | 5′-LCATGTGGTTTAATTCGAAGC-3′ |
| 948f-mut | 5′-CATGTGGTTTAATTCGAAGCTACGTGAAGA-3′ |
| 1063r-GC | 5′-*GACAGCCATGCAGCACCTGT-3′ |
| 5′ GC clamp | 5′-CGGGCCTGCAGCCGGCGCCCCCCGTGCCCCCGCCCCGCCGCCGGCGGCGGCGCC-3′ |
L, fluorescein isothiocyanate label; ∗, location of 5′ GC clamp. Underlining indicates introduced mutations.
Competitive QPCR-CDCE.
Individual phylotypes of symbiont populations were quantified in 13 shipworm specimens (not including those used for the MSL) by a combination of competitive quantitative PCR (QPCR) and CDCE. This technique allows coamplification of templates with group-specific primers and separation and identification (as peaks in CDCE spectra) of individual amplicons based on their different migration behaviors under partially denaturing conditions. Amplifications were performed in a total volume of 20 μl containing 20 ng of bulk nucleic acid from the gills of each shipworm specimen amended with 105 copies of internal competitor DNA (see below), 250 μM each deoxynucleoside triphosphate, 1× buffer, 200 nM each primer (948f and 1063r-GC), and 0.025 U of Taq 2000 (Stratagene, La Jolla, Calif.). A Robocycler (Stratagene, La Jolla, Calif.) thermal cycler was used with the following cycling parameters: 3 min at 94°C, followed by 25 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C, followed by 5 min at 72°C. An aliquot (2 μl) of the PCR product was added to 18 μl of a fresh PCR mixture (final dilution, 1:10), and five cycles of reconditioning PCR were performed (28) with the same cycling parameters. Migration properties of individual amplicons were predicted with MacMelt (Medprobe, AS, Oslo, Norway), and separation conditions were optimized empirically. The identities of CDCE peaks were determined by comigration with PCR products generated from previously sequenced plasmid clones.
Internal competitor DNA was prepared from a linearized plasmid containing ribotype RT5 (clone A01) modified by PCR to contain mutations at positions 968 (A→T) and 972 (C→T). The mutations were introduced by using an extended version of the forward amplification primer (948fmut; Table 1) containing mismatches adjacent to the 948f primer used in QPCR-CDCE. The resulting PCR product was cloned as described above, purified, and linearized by overnight digestion with BamHI (Invitrogen, Carlsbad, CA). Concentration was determined by spectrophotometry after purification with a QIAquick PCR purification kit (QIAGEN, Valencia, CA).
For CDCE analysis, PCR mixtures were diluted 50-fold into distilled and deionized water (Millipore, Billerica, MA) and electroinjected for 45 s at 2 μA into a fused silica capillary (75-μm inside diameter) filled with a replaceable 5% linear polyacrylamide gel matrix (Scientific Polymers, Ontario, NY). Electrophoretic separation was performed at constant current (10 μA) and temperature (75.5°C). A 488-nm argon ion laser (ILT, Salt Lake City, UT) served as the light source for fluorescence detection of the fluorescein isothiocyanate label on individual amplicons, and a photomultiplier tube (Oriel, Stratford, CT) was used for fluorescence detection. The output signal was collected with the Workbench Data Acquisition Program (Strawberry Tree, Inc., Sunnyvale, CA).
To determine whether PCR amplification bias occurred during coamplification of multiple templates, linearized plasmids containing target sequences representative of the five phylotypes examined and the internal competitor were mixed in equal proportions (105 copies of each) and a 25-cycle amplification was performed with primers 948f and 1063r-GC. The PCR product was diluted 1:1,000 and reamplified for 15 cycles for a total of three times. For each target-competitor pair, peak area ratios were compared after each round of amplification. After the final round of amplification (a total of >106-fold), no significant difference in amplification efficiency was observed among target sequences relative to the internal competitor, confirming that QPCR-CDCE could be used to estimate the copy numbers of the identified phylotypes by coamplification from gill samples.
Data analysis.
All QPCRs were performed in triplicate, and replicates were analyzed individually by CDCE. Peak areas were measured with AcqKnowledge 2.1 software (Biopac Systems, Santa Barbara, CA) and were used to estimate target sequence copy number with the formula Pi = (CStd)(APi/AStd)/Wg(Ns/Nt), where Pi is the number of small-subunit rRNA gene copies per gram (wet weight) of gill tissue corresponding to the ith phylotype in the initial symbiont population, CStd is the number of copies of internal competitor added to the reaction mixture, APi is the area of the CDCE peak corresponding to an individual bacterial phylotype, AStd is the area of the peak corresponding to the internal competitor, Wg is the wet weight of the gill tissue, Nt is the total nucleic acid extracted from that tissue, and Ns is the nucleic acid used for amplification. Small-subunit rRNA gene copy numbers are reported as means ± standard deviations (n = 3).
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this work have been submitted to GenBank and assigned accession no. DQ272300 to DQ272317.
RESULTS AND DISCUSSION
To explore the extent of diversity and variation within the intracellular symbiont community of a single bivalve species, we have examined the shipworm L. pedicellatus, which has been reported to harbor as many as four symbiont ribotypes within its gill bacteriocytes (6). Considerable ribotype diversity was observed in both the SSL and the MSL. For both libraries combined, 163 clones were examined by bidirectional sequencing. Of these, 16 were identified as probable chimeras and were excluded from further analyses. Among the remaining 147 clones (73 from the SSL and 74 from the MSL), 18 ribotypes were identified (7 in the SSL and 13 in the MSL). There was little overlap in ribotype identity between the two libraries, with only two ribotypes, RT5 (62 occurrences in the SSL and 38 in the MSL) and RT21 (1 occurrence in the SSL and 7 in the MSL), observed in both. Moreover, the frequency of occurrence of ribotypes within both libraries was highly uneven. A single ribotype (RT5) recurred 100 times in the combined libraries and accounted for 68% of the clones, while the majority of ribotypes appeared only once (50%) or twice (28%) in the combined libraries. Rarefaction analyses suggest that additional sampling would reveal additional ribotypes, and the Chao-1 estimator predicts 12 ± 4 and 29 ± 7 ribotypes in the SSL and combined libraries, respectively.
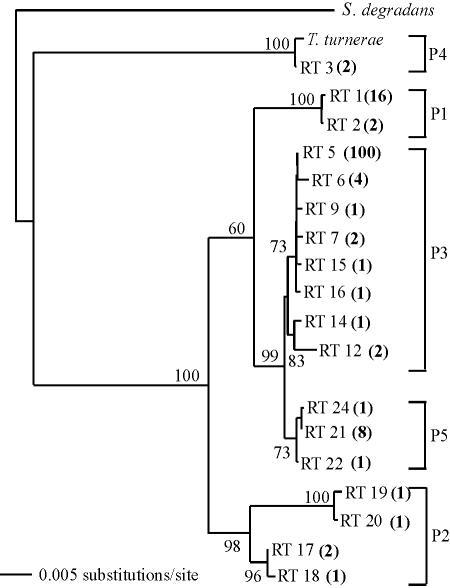
Phylogenetic analyses show that the observed ribotypes form a unique clade within the γ subgroup of proteobacteria whose closest relative among cultivated and characterized bacteria is “Saccharophagus degradans” 2-40, a free-living marine heterotrophic bacterium that degrades a broad spectrum of plant polysaccharides (19). This shipworm symbiont clade, in turn, contains several well-supported internal clades. Hereafter, we refer to these distinct clades or phylogenetic types as phylotypes (Fig. 1). Five of these phylotypes, designated P1 to P5, could be distinguished by CDCE-based analysis with a single variable region of the 16S rRNA gene (see below). Sequence variation between groups, among the five phylotypes, ranged from 0.2 to 8.2% (Phylip 3.6b, DNADIST, uncorrected distance) (Table 2), and ribotype diversity observed within phylotypes ranged from 0 to 1.6%.
FIG. 1.
Phylogenetic relationships among 18 unique symbiont ribotypes (RTs) identified in the combined SSL and MSL. Brackets (P1 to P5) indicate clades (phylotypes) that can be distinguished by CDCE with the primers 948f and 1063r-GC. Bold values in parentheses indicate the numbers of clones in the combined libraries belonging to a specific ribotype. The tree was inferred by an evolutionary-distance method (PAUP 4.0, with minimum evolution optimality criterion and HKY85 correction; 1,234 bp were considered in the analysis). Bootstrap proportions are expressed as a percentage of 1,000 replicates and are indicated on the tree for nodes with values >50%. The reference sequences were T. turnerae T7902T AY028398 and “S. degradans” 2-40 AF055269.
TABLE 2.
Distance matrixa comparing sequence divergences among the 18 unique symbiont ribotypes observed in this study and the “S. degradans” reference sequence
| Organism or ribotype | Distance | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| “S. degradans” | |||||||||||||||||||
| T. turnerae | 8.75 | ||||||||||||||||||
| RT3 | 8.75 | 0.16 | |||||||||||||||||
| RT1 | 8.29 | 7.35 | 7.36 | ||||||||||||||||
| RT2 | 8.35 | 7.30 | 7.31 | 0.08 | |||||||||||||||
| RT5 | 8.76 | 8.01 | 7.92 | 1.57 | 1.51 | ||||||||||||||
| RT6 | 8.96 | 8.21 | 8.11 | 1.74 | 1.68 | 0.16 | |||||||||||||
| RT15 | 8.67 | 8.11 | 8.01 | 1.49 | 1.60 | 0.08 | 0.24 | ||||||||||||
| RT9 | 8.85 | 8.11 | 8.01 | 1.65 | 1.60 | 0.08 | 0.24 | 0.16 | |||||||||||
| RT16 | 8.86 | 8.11 | 8.01 | 1.65 | 1.60 | 0.08 | 0.24 | 0.16 | 0.16 | ||||||||||
| RT7 | 8.85 | 8.11 | 8.01 | 1.65 | 1.60 | 0.08 | 0.24 | 0.16 | 0.16 | 0.16 | |||||||||
| RT14 | 8.86 | 7.92 | 7.82 | 1.57 | 1.51 | 0.33 | 0.49 | 0.41 | 0.41 | 0.41 | 0.41 | ||||||||
| RT12 | 9.34 | 8.30 | 8.20 | 1.82 | 1.77 | 0.57 | 0.65 | 0.65 | 0.65 | 0.65 | 0.65 | 0.49 | |||||||
| RT19 | 9.73 | 7.75 | 7.75 | 3.69 | 3.67 | 3.17 | 3.34 | 3.26 | 3.25 | 3.08 | 3.25 | 3.17 | 3.43 | ||||||
| RT20 | 9.59 | 7.68 | 7.69 | 3.48 | 3.54 | 3.04 | 3.22 | 3.04 | 3.13 | 3.13 | 3.13 | 3.05 | 3.31 | 0.65 | |||||
| RT17 | 9.23 | 7.57 | 7.47 | 3.17 | 3.14 | 1.56 | 1.73 | 1.64 | 1.64 | 1.64 | 1.64 | 1.90 | 2.15 | 1.57 | 1.42 | ||||
| RT18 | 9.17 | 7.50 | 7.40 | 3.48 | 3.45 | 1.75 | 1.93 | 1.84 | 1.84 | 1.84 | 1.84 | 2.02 | 2.38 | 1.58 | 1.42 | 0.09 | |||
| RT24 | 8.95 | 8.20 | 8.02 | 1.57 | 1.51 | 0.41 | 0.57 | 0.49 | 0.49 | 0.32 | 0.49 | 0.41 | 0.65 | 2.83 | 3.05 | 1.98 | 2.20 | ||
| RT22 | 8.95 | 8.02 | 7.83 | 1.57 | 1.51 | 0.41 | 0.57 | 0.49 | 0.49 | 0.49 | 0.49 | 0.41 | 0.65 | 2.83 | 2.87 | 1.81 | 2.02 | 0.16 | |
| RT21 | 8.74 | 7.71 | 7.50 | 1.82 | 1.76 | 0.36 | 0.54 | 0.45 | 0.45 | 0.27 | 0.45 | 0.54 | 0.81 | 3.01 | 3.26 | 1.90 | 2.13 | 0.09 | 0.09 |
Distances are determined based on an alignment of 1,234 nucleotide positions and are expressed as the number of substitutions per nucleotide position (102).
While few ribotypes were common to both libraries, considerable overlap was observed when the ribotypes were clustered into phylotypes. And although ribotypes are undersampled, rarefaction analyses indicate that phylotypes are well sampled in both libraries (Chao-1 estimator predicts five phylotypes). Four of the five phylotypes are represented in the SSL, four occur in the MSL, and three occur in both. Symbiont ribotype consensus sequences identified in a previous study (Lp1, Lp3, and Lp4; GenBank accession numbers AY150583, AY150578, and AY028398) and shown by fluorescence in situ hybridization to be present within gill bacteriocytes of L. pedicellatus (6) can be assigned to three of the five phylotypes (P1, P3, and P4, respectively) in the present study. Phylotype P4 also contains the 16S rRNA gene sequence of the cultivated symbiont of the shipworm T. turnerae. Consensus sequence Lp2 from the previous study was not detected in the present study.
With a combination of CDCE and QPCR (23, 29), symbiont phylotype abundance was estimated in the gills of 13 individual shipworm specimens. Summing peak intensities from all five phylotypes for 11 specimens (2 were omitted from this calculation because gill weights were not determined) yielded estimates of total symbiont rRNA gene copy abundance ranging from 0.81 × 1010 to 8.99 × 1010 g−1 (wet weight) of gill tissue, with a mean of 3.55 × 1010 ± 0.36 × 1010 g−1. These numbers cannot be readily translated into symbiont cell abundance estimates because the average numbers of genome copies per symbiont cell and 16S rRNA operons per symbiont genome are not known. Nonetheless, the16S rRNA gene copy abundances observed here fall within the range of cell abundances (109 to 1011 cells g−1 [wet weight] of symbiont-containing tissue) estimated for the sulfur-oxidizing symbionts of the marine bivalves Solemya reidi, Bathymodiolus thermophilus, and Calyptogena magnifica and the vestimentiferan tubeworm Riftia pachyptila (4, 25).
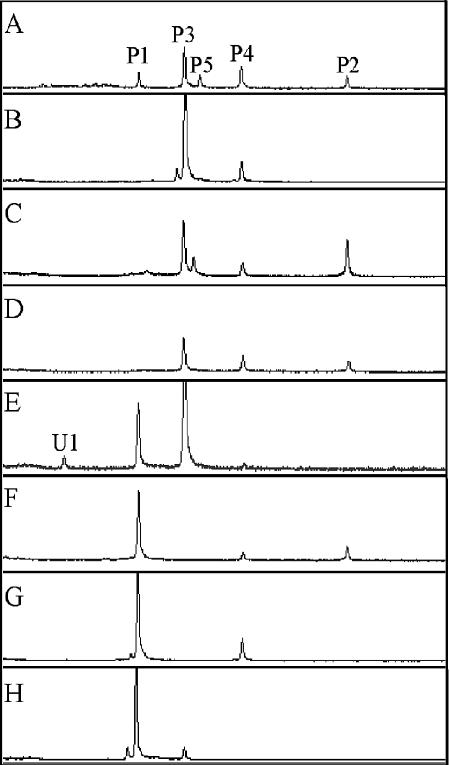
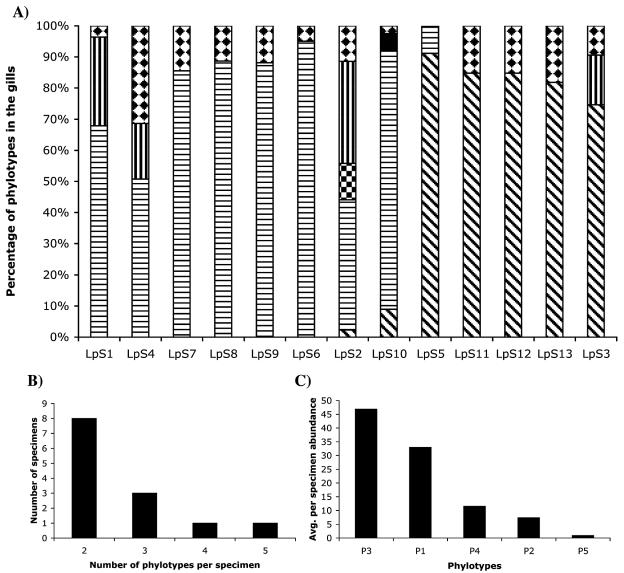
Peak patterns in CDCE spectra varied significantly among the gills of the 13 individual specimens examined (Fig. 2B to H), revealing considerable variation in the composition of symbiont populations (Fig. 3A). The number of phylotypes detected per specimen ranged from two to five and included all phylotypes recovered in the combined libraries plus an additional unidentified target sequence (U1) that appeared as a minor component in one specimen (LpS10) (Fig. 3A). In the majority of specimens (11 of 13), two or three symbiont phylotypes were detected (Fig. 3A). The numerically dominant phylotype among all specimens was P3 (Fig. 3C), and the symbiont phylotype most frequently observed was P4 (Fig. 3A). No correlation was observed between rearing location and pattern of symbiont composition.
FIG. 2.
CDCE profiles obtained from gills of the shipworm L. pedicellatus after PCR amplification with primers 948f and 1063r-GC. (A) Combined gills from four specimens used for the MSL but not included among the 13 individual specimens examined. (B to H) CDCE profiles from gills of seven representative specimens of L. pedicellatus, each showing a different combination of identified peaks. The identities of peaks P1 to P5 were confirmed by comigration with PCR products from sequenced clones spiked into samples after PCR and prior to CDCE. U1 in panel E represents an unidentified peak observed in one specimen only.
FIG. 3.
Phylotype abundance in the gills of 13 individual specimens of L. pedicellatus as determined by QPCR-CDCE expressed as a percentage of the total CDCE peak area for each specimen. (A) Phylotypes are as follows: P1, diagonal stripes; P2, vertical stripes; P3, horizontal stripes; P4, diamonds; P5, checkering; unidentified peak U1, black. (B) Number of phylotypes detected per specimen ranked in order of decreasing frequency. (C) Average abundance of phylotypes expressed as percentages of the total combined CDCE peak area for gills of 13 specimens of L. pedicellatus.
Though symbiont community composition varied widely among specimens, specific correlations were observed. In all specimens, the numerically dominant phylotype was either P1 or P3, and the detection of these two phylotypes was mutually exclusive in most specimens (77%). Phylotypes P1 and P3 were dominant in 5 and 8 of 13 specimens, respectively. These two phylotypes co-occurred in 3 of 13 specimens, but in each case with an approximately 10-fold difference in abundance (P3 was dominant in 2 specimens and P1 in the third) (Fig. 3A). In contrast to the two dominant phylotypes, P4 was detected in nearly all (12 of 13) of the specimens but was present at a relatively low abundance (Fig. 3A and C), averaging 11.5% ± 8.2% for all of the specimens. It should be noted, however, that the QPCR-CDCE method used here has a dynamic range of approximately 100 (28) and so phylotypes present at less than ∼1% of the total population would not be detected. Therefore, it is possible that P4, or any of the other identified symbiont phylotypes, was present in some samples at levels below this detection limit. Thus, in general, the symbiont populations observed in L. pedicellatus are composed of a single dominant phylotype (P1 or P3), a consistent minor phylotype (P4), and in some cases one or more additional minor phylotypes that occur more sporadically. Within each phylotype, there may be considerable ribotype variation and indeed internal clades are present that were not distinguished by the CDCE target sequence used here.
The observation that phylotypes P1 and P3 show inverse covariation with one another but not with P4 indicates that different factors determine the abundance and tissue distribution of these three phylotypes. Thus, at least to this extent, they appear to be ecologically and physiologically differentiated. Indeed, the fact that these two phylotypes are sometimes codetected within the same bacteriocytes (6) suggests that their inverse proportions could be explained by competition for the same intracellular niche. It is interesting, however, that either phylotype may be dominant in a given host and that this pattern holds true even when both phylotypes are detected in the same specimen. Moreover, at least in this small sampling of the host population, dominance by either phylotype occurs with similar frequencies. Thus, a simple dominance hierarchy of the phylotypes does not explain the patterns observed. Instead, other currently unknown factors, such as the timing and mode of symbiont transmission, the physiological state and growth dynamics of the host and symbionts, or the physicochemical parameters of the environment, may influence the composition of symbiont populations.
The question remains to what extent intraspecies variation occurs in other symbiont populations. In recent years, multiple coexisting symbionts have been recognized in a small but growing number of marine animal species. For example, two divergent symbiont ribotypes, one a sulfur oxidizer and the other a methane oxidizer, coexist in the bacteriocytes of certain mussels from deep-sea hydrothermal vents and cold-water seeps (7, 16, 32). Two phylogenetically distinct ribotypes, both presumed to derive from sulfur-oxidizing bacteria, have been reported in separate regions of the gill filaments of a thyasirid clam (17). And in reef-building corals, multiple ribotypes of zooxanthellae (eukaryotic algal endosymbionts) have been detected within individual hosts (2). Multiple coexisting symbiont ribotypes have also been observed in naturally occurring and experimentally manipulated extracellular endosymbioses, including light organ symbioses of sepiolid squid (14, 24) and the subcuticular sulfur-oxidizing, sulfate-reducing symbiont communities in an oligochaete worm (11). In each of these cases, the potential for intraspecies variation in the composition of symbiont populations exists and indeed such variation could have adaptive significance for host survival.
The structure of symbiont communities in L. pedicellatus closely resembles that recently observed within free-living marine bacterioplankton (1) and sediment (21) communities in which clusters of closely related co-occurring ribotypes predominate. In these studies, it has been proposed that these microdiverse ribotype clusters (equivalent to the phylotypes observed here) represent important units of ecological differentiation in these communities and that much of the diversity within such clusters may be selectionally neutral (29). It will be important to determine to what extent the phylotypes observed here represent physiologically and ecologically differentiated populations whose distribution correlates with factors such as the developmental state, condition, or environmental fitness of the host.
Acknowledgments
This work was supported by National Science Foundation grants OCE-0425795, OCE-0221224, and NSF-0129117.
We thank Luisa A. Marcelino and Aoy Tomita-Mitchell for advice and assistance in developing CDCE protocols.
REFERENCES
- 1.Acinas, S. G., V. Klepac-Ceraj, D. E. Hunt, C. Pharino, I. Ceraj, D. L. Distel, and M. F. Polz. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551-554. [DOI] [PubMed] [Google Scholar]
- 2.Baker, A. C. 2001. Reef corals bleach to survive change. Nature 411:765-766. [DOI] [PubMed] [Google Scholar]
- 3.Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D’Souza, Y. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. Shang, N. Yu, and R. R. Gutell. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavanaugh, C. M. 1985. Symbiosis of chemolithotrophic bacteria and marine invertebrates from hydrothermal vents and reducing sediments. Biol. Soc. Wash. Bull. 6:373-388. [Google Scholar]
- 5.Distel, D. L. 2003. The biology of marine wood boring bivalves and their bacterial endosymbionts, p. 253-271. In B. Goodell, D. D. Nicholas, and T. P. Schultz (ed.), Wood deterioration and preservation, vol. 845. American Chemical Society Press, Washington, D.C. [Google Scholar]
- 6.Distel, D. L., D. J. Beaudoin, and W. Morrill. 2002. Coexistence of multiple proteobacterial endosymbionts in the gills of the wood-boring bivalve Lyrodus pedicellatus (Bivalvia: Teredinidae). Appl. Environ. Microbiol. 68:6292-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Distel, D. L., and C. M. Cavanaugh. 1995. Intracellular coexistence of methano- and thioautotrophic bacteria in a hydrothermal vent mussel. Proc. Natl. Acad. Sci. USA 92:9598-9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Distel, D. L., H. Felbeck, and C. M. Cavanaugh. 1994. Evidence for phylogenetic congruence among sulfur-oxidizing chemoautotrophic bacterial endosymbionts and their bivalve hosts. J. Mol. Evol. 38:533-542. [Google Scholar]
- 9.Distel, D. L., D. J. Lane, G. J. Olsen, S. J. Giovannoni, B. Pace, N. R. Pace, D. A. Stahl, and H. Felbeck. 1988. Sulfur-oxidizing bacterial endosymbionts: analysis of phylogeny and specificity by 16S rRNA sequences. J. Bacteriol. 170:2506-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Distel, D. L., W. Morrill, N. MacLaren-Toussaint, D. Franks, and J. Waterbury. 2002. Teredinibacter turnerae gen. nov., sp. nov., a dinitrogen-fixing, cellulolytic, endosymbiotic gamma-proteobacterium isolated from the gills of wood-boring molluscs (Bivalvia: Teredinidae). Int. J. Syst. Evol. Microbiol. 52:2261-2269. [DOI] [PubMed] [Google Scholar]
- 11.Dubilier, N., C. Mulders, T. Ferdelman, D. de Beer, A. Pernthaler, M. Klein, M. Wagner, C. Erseus, F. Thiermann, J. Krieger, O. Giere, and R. Amann. 2001. Endosymbiotic sulphate-reducing and sulphide-oxidizing bacteria in an oligochaete worm. Nature 411:298-302. [DOI] [PubMed] [Google Scholar]
- 12.Durand, P., O. Gros, L. Frenkiel, and D. Prieur. 1996. Phylogenetic characterization of sulfur-oxidizing bacterial endosymbionts in three tropical Lucinidae by 16S rDNA sequence analysis. Mol. Mar. Biol. Biotechnol. 5:37-42. [Google Scholar]
- 13.Eisen, J. A., S. W. Smith, and C. M. Cavanaugh. 1992. Phylogenetic relationships of chemoautotrophic bacterial symbionts of Solemya velum Say (Mollusca: Bivalvia) determined by 16S rRNA gene sequence analysis. J. Bacteriol. 174:3416-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fidopiastis, P. M., S. von Boletzky, and E. G. Ruby. 1998. A new niche for Vibrio logei, the predominant light organ symbiont of squids in the genus Sepiola. J. Bacteriol. 180:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher, C. R. 1990. Chemoautotrophic and methanotrophic symbioses in marine invertebrates. Rev. Aquat. Sci. 2:399-436. [Google Scholar]
- 16.Fisher, C. R., J. M. Brooks, J. S. Vodenicher, J. M. Zande, J. J. Childress, and R. A. Burke, Jr. 1993. The co-occurrence of methanotrophic and chemoautotrophic sulfur-oxidizing bacterial symbionts in a deep-sea mussel. Mar. Ecol. 14:277-289. [Google Scholar]
- 17.Fujiwara, Y., C. Kato, N. Masui, K. Fujikura, and S. Kojima. 2001. Dual symbiosis in the cold-seep thyasirid clam Maorithyas hadalis from the hadal zone in the Japan Trench, western Pacific. Mar. Ecol. Prog. Ser. 214:151-159. [Google Scholar]
- 18.Gallager, S. M., R. D. Turner, and C. J. Berg. 1981. Physiological aspects of wood consumption, growth, and reproduction in the shipworm Lyrodus pedicellatus Quatrefages. J. Exp. Mar. Biol. Ecol. 52:63-77. [Google Scholar]
- 19.Gonzalez, J. M., and R. M. Weiner. 2000. Phylogenetic characterization of marine bacterium strain 2-40, a degrader of complex polysaccharides. Int. J. Syst. Evol. Microbiol. 50(Pt. 2):831-834. [DOI] [PubMed] [Google Scholar]
- 20.Greene, R. V., and S. N. Freer. 1986. Growth characteristics of a novel nitrogen-fixing cellulolytic bacterium. Appl. Environ. Microbiol. 52:982-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klepac-Ceraj, V., M. Bahr, B. C. Crump, A. P. Teske, J. E. Hobbie, and M. F. Polz. 2004. High overall diversity and dominance of microdiverse relationships in salt marsh sulphate-reducing bacteria. Environ. Microbiol. 6:686-698. [DOI] [PubMed] [Google Scholar]
- 22.Lane, D. J. 1991. 16S/23S sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley, New York, N.Y.
- 23.Lim, E. L., A. V. Tomita, W. G. Thilly, and M. F. Polz. 2001. Combination of competitive quantitative PCR and constant-denaturant capillary electrophoresis for high-resolution detection and enumeration of microbial cells. Appl. Environ. Microbiol. 67:3897-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishiguchi, M. K., E. G. Ruby, and M. J. McFall-Ngai. 1998. Competitive dominance among strains of luminous bacteria provides an unusual form of evidence for parallel evolution in sepiolid squid-Vibrio symbioses. Appl. Environ. Microbiol. 64:3209-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Powell, M. A., and G. N. Somero. 1986. Adaptations to sulfide by hydrothermal vent animals. Biol. Bull. (Woods Hole) 171:274-290. [Google Scholar]
- 26.Saiki, R., D. H. Gelfand, S. Stoffel, S. J. Scharf, R. Higuchi, G. T. Horn, K. B. Mullis, and H. A. Erlich. 1987. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487-491. [DOI] [PubMed] [Google Scholar]
- 27.Swofford, D. L. 1997. PAUP* 4.0 (phylogenetic analysis using parsimony). Sinauer, Sunderland, Mass.
- 28.Thompson, J. R., L. A. Marcelino, and M. F. Polz. 2002. Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR’. Nucleic Acids Res. 30:2083-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson, J. R., M. A. Randa, L. A. Marcelino, A. Tomita-Mitchell, E. Lim, and M. F. Polz. 2004. Diversity and dynamics of a North Atlantic coastal Vibrio community. Appl. Environ. Microbiol. 70:4103-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner, R. D. 1966. A survey and illustrated catalogue of the Teredinidae (Mollusca: Bivalvia). The Museum of Comparative Zoology, Harvard University, Cambridge, Mass.
- 31.Waterbury, J. B., C. B. Calloway, and R. D. Turner. 1983. A cellulolytic-nitrogen fixing bacterium cultured from the gland of Deshayes in shipworms (Bivalvia: Teredinidae). Science 221:1401-1403. [DOI] [PubMed] [Google Scholar]
- 32.Won, Y. J., S. J. Hallam, G. D. O’Mullan, I. L. Pan, K. R. Buck, and R. C. Vrijenhoek. 2003. Environmental acquisition of thiotrophic endosymbionts by deep-sea mussels of the genus Bathymodiolus. Appl. Environ. Microbiol. 69:6785-6792. [DOI] [PMC free article] [PubMed] [Google Scholar]