Abstract
Bifidobacterium longum DJO10A is a recent human isolate with probiotic characteristics and contains two plasmids, designated pDOJH10L and pDOJH10S. The complete sequences of both these plasmids have now been determined and consist of two circular DNA molecules of 10,073 and 3,661 bp, with G+C contents of 62.2% and 66.2%, respectively. Plasmid pDOJH10L is a cointegrate plasmid consisting of DNA regions exhibiting very high sequence identity to two other B. longum plasmids, pNAC2 (98%) and pKJ50 (96%), together with another region. Interestingly, the rolling circular replication (RCR) regions of both the pNAC2- and pKJ50-like plasmids were disrupted during the recombination event leading to a further recombination event to acquire a functional replicon. This consists of a new fused rep gene and an RCR-type ori consisting of a conserved DnaA box in an AT-rich region followed by four contiguous repeated sequences consistent with an iteron structure and an inverted repeat. The smaller pDOJH10S had no sequence similarity to any other characterized plasmid from bifidobacteria. In addition, it did not contain any features consistent with RCR, which is the replication mechanism proposed for all the bifidobacteria plasmids characterized to date. It did exhibit sequence similarity with several theta replication-related replication proteins from other gram-positive, high-G+C bacteria, with the closest match from a Rhodococcus rhodochrous plasmid, suggesting a theta mechanism of replication. S1 nuclease analysis of both plasmids in B. longum DJO10A revealed single-strand DNA intermediates for pDOJH10L, which is consistent for RCR, but none were detected for pDOJH10S. As the G+C content of pDOJH10S is similar to that of Rhodococcus rhodochrous (67%) and significantly higher than that of B. longum (60.1%), it may have been acquired through horizontal gene transfer from a Rhodococcus species, as both genera are members of the Actinomycetes and are intestinal inhabitants. An Escherichia coli-B. longum shuttle cloning vector was constructed from pDOJH10S and the E. coli ori region of p15A, a lacZ gene with a multiple cloning site of pUC18, and a chloramphenicol resistance gene (CAT) of pCI372 and was transformed successfully into E. coli and B. longum. It could not be introduced into lactic acid bacteria (Lactococcus and Lactobacillus), showing it was not very promiscuous. It was stably maintained in B. longum in the absence of antibiotic pressure for 92 generations, which is consistent with the segregational stability of theta-replicating plasmids in gram-positive bacteria. This is the first cloning vector for bifidobacteria that does not utilize RCR and should be useful for the stable introduction of heterologous genes into these dominant inhabitants of the large intestine.
Bifidobacteria are gram-positive, non-spore-forming, nonmotile, irregular rod-shaped bacteria that often resemble Y or V shapes (32). While they are generally classified as obligate anaerobes, some isolates, particularly in the species Bifidobacterium lactis, can exhibit significant tolerance to oxygen. They are strictly fermentative organisms that utilize glucose by a very characteristic shunt pathway typified by the enzyme fructose-6-phosphate phosphoketolase and produce primarily acetic and lactic acids. While they are a lactic acid-producing bacteria, they are genetically distinct from the typical lactic acid bacteria, having a higher G+C content of 55 to 67%, and they group in the Actinomycete branch of gram-positive bacteria. The natural habitat for bifidobacteria is the intestines of humans, some animals, and insects. In this habitat they are considered a beneficial organism for the host, with a large number of potential health benefits attributed to them. These include prevention and treatment of diarrhea, establishment of a healthy flora in premature infants, alleviation of constipation and the symptoms of lactose intolerance, enhancement of immune function, suppression of tumorigenesis, and cholesterol reduction (25).
The wide probiotic activities of bifidobacteria point to their vast potential for improving human health. Because of this potential they are frequently included in fermented dairy products as probiotic adjuncts. However, the lack of molecular tools for studying this group of high-G+C gram-positive bacteria has limited our ability to understand what characteristics of these bacteria are important for probiotic activities. Recently, the complete genome sequence of a strain of B. longum was deciphered, shedding light onto their genetic makeup (33). While genome sequences do reveal the gene complement of bacteria, the functional analysis of those genes requires molecular tools for cloning, gene knockout, gene expression, etc. There is also broad interest in using bifidobacteria as potential hosts for the construction of oral vaccines and for the cloning of genes encoding detoxifying activities, such as cholesterol oxidase and bile salt hydrolase (29). Currently there are very few cloning vectors for gene transfer in this genus. A recent E. coli-Bifidobacterium shuttle cloning vector was used to express several tumor-suppressing genes in bifidobacteria for possible treatment of hypoxic tumors (15, 18, 43). This novel approach to gene delivery for cancer therapy via bifidobacteria is an example of the many potential innovative uses for gene cloning and gene expression in these bacteria. To facilitate these potential uses of bifidobacteria, there is a need for development and improvement of cloning vectors and gene transfer systems for these bacteria.
To date, plasmids have only been detected in five species of bifidobacteria: B. longum and B. breve (human inhabitants), B. globosum (pig isolate), and B. indicum and B. asteroides (honeybee isolates) (14, 35, 36, 37). Some plasmids have been characterized at the sequence level (8, 20, 24, 27, 41), and two plasmids were reported to replicate via a rolling circle replication (RCR) mechanism (24, 27). A number of cloning vectors have been constructed with plasmids from Bifidobacterium and E. coli and transformed into both of them by electroporation (19, 20, 21, 27, 29, 30). In all cases, electroporation efficiency in Bifidobacterium was very low, and attempts have been made to optimize it (2, 28).
In this study, we isolated and analyzed two plasmids from the probiotic bacterium B. longum DJO10A, which was isolated in this laboratory. Prior to sequencing the complete genome for this bacterium, the plasmids were sequenced using a shotgun sequencing and contig alignment approach. Following analysis of the plasmid sequences, the smallest plasmid, pDOJH10S, was chosen for the construction of a shuttle cloning vector, and an electroporation procedure was refined for introduction of the vector into bifidobacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bifidobacteria were routinely cultivated at 37°C in Trypticase-peptone-yeast extract (32) or de Man-Rogosa-Sharpe (MRS; Difco, Detroit, MI) broths supplemented with 0.05% (final concentration) l-cysteine · HCl (Sigma, St. Louis, MO) under anaerobic conditions generated using the BBL Anaerobic system (Cockeysville, MD). E. coli was grown aerobically at 37°C in Luria-Bertani broth. Chloramphenicol was used at a concentration of 20 μg/ml and 2 μg/ml for selection in E. coli and B. longum, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Bacterial strains | ||
| Bifidobacterium longum | ||
| DJO10A | Wild-type human isolate; host of pDOJH10L and pDOJH10S | A. Islam, D. D. O’Sullivan, and D. J. O’Sullivan (unpublished) |
| VMK44 | Electroporation host | J. Steeleb |
| Lactobacillus delbrueckii subsp. bulgaricus | Electroporation host | L. L. McKay culture collection |
| E. coli | ||
| XL1-Blue | thi recA1 hsdR17 relA1 gyrA94 α-lacZ complementation; Tcr | 5 |
| Plasmids | ||
| pDOJH10L | 10-kb wild-type plasmid from B. longum DJO10A | This study |
| pDOJH10S | 3.6-kb wild-type plasmid from B. longum DJO10A | This study |
| pDOJHR | E. coli-bifidobacterium shuttle cloning vector; Cmr | This study |
| pUC18 | E. coli cloning vector; source of lacZ and MCS | 23 |
| pACYC184 | E. coli cloning vector; source of p15A ori | 6 |
| pCI372 | E. coli-Lactococcus lactis shuttle cloning vector; Cmr | 12 |
| pDOJ4 | E. coli-Lactobacillus bulgaricus shuttle-cloning vector; Cmr | J. Halgerson, J. H. Kim, J. H. Lee, and D. J. O’Sullivan (unpublished) |
Tcr, tetracycline resistance; Cmr, chloramphenicol resistance; ori, origin of replication; MCS, multiple cloning site.
Department of Food Science, University of Wisconsin.
Plasmid isolation and purification.
Plasmids were isolated from bifidobacteria using the following optimized procedure: 250 ml of cells was harvested and resuspended in 10 ml of TES buffer (30 mM Tris · HCl, pH 8.0, 50 mM NaCl, 5 mM EDTA, pH 8.0, final concentration). Five milliliters of a sucrose-lysozyme solution (30 mg/ml lysozyme in 25% sucrose, 50 mM Tris · HCl, pH 8.0, and 1 mM EDTA, pH 8.0, final concentration) for 1 h at 37°C. After treatment with 10 ml of sodium dodecyl sulfate (SDS) (3% SDS in 0.2 N NaOH, final concentration), the suspension was left to clear for 7 min at room temperature before adding 7.5 ml of ice-cold 3 M sodium acetate, pH 4.8. Following centrifugation, the supernatant was mixed with an equal volume of isopropanol and centrifuged to pellet plasmid DNA. To purify the DNA, the pellet was resuspended in 9.4 ml of water and extracted once with saturated phenol-chloroform/isoamyl alcohol and finally precipitated with −20°C ethanol and resuspended in 250 μl of TER (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, pH 8.0, 0.1 mg/ml RNase A). Small and large plasmid DNA preparations from E. coli were carried out using Mini-prepand Midi-prep kits according to the manufacturer’s recommendations (QIAGEN, Valencia, CA).
Molecular techniques.
DNA was recovered from agarose gels using a GELase kit (Epicenter, Madison, WI). DNA ligations were performed using a Fast-Link Ligation kit (Epicenter). Restriction enzyme digestions were carried out according to standard laboratory procedures (31).
Southern hybridization and single-stranded DNA detection.
Southern hybridization was performed using a DIG DNA Labeling and Detection kit (Roche, Indianapolis, IN) according to the manufacturer’s instructions. The accumulation of single-stranded DNA intermediates, indicative of rolling circle replication, was evaluated by the effect of S1 nuclease digestion on B. longum DJO10A plasmids, as previously described (24, 27).
DNA sequencing and analysis.
A HydroShear DNA shearing machine (Genomic Solutions, Ann Arbor, MI) was used to randomly shear plasmid DNA for library construction. Sequencing of library clones was performed using a Big-Dye terminator and an ABI Prism 3100 Auto sequencer (Applied Biosystems, Foster City, CA). DNASTAR (DNASTAR, Inc., Madison, WI) was used for assembly of contigs, and primer walking was used to fill in gaps in the plasmid sequences. Basic DNA and amino acid sequence analyses were performed using DNASTAR and OMIGA (Accelrys, San Diego, CA) software programs. The BLAST server at the National Center for Biotechnology Information was used for sequence similarity searches (1). The GENEMARK server at University of Washington was used for open reading frame (ORF) predictions. The PFAM database at the Sanger Institute (3) and PROSITE at the SWISS-PROT server (4) were used for conserved domain searches. The TMHMM server was used for trans-membrane predictions (38), and multiple sequence alignments were performed using CLUSTAL X (42).
PCR.
All PCRs were performed using a Robocycler (Stratagene, La Jolla, CA). The reaction mixtures (final volume, 50 μl) contained 2 μl of template, 1 μl of each primer (30 μM), 1 μl of deoxynucleoside triphosphates (each at a concentration of 10 mM), and 0.5 μl of Pfu turbo DNA polymerase (Stratagene, La Jolla, CA). The amplification conditions were as follows: 1 cycle of 92°C for 4 min, 30 cycles of 92°C for 1 min, 30 cycles of 52°C for 5 min, 30 cycles of 72°C for 1 min, and 1 cycle of 72°C for 10 min.
Plasmid transfer.
Plasmids were introduced into E. coli using standard heat-shock transformation (31), and electroporation was used for plasmid transfer into bifidobacteria and Lactobacillus. Electrocompetent cells of B. longum VMK44 and Lactobacillus bulgaricus were prepared as follows. Cultures were incubated anaerobically in 250 ml of MRS broth supplemented with 0.5 M sucrose until an optical density at 600 nm of 0.35. Cells were then pelleted by centrifugation and washed four times with 500 ml of 0.5 M sucrose containing 10% glycerol. Pellets were then resuspended in 400 μl of 0.5 M sucrose containing 10% glycerol. Electroporation was performed with a Gene-Pulser (Bio-Rad, Hercules, CA) using 0.2-mm cuvettes with the pulser settings 2.2 kV/cm, 200 Ω, and 25 μF. Plasmids were selected using MRS agar containing 2 μg/ml chloramphenicol.
Segregation stability of vector in bifidobacteria.
B. longum VMK44 containing pDOJHR was first cultured in MRS broth containing 2 μg/ml chloramphenicol. Cells were then subinoculated in fresh MRS broth without antibiotic selection every 12 h for a total of 92 generations. To monitor vector segregation stability, the culture was periodically plated out for isolated colonies and 20 were spot inoculated onto fresh MRS agar plates with and without 2 μg/ml chloramphenicol and incubated at 37°C for 24 h.
Nucleotide sequence accession numbers.
GenBank accession numbers for the sequences of pDOJH10L, pDOJH10S, and pDOJHR are NC004252, NC004253, and DQ123820, respectively.
RESULTS
Plasmid analysis of Bifidobacterium longum DJO10A.
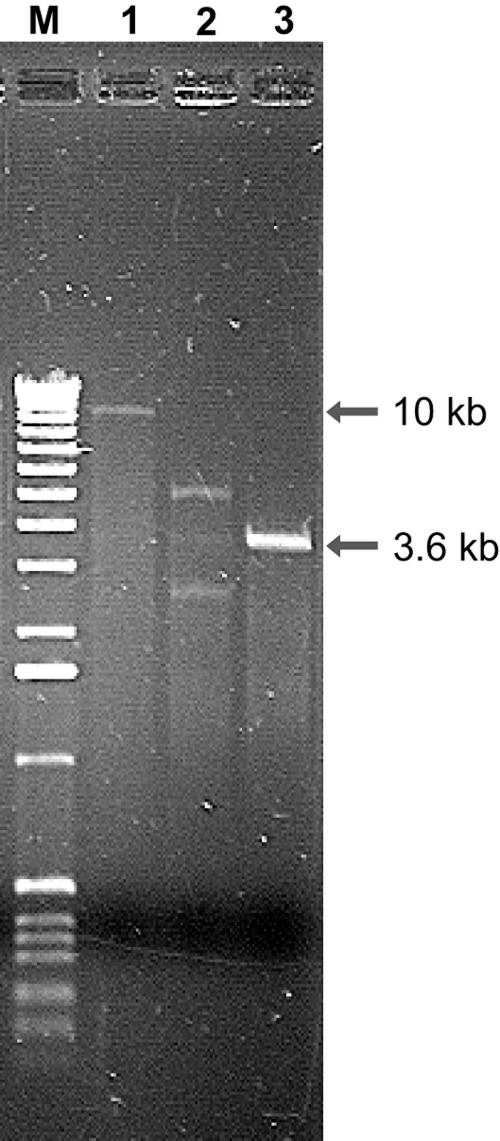
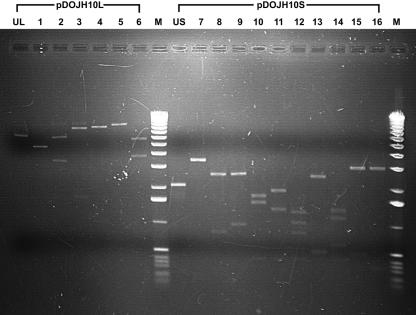
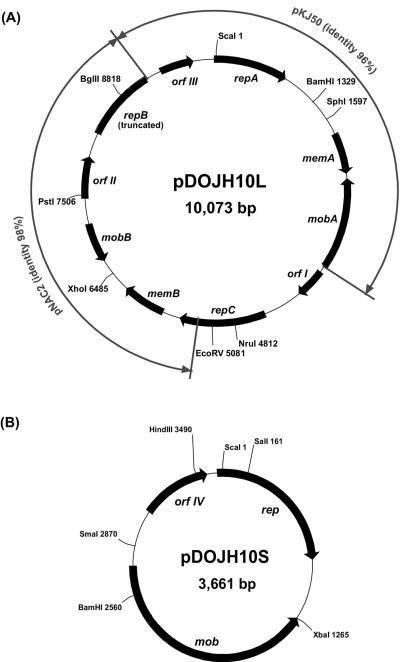
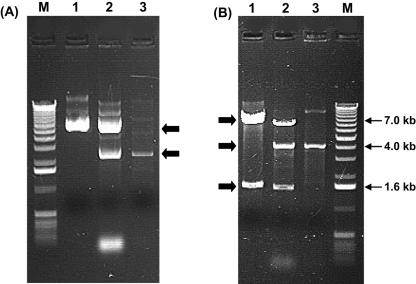
Two plasmids were detected in B. longum DJO10A by agarose gel electrophoresis (Fig. 1). Both plasmids were linearized using ScaI digestion to estimate their size, revealing a large plasmid designated pDOJH10L (10 kb) and a smaller one, pDOJH10S (3.6 kb). The complete sequence of the two plasmids was obtained using shotgun sequencing and primer walking strategies. This resulted in a sequence coverage of 9.2×, and the sequences were manually checked to eliminate errors. Furthermore, restriction enzyme digestion was used to verify the predicted sequences of both plasmids (Fig. 2). Restriction maps of the plasmids revealed several unique sites in both plasmids, with ScaI, BamHI, SphI, BglII, NruI, EcoRV, XhoI, and PstI unique to pDOJH10L and ScaI, XbaI, BamHI, HindIII, SmaI, and SalI unique to pDOJH10S (Fig. 3).
FIG. 1.
Agarose gel electrophoresis of plasmids isolated from B. longum DJO10A (lane 2). Lane 1, ScaI digestion of the large plasmid, pDOJH10L; lane 3, ScaI digestion of the small plasmid, pDOJH10S. M, 1-kb DNA ladder (Invitrogen).
FIG. 2.
Verification of predicted restriction enzyme sites in plasmids pDOJH10L and pDOJH10S. Lanes: UL, uncut pDOJH10L; 1, EcoRV plus ScaI (5 kb, 5 kb, respectively); 2, EcoRV plus SphI (3.5 kb, 6.5 kb); 3, ScaI plus SphI (1.6 kb, 8.4 kb); 4, ScaI plus SacII (1 kb, 8.8 kb, 0.2 kb); 5, SphI plus SacII (9.5 kb, 0.3 kb, 0.2 kb); 6, EcoRV plus SacII (3.9 kb, 5.9 kb, 0.2 kb); M, 1-kb DNA ladder (Invitrogen); US, uncut pDOJH10S; 7, ScaI plus HindIII (3.5 kb, 0.2 kb); 8, ScaI plus NruI (2.7 kb, 0.8 kb, 0.2 kb); 9, HindIII plus NruI (2.7 kb, 1 kb); 10, ScaI plus SphI (1.8 kb, 1.6 kb, 0.3 kb); 11, HindIII plus SphI (2 kb, 1.4 kb, 0.3 kb); 12, NruI plus SphI (1.3 kb, 1.1 kb, 0.9 kb, 0.4 kb); 13, NruI plus SacII (2.6 kb, 0.5 kb, 0.5 kb, 0.1 kb); 14, SphI plus ScaII (1.4 kb, 1.2 kb, 0.6 kb, 0.4 kb); 15, HindIII plus SacII (3 kb, 0.5 kb, 0.2 kb); 16, ScaI plus SacII (3.1 kb, 0.3 kb, 0.3 kb).
FIG. 3.
ORF and restriction enzyme maps of plasmids pDOJH10L (A) and pDOJH10S (B). Outer arrows indicate regions of high sequence identity between pDOJH10L and two other bifidobacterial plasmids, pKJ50 (27) and pNAC2 (8).
Sequence analysis of pDOJH10L.
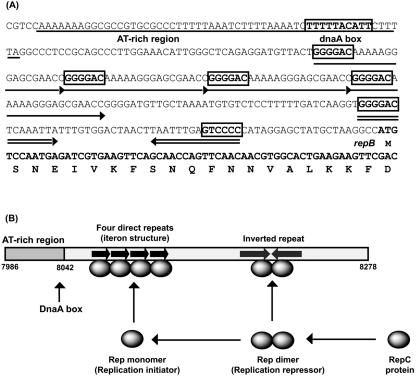
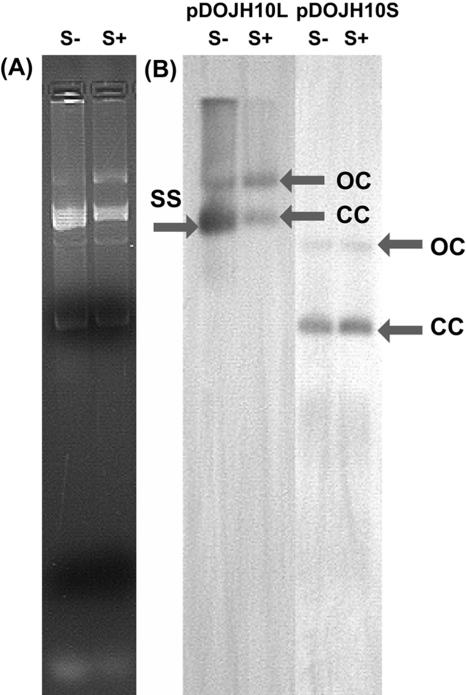
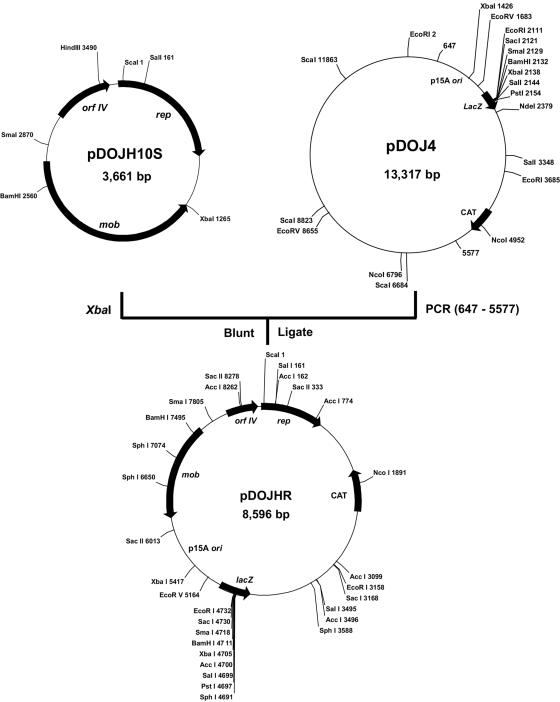
The complete plasmid sequence of pDOJH10L consisted of 10,073 bp, with a G+C content of 62.2%. It was predicted to encode 10 putative (ORFs), 3 involved in replication (Rep), 2 with transmembrane domains, 2 of mobilization genes, and 3 of unknown function (Table 2). DNA sequence comparisons suggest it was formed via a cointegration involving plasmids very similar to those from two other B. longum strains, pKJ50 (96% identity) from B. longum KJ (27) and pNAC2 (98% identity) from B. longum RW041 (8) (Fig. 3). While pDOJH10L contains three genes encoding Rep proteins, only one appears to be functional, as repB is truncated and the ori sequences for repA are not present. The repC gene is a fusion of a rep gene from another source (the closest database match was a distant relationship to a replicase gene on pAP1 from B. asteroides [P. Kaufmann, A. Pfefferkorn, M. Teuber, and L. Meile, GenBank accession number Y11549]), with the 3′ end of repB that occurred during the cointegration event leading to the formation of pDOJH10L (Fig. 3). The ori sequences upstream from repB consist of all the features necessary for functional plasmid replication based on Rep proteins (9) (Fig. 4A). All the features are capable of functioning based on the proposed model for iteron plasmid replication (26) (Fig. 4B). The ori region from the pKJ50-like plasmid got separated from repA during the cointegration event that likely occurred, resulting in the loss of one of the four iteron repeats and also the AT-rich region. It is likely that pDOJH10L replicates via rolling circle replication (RCR) utilizing RepC and the complete ori structure positioned upstream from repB (Fig. 4B). To verify that pDOJH10L replicates via RCR, plasmids from B. longum DJO10A were subjected to S1 nuclease, and, following Southern hybridization with a pDOJH10L probe, a single-stranded DNA band disappeared, which is consistent with RCR (Fig. 5).
TABLE 2.
ORF analysis of the two native plasmids from B. longum DJO10A
| Plasmid and protein or ORF | Position | MWa | pIb | Identityc (%) | Best BLAST match | GenBank accession no. |
|---|---|---|---|---|---|---|
| pDOJH10L | ||||||
| RepA | 10,053-890 | 34,216 | 8.88 | 99 | RepA of pKJ50, B. longum | U76614 |
| RepB | 8,278-9,171 | 34,002 | 9.06 | 99 | RepB of pMG1, B. longum | AY210701 |
| RepC | 4,427-5,485 | 40,343 | 9.35 | 54 | Replicase of pAP1, B. asteroids | Y11549 |
| MobA | 3,481-2,369 | 41,997 | 9.84 | 93 | MobA of pKJ50, B. longum | U76614 |
| MobB | 7,150-6,668 | 17,502 | 12.58 | 99 | MobA of pTB6, B. longum | AB187597 |
| MemA | 1,802-2,305 | 17,322 | 4.19 | 99 | MemA of pKJ50, B. longum | U76614 |
| MemB | 5,708-6,235 | 18,404 | 11.05 | 99 | Unknown ORF of pB44, B. longum | AY066026 |
| ORF I | 3,573-3,941 | 13,557 | 4.18 | 82 | Orf3 of pNAC2, B. longum | NC004769 |
| ORF II | 7,468-7,980 | 19,303 | 4.98 | 98 | Orf3 of pNAC2, B. longum | NC004769 |
| ORF III | 9,373-9,792 | 16,083 | 10.13 | 99 | Unknown ORF of pKJ50, B. longum | U76614 |
| pDOJH10S | ||||||
| Rep | 3,626-878 | 34,618 | 6.10 | 61 | RepA (theta replicase) of pRC4, Rhodococcus rhodochrous | AB040101 |
| Mob | 2,743-1,265 | 54,863 | 9.77 | 63 | MobB of pKJ36, B. longum | NC002635 |
| ORF IV | 3,103-3,558 | 16,907 | 10.49 | 36 | Orf3 of pNAC2, B. longum | NC004769 |
MW, molecular weight.
pI, isoelectric point.
Amino acid sequence identity.
FIG. 4.
(A) Proposed functional ori region of pDOJH10L containing all the required components for rolling circular replication, which are an AT-rich region (underlined) containing a DnaA box, an iteron structure composed of four direct repeats (single arrow) with the proposed RepC binding site boxed, and an inverted repeat (double arrow). (B) Model for replication initiation and copy number control for pDOJH10L based on the proposed model for RCR plasmids (17).
FIG. 5.
S1 nuclease analysis of plasmids from B. longum DJO10A to investigate the existence of single-stranded DNA intermediates. (A) Agarose gel electrophoresis of plasmids from B. longum DJO10A. (B) Southern hybridization of the plasmids from panel A with probes from pDOJH10L and pDOJH10S. S−, not treated with S1 nuclease; S+, treated with S1 nuclease; CC, closed circular plasmid DNA; OC, open circular plasmid DNA; SS, single-stranded DNA intermediates.
The plasmid pDOJH10L encodes two Mob proteins and contains a putative oriT structure upstream of MobB. The putative oriT consists of an inverted repeat and a conserved DNA sequence (5′-TAAGTGCGCCCT-3′), consistent with several other mobilizable plasmids (7) (Fig. 6). The other mob gene on pDOJH10L is contained on the region with sequence similarity to pKJ50. However, this mob gene does not have a cognate oriT. Interestingly, this mob gene in pKJ50 does have a putative oriT, but this DNA region is not present in pDOJH10L eventhough DNA regions on both sides of it are present. Eachpredicted Mob protein has a highly conserved motif (xPHuHuuuxxu) that contains one proline and two histidine residues (in boldface) embedded in a highly hydrophobic sequence. This motif is believed to be involved in the nicking of oriT as a metal center (13).
FIG. 6.
Alignment of the putative oriT regions of pDOJH10L and pDOJH10S with the predicted oriT regions of other mobile plasmids.
The two genes encoding putative transmembrane proteins on pDOJH10L, memA and memB, are also on the respective homologous regions from pKJ50 and pNAC2. They are predicted to encode transmembrane proteins based on sequence similarity to other known transmembrane proteins. However, while MemA has a low pI, consistent with transmembrane proteins, MemB has a very high pI value (Table 2). The other three ORFs on the plasmid contained similarities to hypothetical genes from other sequenced B. longum plasmids (Table 2).
Sequence analysis of pDOJH10S.
Plasmid pDOJH10S consists of 3,661 bp with an average G+C content of 66.2%. Unlike pDOJH10L, its DNA sequence exhibits very low homology to other characterized bifidobacteria plasmids. The Rep protein in pDOJH10S has significant sequence identity to theta replication-related Rep proteins, with the highest identity (61%) to a Rep protein from a plasmid in Rhodococcus rhodochrous (Table 2). This would suggest that the plasmid likely replicates via theta replication. In addition, the plasmid does not have structures normally found on plasmids that replicate by rolling circular replication (RCR), such as an AT-rich region with a DnaA box and iteron structures, further supporting that it most likely replicates by theta replication. To verify that no single-strand intermediates were present, plasmids from B. longum DJO10A were subjected to S1 nuclease and compared to undigested plasmids using Southern hybridization with a probe from pDOJH10S. No single-stranded intermediates were detected, supporting that the plasmid did not replicate via RCR (Fig. 6).
Sequence analysis of the mob region on pDOJH10S suggests that this is a mobilizable plasmid. It contains all the relevant motifs for Mob proteins and a putative oriT (Fig. 6). Interestingly, the Mob protein exhibits very low sequence similarity to other Mob proteins encoded on plasmids from bifidobacteria. A third ORF on the plasmid encoded a hypothetical protein with some sequence identity (36%) to another hypothetical protein from another B. longum plasmid (8).
Construction of a shuttle cloning vector.
Plasmid pDOJH10S was chosen as the basis for a cloning vector for bifidobacteria, as its likely theta replication is more desirable for structural stability than RCR. The E. coli replicon used was from pACYC184, which is termed p15A, and was obtained on a 4.9-kb region from an E. coli-Lactobacillus bulgaricus shuttle cloning vector, pDOJ4 (J. S. Halgerson, J. H. Kim, J. H. Lee, and D. J. O’Sullivan, unpublished data) and also included a multiple-cloning site from pUC18 and a chloramphenicol resistance gene (CAT) from pCI372. This region was amplified by PCR using the primers pDOJ4-F (5′-GTGGCAGGAGAAAAAAGG-3′, positions 647 to 664) and pDOJ4-R (5′-GGAGGCAGACAAGGTATAGG-3′, positions 5,577 to 5,558). This fragment was then cloned into a blunted XbaI site of pDOJH10S to construct the shuttle cloning vector pDOJHR (Fig. 7). To confirm that the final plasmid construction occurred as anticipated, the cojoined regions were sequenced using primers SHUT-F (5′-ATTTCCACCTTTCTCTGTCC-3′, positions 1,075 to 1,094 in pDOJH10S) and SHUT-R (5′-CAGCAAACAGCTCAAACG-3′, positions 1,397 to 1,380 in pDOJH10S).
FIG. 7.
Construction strategy and restriction enzyme map of the E. coli-B. longum shuttle vector, pDOJHR.
This shuttle vector was successfully introduced into B. longum strain VMK44 by electroporation, albeit at a low transfer efficiency of 2.0 × 101 CFU/μg DNA. Plasmid analysis of B. longum VMK44 transformants confirmed that it was introduced without any structural rearrangements occurring (Fig. 8). It could not be electroporated into a Lactobacillus bulgaricus strain using a protocol previously devised for this strain, suggesting it is not a very broad-host-range vector. In another study, the plasmid was successfully introduced into B. longum DJO10A but could not be introduced into Lactococcus lactis, further substantiating the limited host range (J. Yang and D. J. O’Sullivan, unpublished data).
FIG. 8.
Confirmation of vector introduction into B. longum VMK44 using agarose gel electrophoresis. (A) Uncut plasmids; (B) EcoRI-digested plasmids. Lane 1, pDOJHR; lane 2, B. longum VMK44(pDOJHR); lane 3, B. longum VMK44; M, 1-kb DNA ladder (Invitrogen).
Segragational stability of pDOJHR in B. longum VMK44.
Evaluation of the segregational stability of pDOJHR in B. longum VMK44 was monitored over 92 generations without chloramphenicol selection. No plasmid loss was detected in this time, indicating that the vector was very stable without antibiotic selection.
DISCUSSION
Bifidobacterium longum DJO10A is a dominant human isolate with broad-spectrum antimicrobial capabilities (A. Islam, D. D. O’Sullivan, and D. J. O’Sullivan, unpublished data). As this strain has potentially very interesting characteristics, it was chosen for an in-depth analysis. Functional analysis of its characteristics requires molecular tools, and these are not plentiful for bifidobacteria. It was therefore the objective of this study to analyze the two native plasmids of this strain and construct a shuttle cloning vector for bifidobacteria.
The mode of replication of the large plasmid in B. longum DJO10A, pDOJH10L, was predicted to be rolling circular replication. This plasmid contains DNA iterons, which have been shown to be involved in plasmid replication and copy number (22). It also contains a DnaA box that is involved in the initiation of replication in rolling circular plasmids (26, 39). To replicate in bifidobacteria, it is likely that the host DnaA protein binds to the DnaA box region, causing an open complex formation of duplex DNA at the AT-rich region, thus enhancing the unwinding of the origin for initiation of replication. Replication initiation also involves a Rep protein encoded by pDOJH10L. Interestingly, this plasmid contains three rep genes, but repB became truncated during the cointegration event, leading to the formation of pDOJH10L, and the iteron structures corresponding to RepA are incomplete. RepC, which originated from a gene fusion event during the formation of pDOJH10L, is likely the functional Rep gene of this plasmid. This Rep protein likely functions with the complete replication region (iteron structure, DnaA box, and AT-rich region) located upstream from the truncated repB gene during the replication of this plasmid. Mechanistic studies have shown that Rep proteins function as initiators of replication when in a monomer formation and as repressors when in a dimer formation (17). The RepC protein of pDOJH10L would therefore likely have a central role in initiating its replication and controlling its copy number.
All sequenced plasmids from bifidobacteria to date have been proposed to replicate by rolling circular replication. Interestingly, the small plasmid from B. longum DJO10A replicates via a different mechanism, having all the characteristics of a theta replicating plasmid. In addition, the DNA sequence of pDOJH10S did not show any significant homology to previously sequenced bifidobacterial plasmids, suggesting it may be a novel plasmid type for bifidobacteria. The Rep protein ofpDOJH10S has high homology to several known Rep proteins, with the highest homology to the theta replicase from Rhodococcus rhodochrous. Interestingly, the G+C content of pDOJH10S (66.2%) is significantly higher than that of the genome of B. longum DJO10A (60.1%), further substantiating a recent acquisition of this plasmid in bifidobacteria by horizontal gene transfer. As its G+C content is similar to that of Rhodococcus rhodochrous, which is listed as 67 to 70% (11), it is likely that Rhodococcus is the original origin of the plasmid. Both Rhodococcus and Bifidobacterium are closely related, being members of the Actinomycetes. The opportunity for horizontal gene transfer from Rhodococcus also exists as members of this genus are frequently found in the feces of animals (40).
A probable mechanism for the transfer of this plasmid from Rhodococcus into Bifidobacterium is via conjugation. Plasmid pDOJH10S contains a Mob and a putative oriT region that contain all the components necessary for plasmid mobilization. While neither pDOJH10S nor pDOJH10L contain tra genes, Tra functions can be supplied in trans to facilitate plasmid mobilization. Plasmid pDOJH10L has two Mob proteins, but it has only one putative oriT upstream of MobB. The lack of a putative oriT upstream of MobA is most likely the result of a cointegration event during the formation of pDOJH10L from at least two other bifidobacterial plasmids.
The evidence for a cointegration event during the formation of pDOJH10L is very convincing, given the more than 96 and 98% identity with two other plasmids from another B. longum strain (Fig. 3). To date, this is the first cointegration event reported for a plasmid in bifidobacteria, suggesting it may not be a very frequent event. Analysis of the cointegration event indicates that it involved at least one other plasmid during pDOJH10L formation, as an extra rep gene is also present. The selective pressure for obtaining this third rep gene appears to be the disruption of both the RepA and RepB replication regions during the formation of pDOJH10L. Thus, RepC is likely involved in replicating the plasmid via the complete replication region upstream of the truncated repB gene.
Several Bifidobacterium-E. coli shuttle vectors have been constructed, and all are based on RCR plasmids from bifidobacteria (19, 20, 21, 27, 29, 30). However, plasmids that utilize RCR display both structural and segregational instabilities in gram-positive bacteria, while theta replicating plasmids display much greater stabilities (10, 16, 34). In this study, the theta replicating pDOJH10S plasmid was used as the basis for a shuttle cloning vector, as this would likely lead to a more stable cloning vector. It was successfully electroporated into B. longum and E. coli but could not be introduced into Lactobacillus bulgaricus. It also does not replicate in Lactococcus lactis (J. Yang and D. J. O’Sullivan, unpublished data), illustrating the limited hosts capable of replicating this vector. This is consistent with other studies on plasmid replication origins from bifidobacteria, as they have not been able to replicate in lactic acid bacteria (19, 21, 27, 29, 30). The availability of a theta-replicating vector for stable gene cloning in bifidobacteria will facilitate functional genomic analysis of this gut-friendly inhabitant.
Acknowledgments
This work was supported in part by Dairy Management, Inc., and the Minnesota Agricultural Experimental Station.
We thank Jamie Halgerson for technical assistance with plasmid sequencing.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Argnani, A., R. J. Leer, N. van Luijk, and P. H. Pouwels. 1996. A convenient and reproducible method to genetically transform bacteria of the genus Bifidobacterium. Microbiology 142:109-114. [DOI] [PubMed] [Google Scholar]
- 3.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bucher, P., and A. Bairoch. 1994. A generalized profile syntax for biomolecular sequence motifs and its function in automatic sequence interpretation. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:53-61. [PubMed] [Google Scholar]
- 5.Bullock, W. O., J. M. Fernandez, and J. M. Stuart. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques 5:376-379. [Google Scholar]
- 6.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Climo, M. W., V. K. Sharma, and G. L. Archer. 1996. Identification and characterization of the origin of conjugative transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease on the staphylococcal plasmid pGO1. J. Bacteriol. 178:4975-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corneau, N., E. Emond, and G. LaPointe. 2004. Molecular characterization of three plasmids from Bifidobacterium longum. Plasmid 51:87-100. [DOI] [PubMed] [Google Scholar]
- 9.Doran, K. S., D. R. Helinski, and I. Konieczny. 1999. A critical DnaA box directs the cooperative binding of the Escherichia coli DnaA protein to the plasmid RK2 replication origin. J. Biol. Chem. 274:17918-17923. [DOI] [PubMed] [Google Scholar]
- 10.Ehrlich, S. D., C. Bruand, S. Sozhamannan, P. Dabert, M. F. Gros, L. Janniere, and A. Gruss. 1991. Plasmid replication and structural stability in Bacillus subtilis. Res. Microbiol. 142:869-873. [DOI] [PubMed] [Google Scholar]
- 11.Goodfellow, M. 1986. Genus Rhodococcus Zopf 1891, 28AL, p. 1472-1481. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey’s manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 12.Hayes, F., P. Vos, G. F. Fitzgerald, W. M. de Vos, and C. Daly. 1991. Molecular organization of the minimal replicon of novel, narrow-host-range, lactococcal plasmid pCI305. Plasmid 25:16-26. [DOI] [PubMed] [Google Scholar]
- 13.Ilyina, T. V., and E. V. Koonin. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 20:3279-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwata, M., and T. Morishita. 1989. The presence of plasmids in Bifidobacterium breve. Lett. Appl. Microbiol. 9:165-168. [Google Scholar]
- 15.Kanamura, T., T. Sasaki, M. Fujimori, K. Yazawa, Y. Kano, J. Amano, and S. Tanicuchi. 2002. Cloned cytosine deaminase gene expression of Bifidobacterium longum and application to enzyme/pro-drug therapy of hypoxic solid tumors. Biosci. Biotechnol. Biochem. 66:2362-2366. [DOI] [PubMed] [Google Scholar]
- 16.Kiewiet, R., S. Bron, K. de Jonge, G. Venema, and J. F. Seegers. 1993. Theta replication of the lactococcal plasmid pWVO2. Mol. Microbiol. 10:319-327. [PubMed] [Google Scholar]
- 17.Komori, H., F. Matsunaga, Y. Higuchi, M. Ishiai, C. Wada, and K. Miki. 1999. Crystal structure of a prokaryotic replication initiator protein bound to DNA at 2.6 Å resolution. EMBO J. 18:4597-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, X., G. F. Fu, Y. R. Fan, W. H. Liu, X. J. Liu, J. J. Wang, and G. X. Xu. 2003. Bifidobacterium adolescentis as a delivery system of endostatin for cancer gene therapy: selective inhibitor of angiogenesis and hypoxic tumor growth. Cancer Gene Ther. 10:105-111. [DOI] [PubMed] [Google Scholar]
- 19.Matsumura, H., A. Takeuchi, and Y. Kano. 1997. Construction of Escherichia coli-Bifidobacterium longum shuttle vector transforming B. longum 105-A and 108-A. Biosci. Biotechnol. Biochem. 61:1211-1212. [DOI] [PubMed] [Google Scholar]
- 20.Matteuzzi, D., P. Brigidi, M. Rossi, and D. Di. 1990. Characterization and molecular cloning of Bifidobacterium longum cryptic plasmid pMB1. Lett. Appl. Microbiol. 11:220-223. [DOI] [PubMed] [Google Scholar]
- 21.Missich, R., B. Sgorbati, and D. J. LeBlanc. 1994. Transformation of Bifidobacterium longum with pRM2, a constructed Escherichia coli-B. longum shuttle vector. Plasmid 32:208-211. [DOI] [PubMed] [Google Scholar]
- 22.Nordstrom, K. 1990. Control of plasmid replication-how do DNA iterons set the replication frequency? Cell 63:1121-1124. [DOI] [PubMed] [Google Scholar]
- 23.Norrander, J., T. Kempe, and J. Messing. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26:101-106. [DOI] [PubMed] [Google Scholar]
- 24.O’Riordan, K., and G. F. Fitzgerald. 1999. Molecular characterisation of a 5.75-kb cryptic plasmid from Bifidobacterium breve NCFB 2258 and determination of mode of replication. FEMS Microbiol. Lett. 174:285-294. [DOI] [PubMed] [Google Scholar]
- 25.O’Sullivan, D. J. 2001. Screening of intestinal microflora for effective probiotic bacteria. J. Agric. Food Chem. 49:1751-1760. [DOI] [PubMed] [Google Scholar]
- 26.Pacek, M., G. Konopa, and I. Konieczny. 2001. DnaA box sequences as the site for helicase delivery during plasmid RK2 replication initiation in Escherichia coli. J. Biol. Chem. 276:23639-23644. [DOI] [PubMed] [Google Scholar]
- 27.Park, M. S., D. W. Shin, K. H. Lee, and G. E. Ji. 1999. Sequence analysis of plasmid pKJ50 from Bifidobacterium longum. Microbiology 145:585-592. [DOI] [PubMed] [Google Scholar]
- 28.Rossi, M., P. Bridigi, and D. Matteuzzi. 1997. An efficient transformation system for Bifidobacterium spp. Lett. Appl. Microbiol. 24:33-36. [Google Scholar]
- 29.Rossi, M., P. Brigidi, y. R. A. Gonzalez Vara, and D. Matteuzzi. 1996. Characterization of the plasmid pMB1 from Bifidobacterium longum and its use for shuttle vector construction. Res. Microbiol. 147:133-143. [DOI] [PubMed] [Google Scholar]
- 30.Rossi, M., P. Brigidi, and D. Matteuzzi. 1998. Improved cloning vectors for Bifidobacterium spp. Lett. Appl. Microbiol. 26:101-104. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 32.Scardovi, V. 1986. The genus Bifidobacterium Orla-jensen 1924, 472AL, p. 1418-1434. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey’s manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 33.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seegers, J. F., C. M. Franke, R. Kiewiet, G. Venema, and S. Bron. 1995. Use of continuous culture for the selection of plasmids with improved segregational stability. Plasmid 33:71-77. [DOI] [PubMed] [Google Scholar]
- 35.Sgorbati, B., V. Scardovi, and D. J. Leblanc. 1982. Plasmids in the genus Bifidobacterium. J. Gen. Microbiol. 128:2121-2131. [DOI] [PubMed] [Google Scholar]
- 36.Sgorbati, B., V. Scardovi, and D. J. Leblanc. 1986. Related structures in the plasmid profiles of Bifidobacterium asteroides, B. indicum and B. globosum. Microbiologica 9:443-454. [PubMed] [Google Scholar]
- 37.Sgorbati, B., V. Scardovi, and D. J. Leblanc. 1986. Related structures in the plasmid profiles of Bifidobacterium longum. Microbiologica 9:415-422. [PubMed] [Google Scholar]
- 38.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175-182. [PubMed] [Google Scholar]
- 39.Speck, C., and W. Messer. 2001. Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 20:1469-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takai, S., M. Syakalima, J. Yasuda, Y. Sasaki, H. Tsutsumi, E. Miyagawa, K. Wada, T. Kakuda, S. Tsubaki, and C. Sugimoto. 2004. Isolation of Rhodococcus equi from the feces of indigenous animals and soil from the Lower Zambezi National Park and Lochinvar National Park, Zambia. J. Vet. Med. Sci. 66:743-746. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka, K., K. Samura, and Y. Kano. 2005. Structural and functional analysis of pTB6 from Bifidobacterium longum. Biosci. Biotechnol. Biochem. 69:422-425. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J. D., F. Plewniak, and O. Poch. 1999. A comprehensive comparison of multiple sequence alignment programs. Nucleic Acids Res. 27:2682-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yazawa, K., M. Fujimori, J. Amano, Y. Kano, and S. Taniguchi. 2000. Bifidobacterium longum as a delivery system for cancer gene therapy: selective localization and growth in hypoxic tumors. Cancer Gene Ther. 7:269-274. [DOI] [PubMed] [Google Scholar]